Assessing the Genetic Diversity of Wild and Commercial Feijoa sellowiana Accessions Using AFLPs
Abstract
:1. Introduction
2. Materials and Methods
2.1. Feijoa sellowiana Sampling
2.2. DNA Isolation and AFLP Protocol
2.3. Data Analysis
3. Results
3.1. AFLP Genetic Parameters
3.2. Genetic Diversity of Wild Types and Cultivars
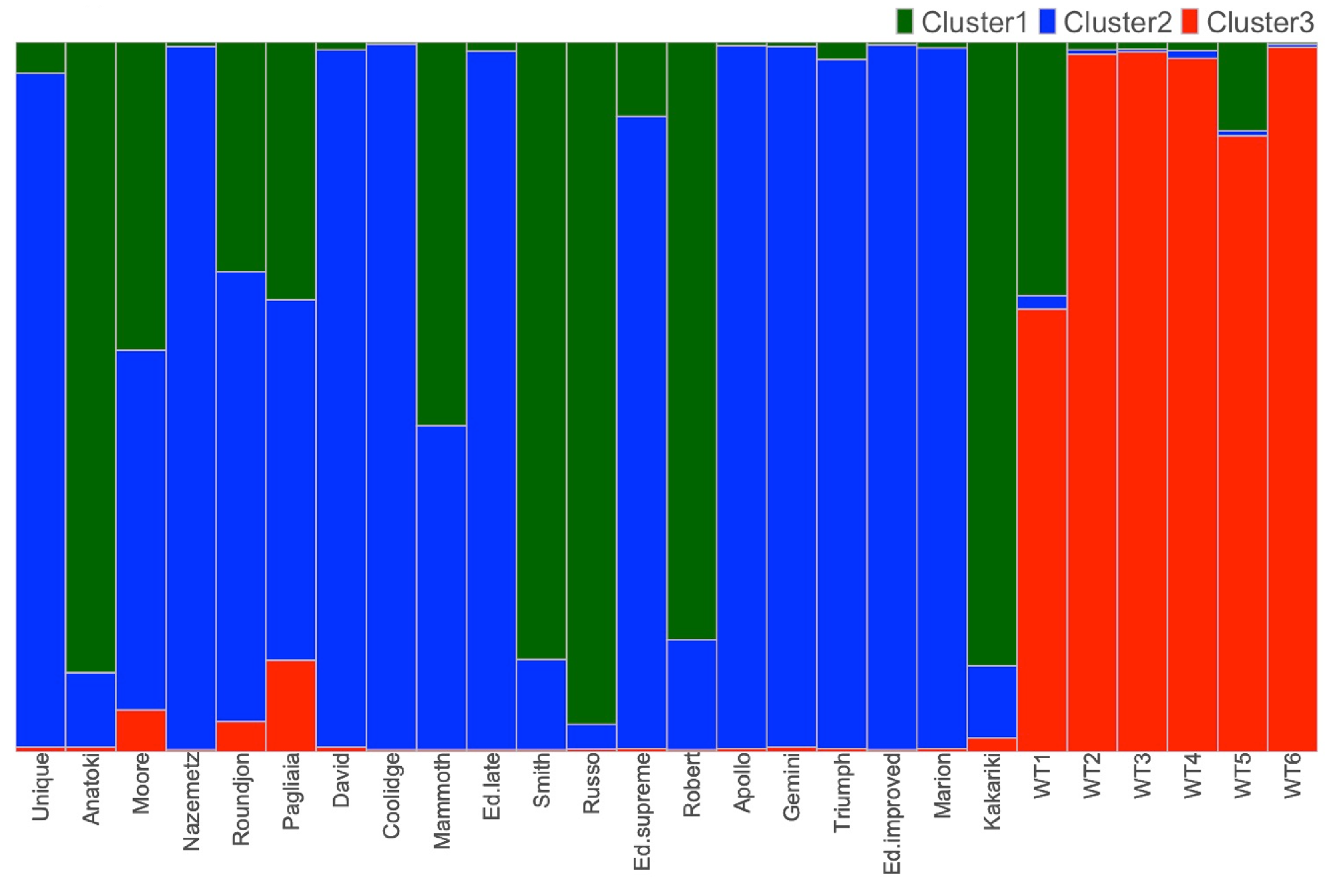
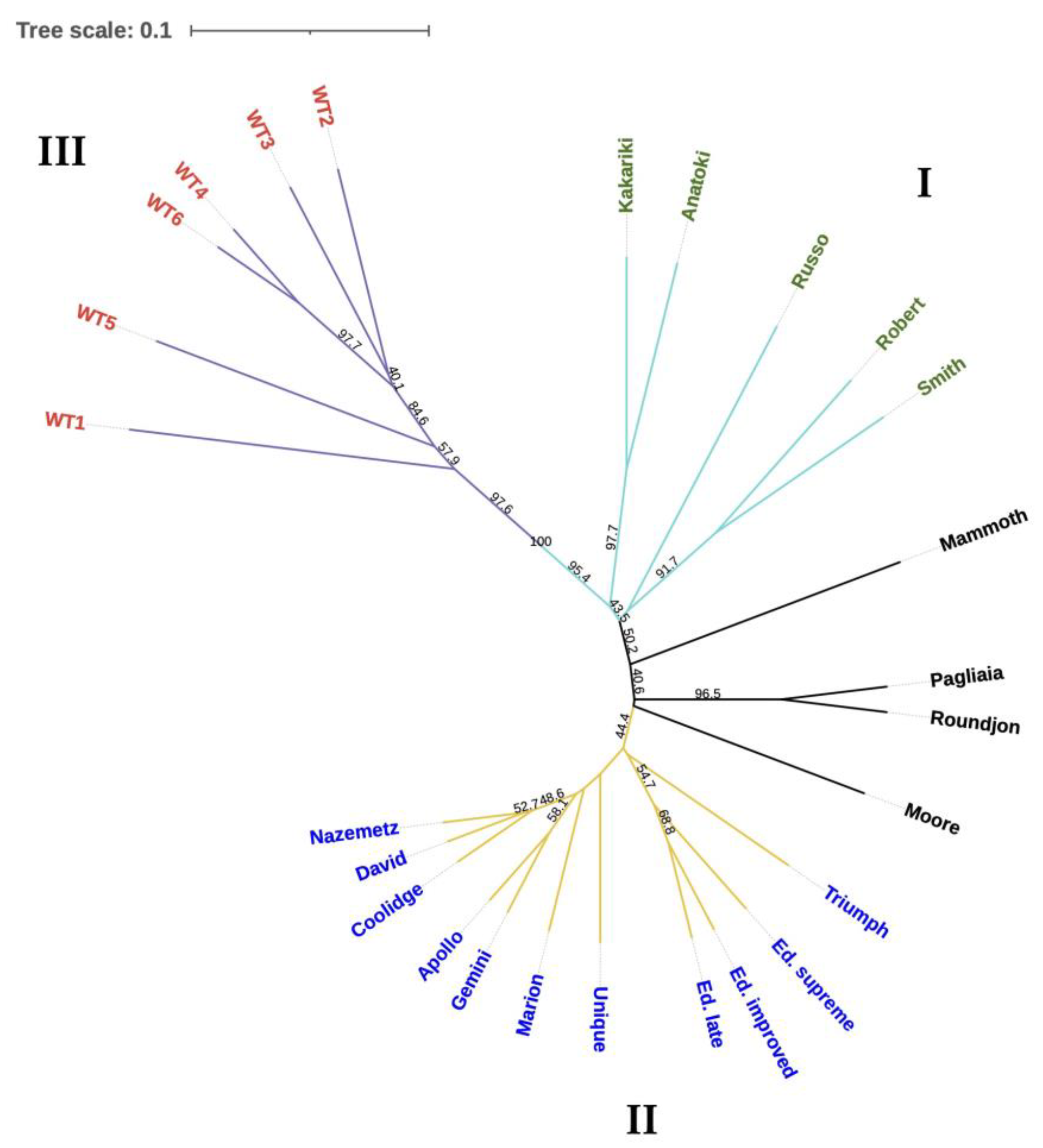
3.3. Population Structure
Clustering Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Weston, R.J. Bioactive Products from Fruit of the Feijoa (Feijoa sellowiana, Myrtaceae): A Review. Food Chem. 2010, 121, 923–926. [Google Scholar] [CrossRef]
- Tuncel, N.B.; Yılmaz, N. Optimizing the Extraction of Phenolics and Antioxidants from Feijoa (Feijoa sellowiana, Myrtaceae). J. Food Sci. Technol. 2015, 52, 141–150. [Google Scholar] [CrossRef]
- Ramírez, F.; Kallarackal, J. Feijoa [Acca sellowiana (O. Berg) Burret] Pollination: A Review. Sci. Hortic. 2017, 226, 333–341. [Google Scholar] [CrossRef]
- Zhu, F. Chemical and Biological Properties of Feijoa (Acca sellowiana). Trends Food Sci. Technol. 2018, 81, 121–131. [Google Scholar] [CrossRef]
- Thorp, G.; Bieleski, R. Feijoas: Origins, Cultivars and Uses; David Bateman Ltd.: Auckland, New Zealand, 2002. [Google Scholar]
- Álvarez-Herrera, J.G.; Tovar-Escobar, J.; Ruiz, H.D. Postharvest behavior of feijoa fruit (Acca sellowiana Berg) subjected to different 1-MCP doses and storage temperatures. Rev. Bras. Frutic. 2023, 45. Available online: https://www.scielo.br/j/rbf/a/pqCfZWvcMJwpc3TZw9XZxrt/ (accessed on 1 January 2024).
- Rupavatharam, S.; East, A.R.; Heyes, J.A. Re-Evaluation of Harvest Timing in ‘Unique’Feijoa Using 1-MCP and Exogenous Ethylene Treatments. Postharvest Biol. Technol. