Identification of Eggplant SmMPK Gene Family and Functional Verification of SmMPK4.1
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material, Strains, and Vectors
2.2. Analysis of the SmMPKs Gene Family in Eggplant
2.3. RNA Extraction and qPCR
2.4. Gene Cloning and Plasmids Construction
2.5. Treatment Conditions for Induced Expression Analysis
2.6. Tobacco Subcellular Localization
2.7. Eggplant Stabilizes Genetic Transformation
2.8. Determination of Anthocyanin Content
2.9. Widely-Targeted Metabolomic Analysis of SmMPK4-OE Eggplant Pulp
2.10. Yeast Two-Hybrid Assays
2.11. Tobacco Transient Conversion
2.12. Dual Luciferase Assay
2.13. Primers
2.14. Statistical Analysis
3. Results
3.1. Analysis of the MPK Gene Family in Eggplant
3.2. Subcellular Localization of SmMPK4.1 Protein
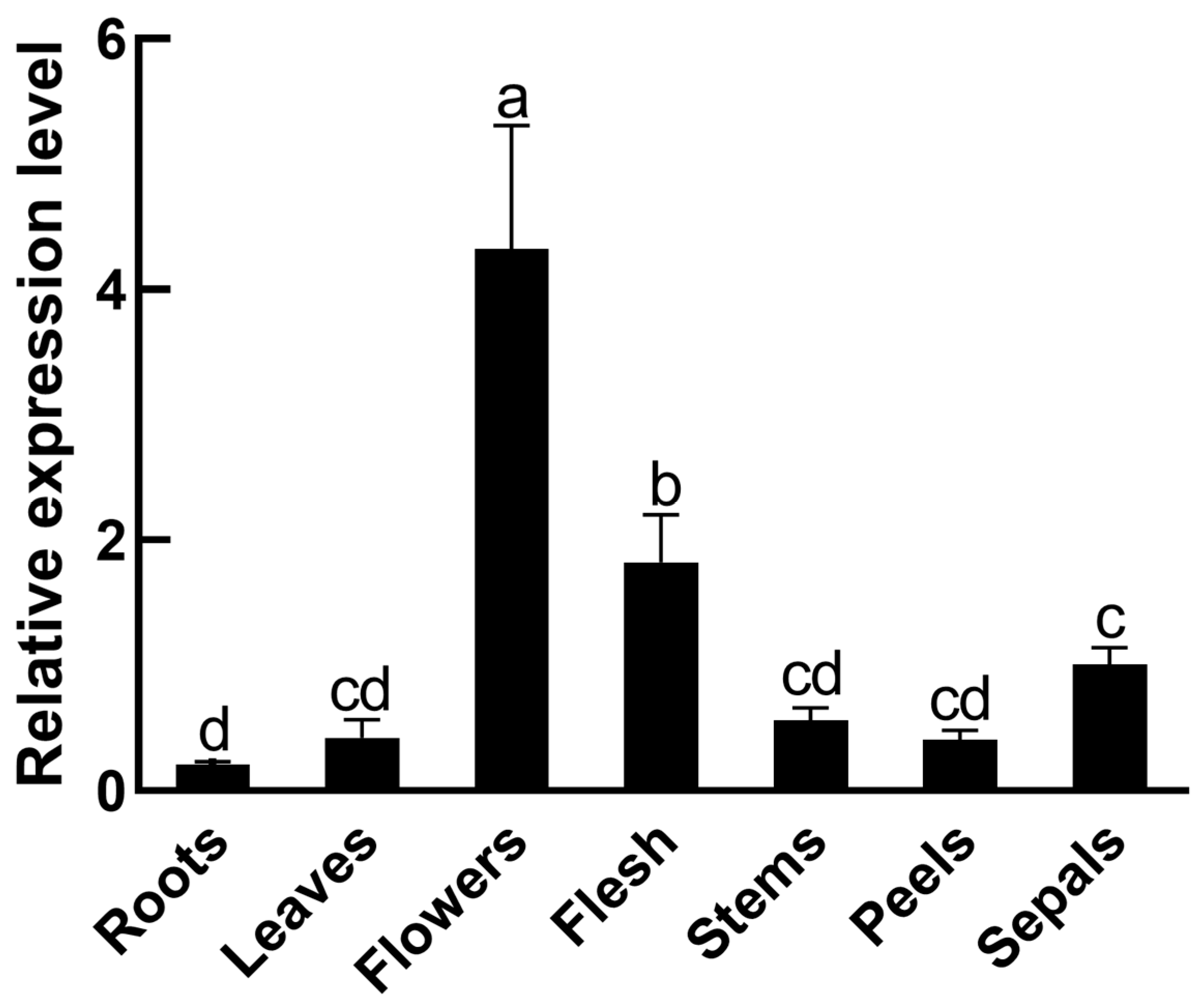
3.3. Tissue-Specific Expression of SmMPK4.1
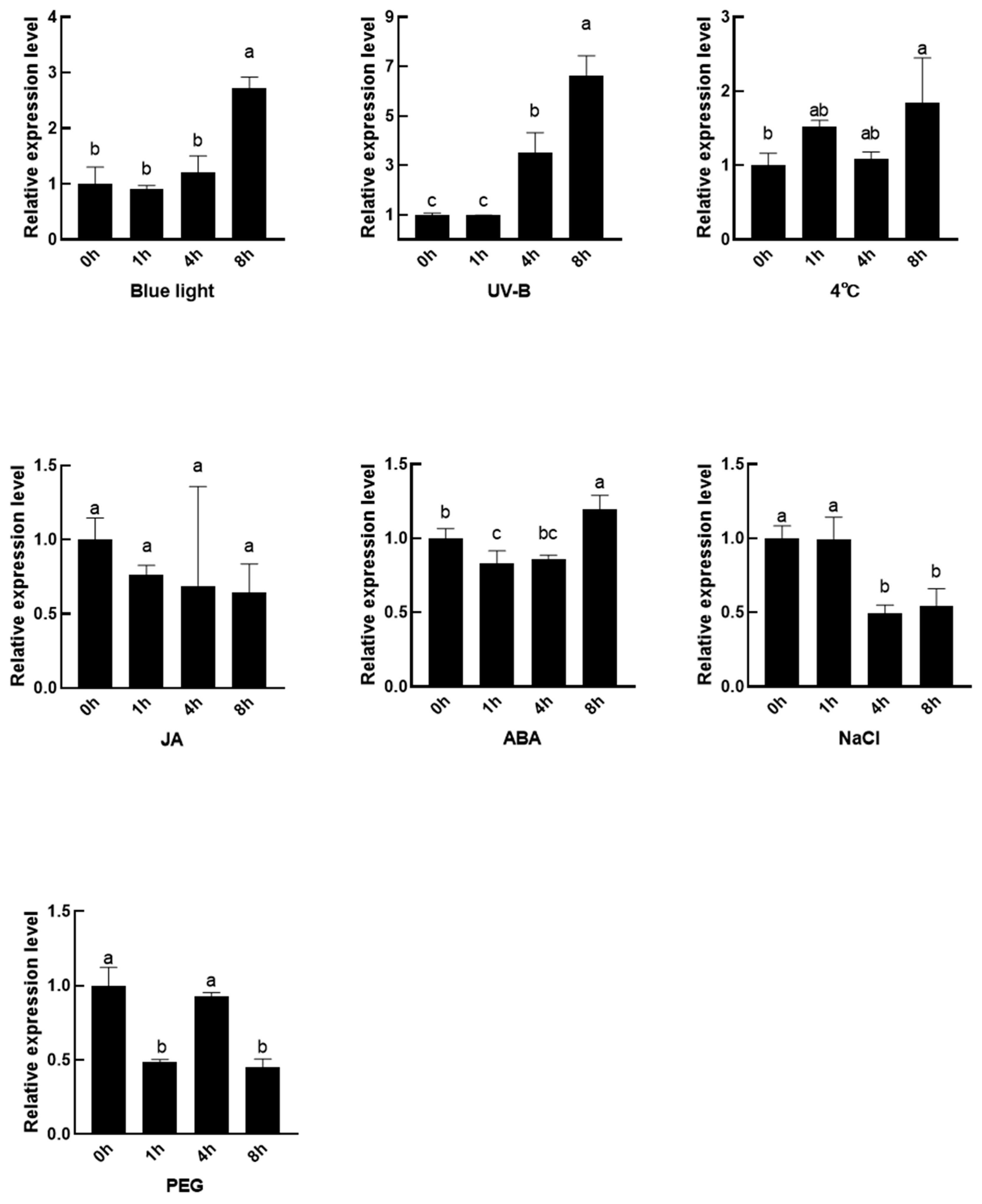
3.4. Analysis of SmMPK4.1 Induced Expression
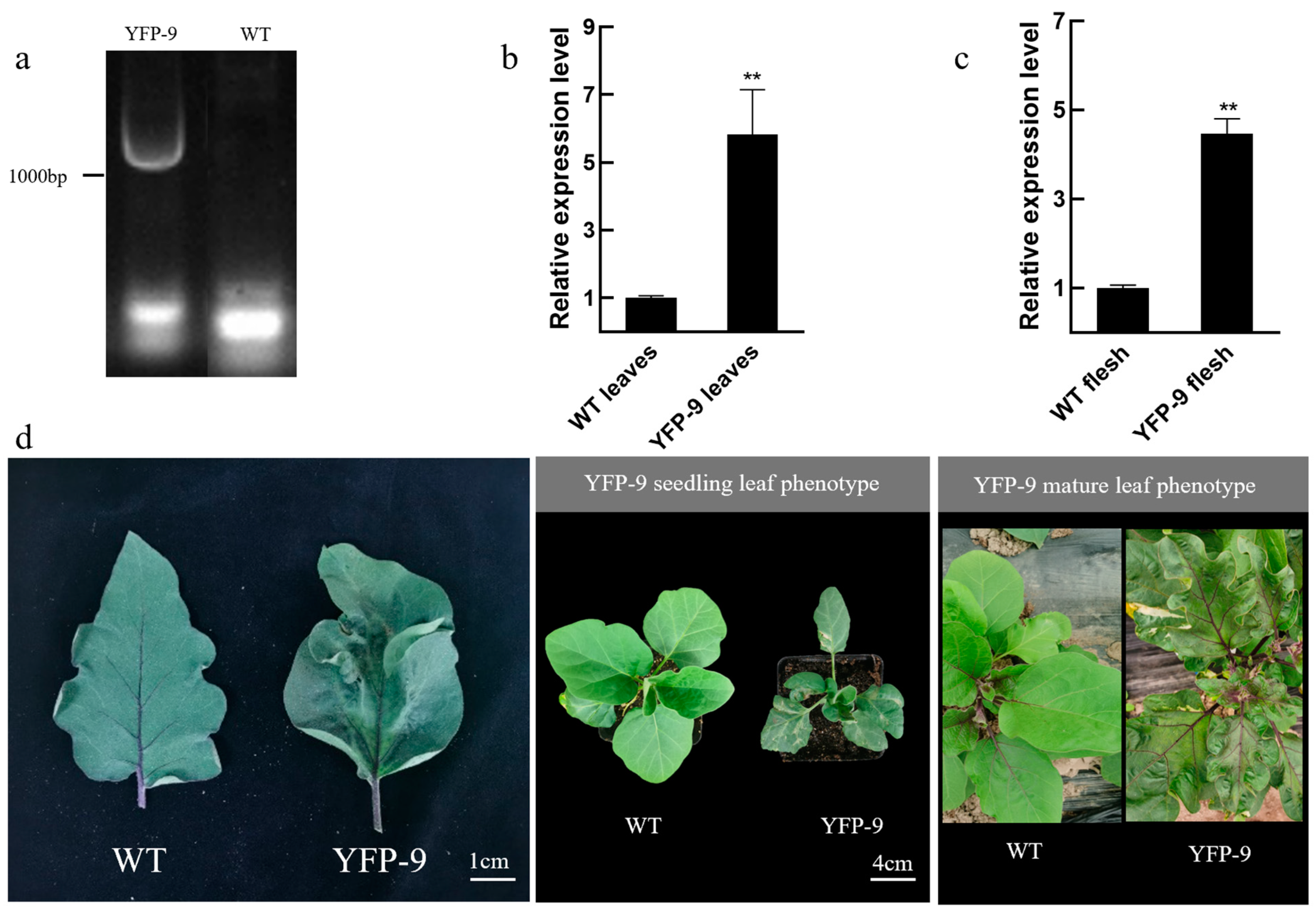
3.5. Leaf Phenotype and Gene Expression Characterization of SmMPK4.1 Overexpression Plants
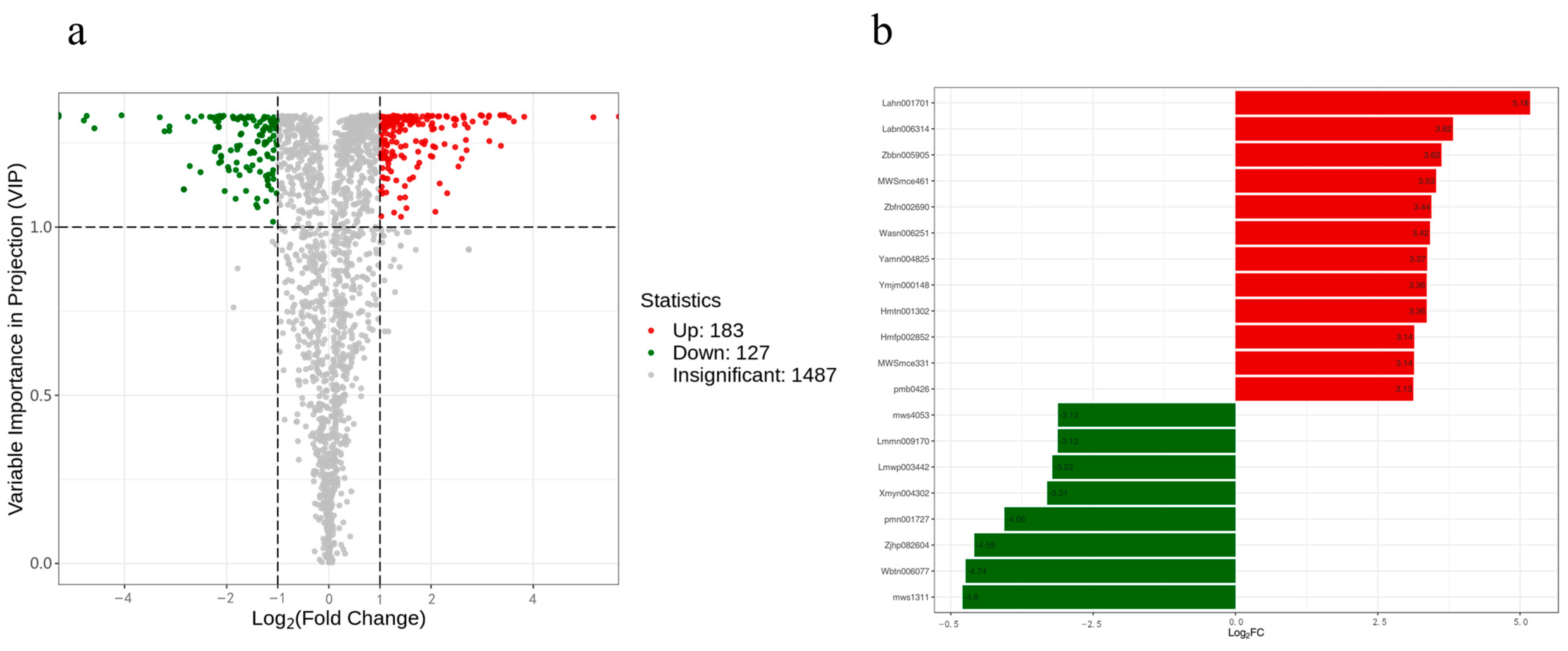
3.6. Widely-Targeted Metabolome Analysis of SmMPK4.1 Overexpression Plants
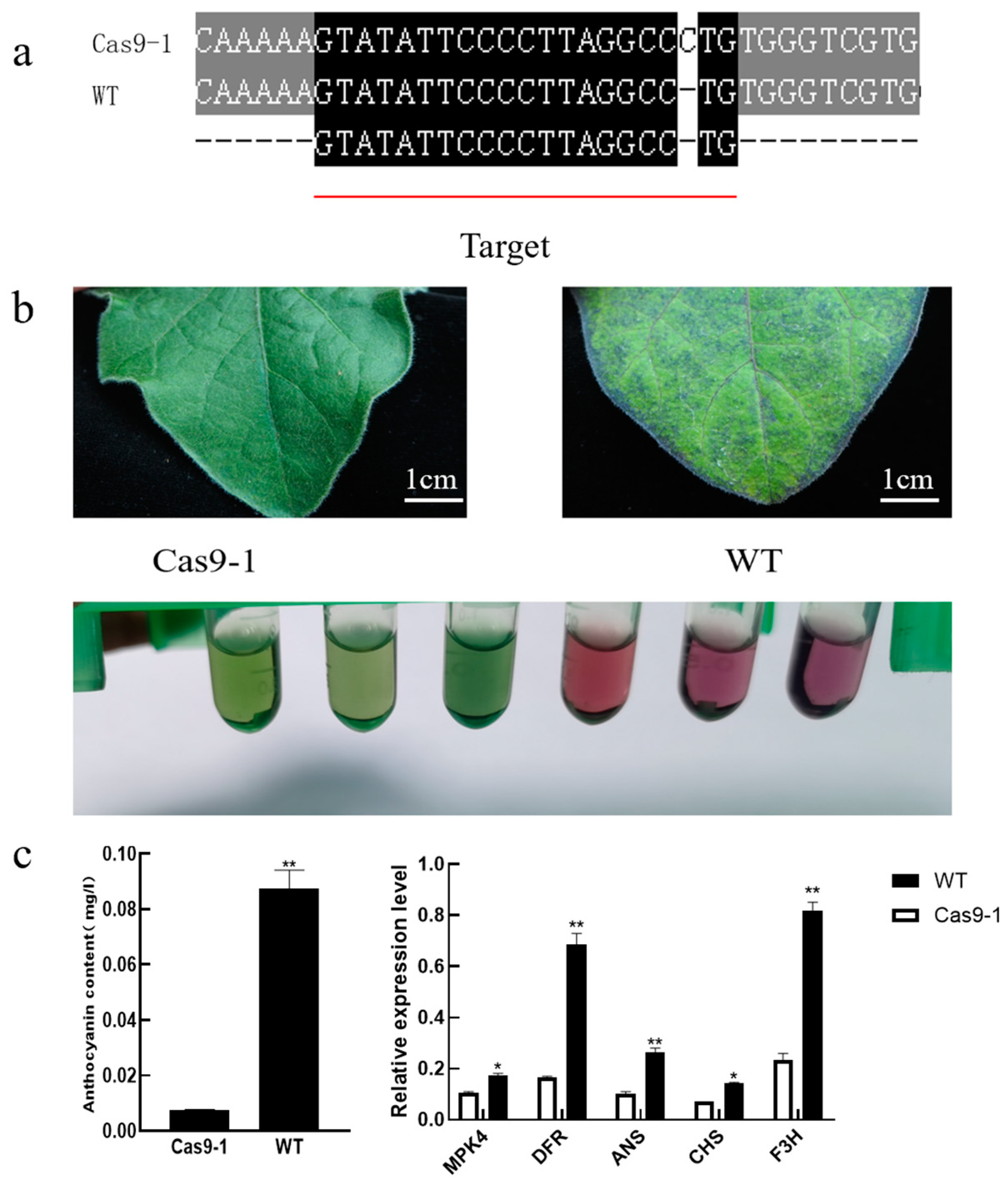
3.7. SmMPK4.1 Knockout Plants Fail to Respond to High Light-Induced Anthocyanin Synthesis Process
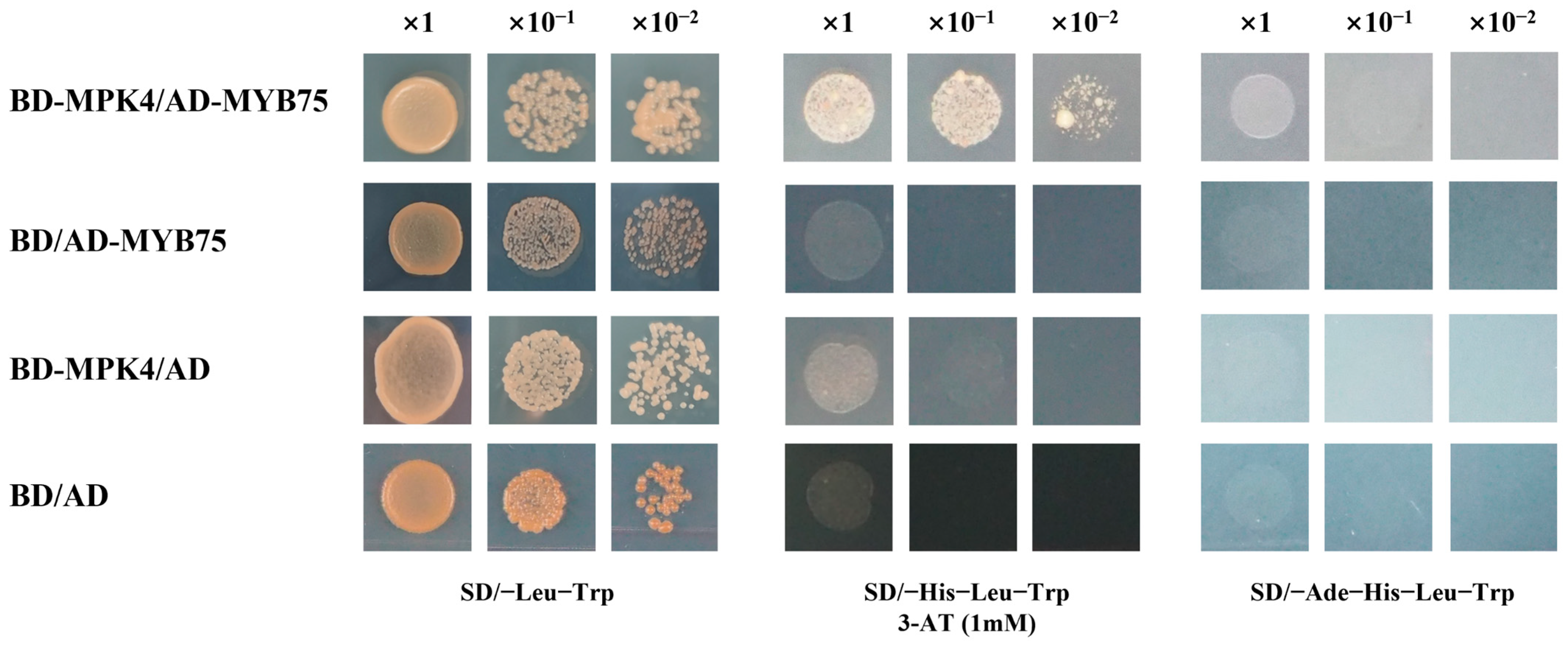
3.8. Yeast Two-Hybrid Demonstration of SmMPK4.1 Interactions with SmMYB75
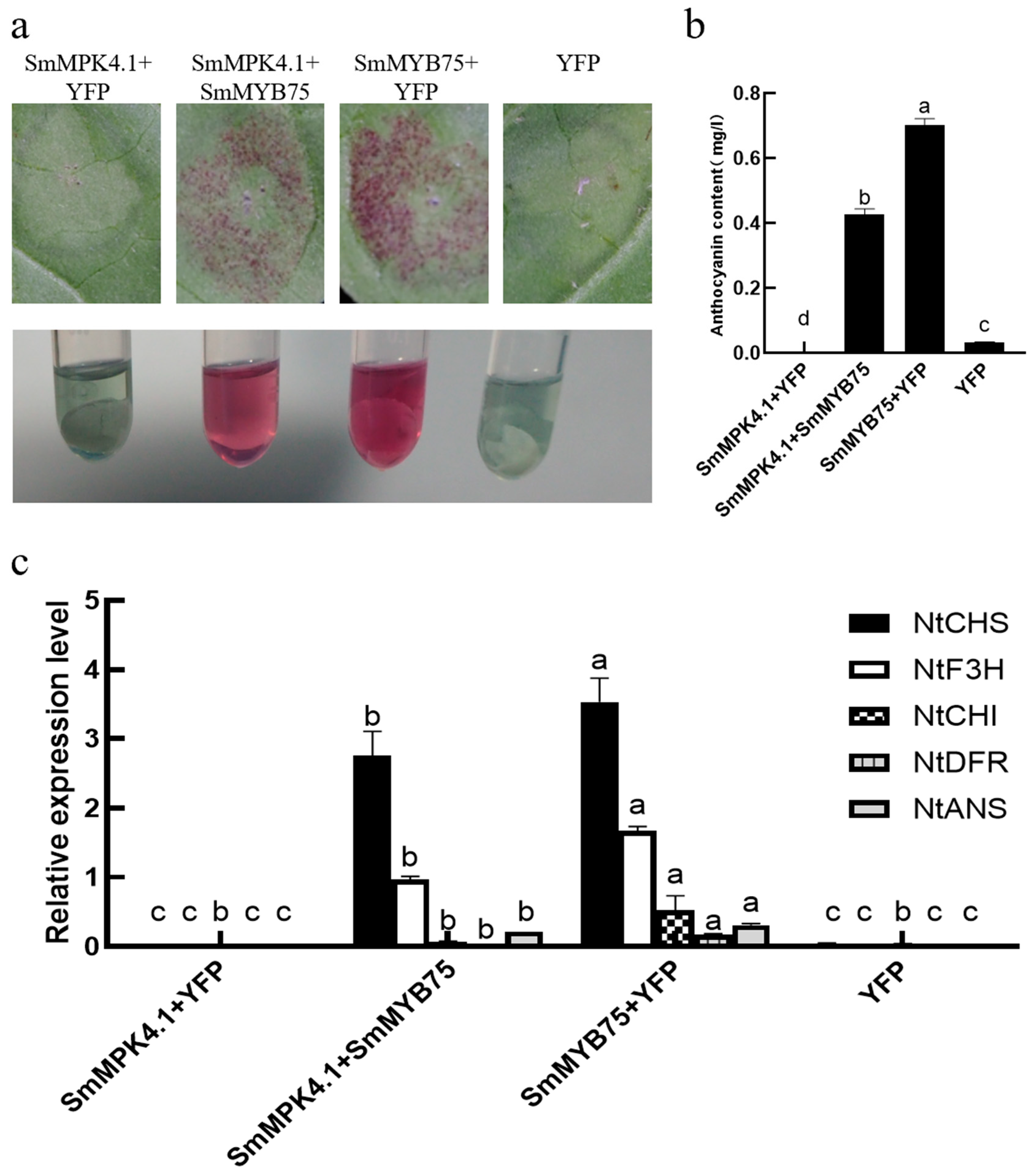
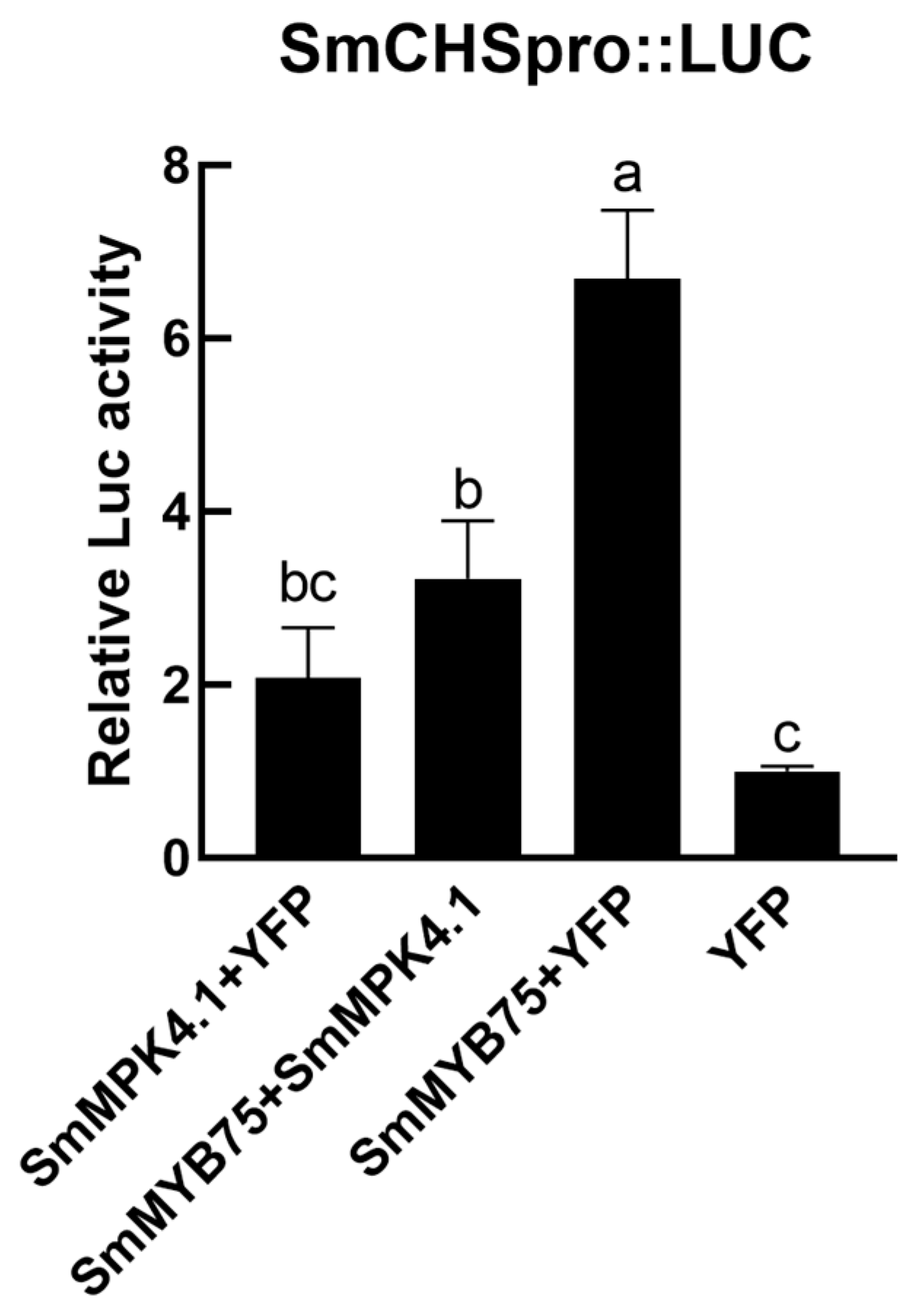
3.9. SmMPK4.1 Can Inhibit SmMYB75 to Promote Anthocyanin Synthesis
3.10. SmMPK4.1 Negatively Regulates the Transcriptional Activation Function of SmMYB75
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Rasmussen, M.W.; Roux, M.; Petersen, M.; Mundy, J. MAP kinase cascades in Arabidopsis innate immunity. Front. Plant Sci. 2012, 3, 169. [Google Scholar] [CrossRef]
- Beck, M.; Komis, G.; Mueller, J.; Menzel, D.; Samaj, J. Arabidopsis homologs of nucleus- and phragmoplast-localized kinase 2 and 3 and mitogen-activated protein kinase 4 are essential for microtubule organization. Plant Cell 2010, 22, 755–771. [Google Scholar] [CrossRef]
- Beck, M.; Komis, G.; Ziemann, A.; Menzel, D.; Samaj, J. Mitogen-activated protein kinase 4 is involved in the regulation of mitotic and cytokinetic microtubule transitions in Arabidopsis thaliana. N. Phytol. 2011, 189, 1069–1083. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; Liu, J.; Bi, D.; Zhang, Z.; Cheng, F.; Chen, S.; Zhang, Y. MEKK1, MKK1/MKK2 and MPK4 function together in a mitogen-activated protein kinase cascade to regulate innate immunity in plants. Cell Res. 2008, 18, 1190–1198. [Google Scholar] [CrossRef] [PubMed]
- Petersen, M.; Brodersen, P.; Naested, H.; Andreasson, E.; Lindhart, U.; Johansen, B.; Nielsen, H.B.; Lacy, M.; Austin, M.J.; Parker, J.E.; et al. Arabidopsis MAP kinase 4 negatively regulates systemic acquired resistance. Cell 2000, 103, 1111–1120. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Q.; Chen, J.-G.; Ellis, B.E. AtMPK4 is required for male-specific meiotic cytokinesis in Arabidopsis. Plant J. 2011, 67, 895–906. [Google Scholar] [CrossRef]
- Witon, D.; Sujkowska-Rybkowska, M.; Dabrowska-Bronk, J.; Czarnocka, W.; Bernacki, M.; Szechynska-Hebda, M.; Karpinski, S. Mitogen-Activated Protein Kinase 4 impacts leaf development, temperature, and stomatal movement in hybrid aspen. Plant Physiol. 2021, 186, 2190–2204. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Bosmans, K.C.; Hsu, P.K.; Paul, K.; Seitz, C.; Yeh, C.Y.; Wang, Y.S.; Yarmolinsky, D.; Sierla, M.; Vahisalu, T.; et al. Stomatal CO2/bicarbonate sensor consists of two interacting protein kinases, Raf-like HT1 and non-kinase-activity activity requiring MPK12/MPK4. Sci. Adv. 2022, 8, eabq6161. [Google Scholar] [CrossRef]
- Liu, Y.; Li, Y. MPK4 negatively regulates the l-arabinose synthesis of cell wall in Arabidopsis. Biochem. Biophys. Res. Commun. 2022, 613, 7–11. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Lott, A.A.; Zhu, W.; Dufresne, C.P.; Chen, S. Mitogen-Activated Protein Kinase 4-Regulated Metabolic Networks. Int. J. Mol. Sci. 2022, 23, 880. [Google Scholar] [CrossRef]
- Wang, S.; Wang, T.; Li, Q.; Xu, C.; Tian, J.; Wang, Y.; Zhang, X.; Xu, X.; Han, Z.; Wu, T. Phosphorylation of MdERF17 by MdMPK4 promotes apple fruit peel degreening during light/dark transitions. Plant Cell 2022, 34, 1980–2000. [Google Scholar] [CrossRef]
- Yang, T.; Ma, H.Y.; Li, Y.; Zhang, Y.; Zhang, J.; Wu, T.; Song, T.T.; Yao, Y.C.; Tian, J. Apple MPK4 mediates phosphorylation of MYB1 to enhance light-induced anthocyanin accumulation. Plant J. 2021, 106, 1728–1745. [Google Scholar] [CrossRef]
- Li, S.; Wenyi, W.; Gao, J.; Yin, K.; Wang, R.; Wang, C.; Petersen, M.; Mundy, J.; Qiu, J.-L. MYB75 Phosphorylation by MPK4 Is Required for Light-Induced Anthocyanin Accumulation in Arabidopsis. Plant Cell 2016, 28, 2866–2883. [Google Scholar] [CrossRef]
- Furuya, T.; Matsuoka, D.; Nanmori, T. Membrane rigidification functions upstream of the MEKK1-MKK2-MPK4 cascade during cold acclimation in Arabidopsis thaliana. FEBS Lett. 2014, 588, 2025–2030. [Google Scholar] [CrossRef]
- Du, X.Z.; Jin, Z.P.; Liu, Z.Q.; Liu, D.M.; Zhang, L.P.; Ma, X.L.; Yang, G.D.; Liu, S.; Guo, Y.R.; Pei, Y.X. H2S Persulfidated and Increased Kinase Activity of MPK4 to Response Cold Stress in Arabidopsis. Front. Mol. Biosci. 2021, 8, 635470. [Google Scholar] [CrossRef]
- Sun, Y.H.; Zhao, J.; Li, X.Y.; Li, Y.Z. E2 conjugases UBC1 and UBC2 regulate MYB42-mediated SOS pathway in response to salt stress in Arabidopsis. N. Phytol. 2020, 227, 455–472. [Google Scholar] [CrossRef]
- Takahama, U. Oxidation of vacuolar and apoplastic phenolic substrates by peroxidase: Physiological significance of the oxidation reactions. Phytochem. Rev. 2004, 3, 207–219. [Google Scholar] [CrossRef]
- Stracke, R.; Werber, M.; Weisshaar, B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001, 4, 447–456. [Google Scholar] [CrossRef] [PubMed]
- Jaakola, L. New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 2013, 18, 477–483. [Google Scholar] [CrossRef] [PubMed]
- Hemm, M.R.; Herrmann, K.M.; Chapple, C. AtMYB4: A transcription factor general in the battle against UV. Trends Plant Sci. 2001, 6, 135–136. [Google Scholar] [CrossRef] [PubMed]
- Kiferle, C.; Fantini, E.; Bassolino, L.; Povero, G.; Spelt, C.; Buti, S.; Giuliano, G.; Quattrocchio, F.; Koes, R.; Perata, P.; et al. Tomato R2R3-MYB Proteins SlANT1 and SlAN2: Same Protein Activity, Different Roles. PLoS ONE 2015, 10, e0136365. [Google Scholar] [CrossRef]
- Jian, W.; Cao, H.; Yuan, S.; Liu, Y.; Lu, J.; Lu, W.; Li, N.; Wang, J.; Zou, J.; Tang, N.; et al. SlMYB75, an MYB-type transcription factor, promotes anthocyanin accumulation and enhances volatile aroma production in tomato fruits. Hortic. Res. 2019, 6, 22. [Google Scholar] [CrossRef]
- Zhang, Y.; Hu, Z.; Chu, G.; Huang, C.; Tian, S.; Zhao, Z.; Chen, G. Anthocyanin Accumulation and Molecular Analysis of Anthocyanin Biosynthesis-Associated Genes in Eggplant (Solanum melongena L.). J. Agric. Food Chem. 2014, 62, 2906–2912. [Google Scholar] [CrossRef]
- Li, L.; Li, S.; Ge, H.; Shi, S.; Li, D.; Liu, Y.; Chen, H. A light-responsive transcription factor SmMYB35 enhances anthocyanin biosynthesis in eggplant (Solanum melongena L.). Planta 2022, 255, 12. [Google Scholar] [CrossRef]
- Shi, S.; Liu, Y.; He, Y.; Li, L.; Li, D.; Chen, H. R2R3-MYB transcription factor SmMYB75 promotes anthocyanin biosynthesis in eggplant (Solanum melongena L.). Sci. Hortic. 2021, 282, 110020. [Google Scholar] [CrossRef]
- Li, L.; He, Y.; Ge, H.; Liu, Y.; Chen, H. Functional characterization of SmMYB86, a negative regulator of anthocyanin biosynthesis in eggplant (Solanum melongena L.). Plant Sci. 2021, 302, 110696. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; He, Y.-J.; Zhou, L.; Liu, Y.; Jiang, M.; Ren, L.; Chen, H. Transcriptome profiling of genes related to light-induced anthocyanin biosynthesis in eggplant (Solanum melongena L.) before purple color becomes evident. BMC Genom. 2018, 19. [Google Scholar] [CrossRef]
- Cominelli, E.; Gusmaroli, G.; Allegra, D.; Galbiati, M.; Wade, H.K.; Jenkins, G.I.; Tonelli, C. Expression analysis of anthocyanin regulatory genes in response to different light qualities in Arabidopsis thaliana. J. Plant Physiol. 2008, 165, 886–894. [Google Scholar] [CrossRef] [PubMed]
- Leyva, A.; Jarillo, J.A.; Salinas, J.; Martinez-Zapater, J.M. Low temperature induces the accumulation of phenylalanine ammonia-lyase and chalcone synthase mRNAs of Arabidopsis thaliana in a light-dependent manner. Plant Physiol. 1995, 108, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.S.; Jalaluddin, M.; Garner, J.O.; Yoshimoto, M.; Yamakawa, O. Artificial shading and temperature influence on anthocyanin compositions in sweetpotato leaves. Hortscience 2005, 40, 176–180. [Google Scholar] [CrossRef]
- An, X.-H.; Tian, Y.; Chen, K.-Q.; Liu, X.-J.; Liu, D.-D.; Xie, X.-B.; Cheng, C.-G.; Cong, P.-H.; Hao, Y.-J. MdMYB9 and MdMYB11 are involved in the regulation of the JA-induced biosynthesis of anthocyanin and proanthocyanidin in apples. Plant Cell Physiol. 2015, 56, 650–662. [Google Scholar] [CrossRef] [PubMed]
- Gagné, S.; Cluzet, S.; Mérillon, J.-M.; Gény, L. ABA Initiates Anthocyanin Production in Grape Cell Cultures. J. Plant Growth Regul. 2011, 30, 1–10. [Google Scholar] [CrossRef]
- Li, X.; Zhang, J.; Zhou, L.; Sheng, L.; Chen, H. Establishment of Agrobacterium tumefaciens-mediated genetic transformation system for eggplant cotyledons. J. Shanghai Jiao Tong Univ. Agric. Sci. 2018, 36, 1–6. [Google Scholar] [CrossRef]
- Wang, D.; Wang, M.; Liu, J.; Zhou, X.H.; Liu, S.Y.; Yang, Y.; Zhuang, Y. Cloning of U6 Promoters and Establishment of CRISPR/Cas9 Mediated Gene Editing System in Eggplant. Hortic. Plant J. 2022, 49, 791–800. [Google Scholar] [CrossRef]
- Lee, J.; Durst, R.W.; Wrolstad, R.E. Determination of total monomeric anthocyanin pigment content of fruit juices, beverages, natural colorants, and wines by the pH differential method: Collaborative study. J. AOAC Int. 2005, 88, 1269–1278. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wen, L.; Chen, Z.; Zhang, Z.; Pang, X.; Deng, Z.; Liu, T.; Guo, Y. Study on metabolic variation in whole grains of four proso millet varieties reveals metabolites important for antioxidant properties and quality traits. Food Chem. 2021, 357, 129791. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Lee, H.K.; Kim, C.Y.; Hong, Y.J.; Choe, C.M.; You, T.W.; Seong, G.J. Purified high-dose anthocyanoside oligomer administration improves nocturnal vision and clinical symptoms in myopia subjects. Br. J. Nutr. 2005, 93, 895–899. [Google Scholar] [CrossRef]
- Nguyen, X.C.; Hoang, M.H.T.; Kim, H.S.; Lee, K.; Liu, X.-M.; Kim, S.H.; Bahk, S.; Park, H.C.; Chung, W.S. Phosphorylation of the transcriptional regulator MYB44 by mitogen activated protein kinase regulates Arabidopsis seed germination. Biochem. Biophys. Res. Commun. 2012, 423, 703–708. [Google Scholar] [CrossRef]
- Mao, G.; Meng, X.; Liu, Y.; Zheng, Z.; Chen, Z.; Zhang, S. Phosphorylation of a WRKY Transcription Factor by Two Pathogen-Responsive MAPKs Drives Phytoalexin Biosynthesis in Arabidopsis. Plant Cell 2011, 23, 1639–1653. [Google Scholar] [CrossRef]
- Meng, X.; Zhang, S. MAPK Cascades in Plant Disease Resistance Signaling. Annu. Rev. Phytopathol. 2013, 51, 245–266. [Google Scholar] [CrossRef]
- Lee, H. Mitogen-Activated Protein Kinase Kinase 3 Is Required for Regulation during Dark-Light Transition. Mol. Cells 2015, 38, 651–656. [Google Scholar] [CrossRef]
- Sethi, V.; Raghuram, B.; Sinha, A.K.; Chattopadhyay, S. A Mitogen-Activated Protein Kinase Cascade Module, MKK3-MPK6 and MYC2, Is Involved in Blue Light-Mediated Seedling Development in Arabidopsis. Plant Cell 2014, 26, 3343–3357. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.-Y.; Zhang, C.; Li, Z.-C.; Wang, Z.-R.; Jiang, X.-X.; Shi, Y.-F.; Tian, S.-N.; Braun, E.; Mei, Y.; Qiu, W.-L.; et al. The MAPK Kinase Kinase GmMEKK1 Regulates Cell Death and Defense Responses. Plant Physiol. 2018, 178, 907–922. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, J.; Xu, L.; Wang, Q.; Lou, Y. Expressing OsMPK4 Impairs Plant Growth but Enhances the Resistance of Rice to the Striped Stem Borer Chilo suppressalis. Int. J. Mol. Sci. 2018, 19, 1182. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Schneider, J.D.; Lin, C.; Geng, S.; Ma, T.; Lawrence, S.R.; Dufresne, C.P.; Harmon, A.C.; Chen, S. MPK4 Phosphorylation Dynamics and Interacting Proteins in Plant Immunity. J. Proteome Res. 2019, 18, 826–840. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wang, B.; Xin, X.; Ren, D. Protein Kinases in Shaping Plant Architecture. Curr. Protein Pept. Sci. 2018, 19, 390–400. [Google Scholar] [CrossRef] [PubMed]
- Hamel, L.-P.; Nicole, M.-C.; Sritubtim, S.; Morency, M.-J.; Ellis, M.; Ehlting, J.; Beaudoin, N.; Barbazuk, B.; Klessig, D.; Lee, J.; et al. Ancient signals: Comparative genomics of plant MAPK and MAPKK gene families. Trends Plant Sci. 2006, 11, 192–198. [Google Scholar] [CrossRef] [PubMed]
- Rao, K.P.; Richa, T.; Kumar, K.; Raghuram, B.; Sinha, A.K. In Silico Analysis Reveals 75 Members of Mitogen-Activated Protein Kinase Kinase Kinase Gene Family in Rice. DNA Res. 2010, 17, 139–153. [Google Scholar] [CrossRef]
- Jiao, J.; Liu, X.; Wu, J.; Xu, G.; Zhang, S. Characterization of the MAPK Gene Family and PbrMAPK13 Response to Hormone and Temperature Stresses via Different Expression Pattern in Pyrus × bretschneideri Pollen. J. Am. Soc. Hortic. Sci. 2017, 142, 163–174. [Google Scholar] [CrossRef]
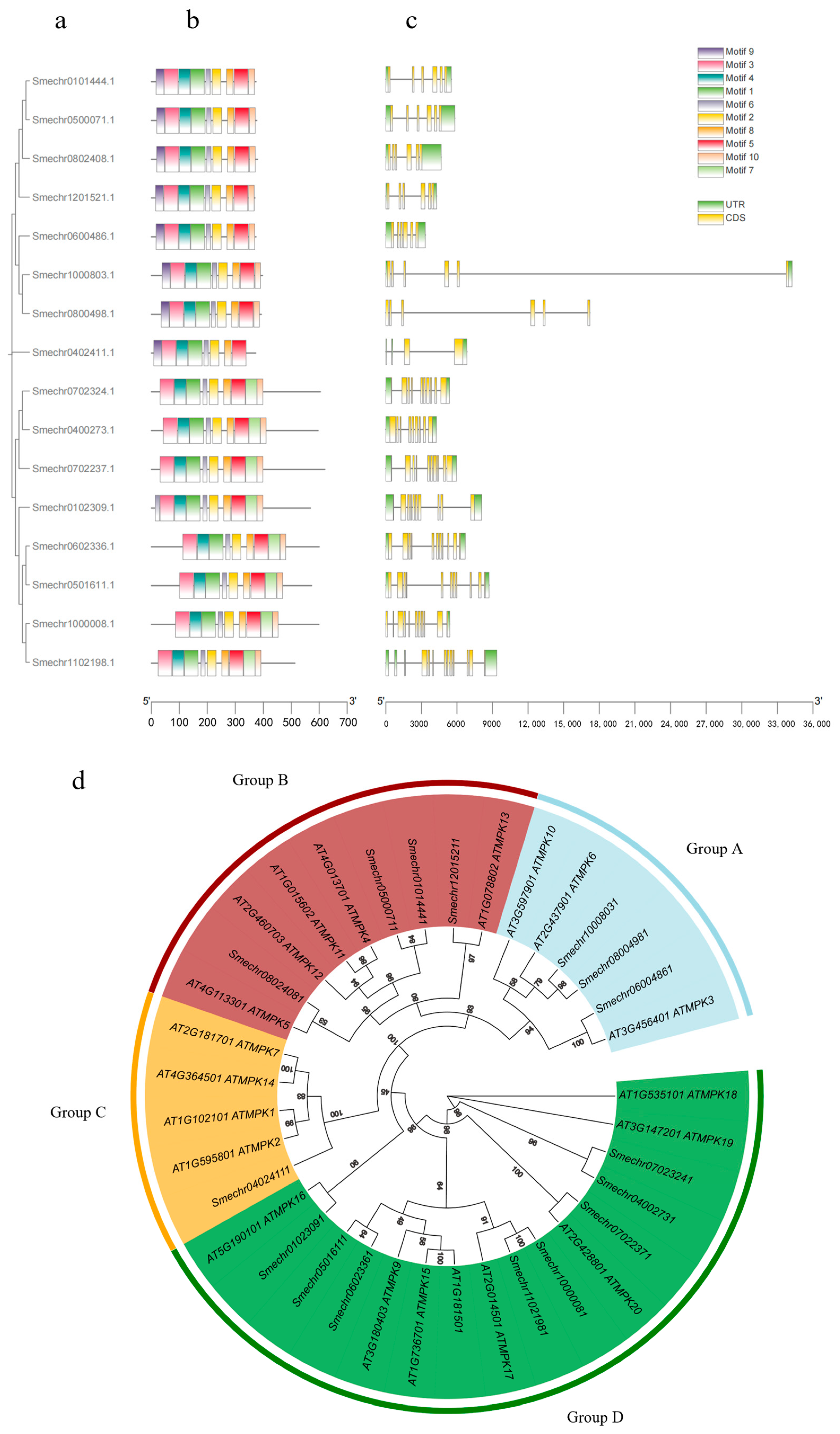

| Gene Name | Amino Acid Length | Molecular Weight (kDa) | pI | Instability Index | Aliphatic Index | Grand Average of Hydropathicity | Cellular Localization Prediction |
|---|---|---|---|---|---|---|---|
| Smechr0602336.1 | 599 | 68,206.41 | 6.81 | 40.29 | 79.78 | −0.542 | chloroplast |
| Smechr0102309.1 | 568 | 64,702.95 | 8.9 | 40.47 | 77.62 | −0.539 | cytosol |
| Smechr0702237.1 | 619 | 70,276.51 | 9.22 | 38.26 | 78.01 | −0.517 | cytosol |
| Smechr0501611.1 | 572 | 64,935.83 | 6.57 | 47.56 | 82.36 | −0.505 | chloroplast |
| Smechr0702324.1 | 603 | 68,417.71 | 9.42 | 35.05 | 82.17 | −0.422 | cytosol |
| Smechr1102198.1 | 512 | 58,418.06 | 7.85 | 39.2 | 77.93 | −0.418 | nucleus |
| Smechr0101444.1 | 373 | 42,977.83 | 6.02 | 42.24 | 85.2 | −0.411 | cytosol |
| Smechr0500071.1 | 376 | 43,136.23 | 6.6 | 43.95 | 90.24 | −0.393 | cytosol |
| Smechr0400273.1 | 595 | 67,249.37 | 9.21 | 32.35 | 83.92 | −0.362 | chloroplast |
| Smechr1000803.1 | 396 | 45,656.19 | 5.57 | 44.32 | 86.74 | −0.361 | nucleus |
| Smechr1000008.1 | 598 | 68,571.43 | 6.33 | 39.93 | 85.95 | −0.312 | cytosol |
| Smechr0800498.1 | 392 | 45,015.51 | 5.47 | 43.77 | 89.11 | −0.306 | nucleus |
| Smechr0802408.1 | 379 | 43,626.87 | 5.79 | 41.98 | 91.56 | −0.301 | chloroplast |
| Smechr1201521.1 | 369 | 42,523.68 | 5.06 | 40.35 | 94.88 | −0.298 | cytosol |
| Smechr0600486.1 | 373 | 42,948.28 | 5.46 | 36.65 | 94.13 | −0.266 | cytosol |
| Smechr0402411.1 | 372 | 42,699.36 | 6.32 | 43.59 | 94.35 | −0.26 | nucleus |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liao, J.; Dong, Y.; Hua, Z.; Hao, J.; Zhao, N.; Li, S.; Chen, H. Identification of Eggplant SmMPK Gene Family and Functional Verification of SmMPK4.1. Horticulturae 2024, 10, 239. https://doi.org/10.3390/horticulturae10030239
Liao J, Dong Y, Hua Z, Hao J, Zhao N, Li S, Chen H. Identification of Eggplant SmMPK Gene Family and Functional Verification of SmMPK4.1. Horticulturae. 2024; 10(3):239. https://doi.org/10.3390/horticulturae10030239
Chicago/Turabian StyleLiao, Jielei, Yanxiao Dong, Ziyi Hua, Jiangnan Hao, Na Zhao, Shaohang Li, and Huoying Chen. 2024. "Identification of Eggplant SmMPK Gene Family and Functional Verification of SmMPK4.1" Horticulturae 10, no. 3: 239. https://doi.org/10.3390/horticulturae10030239





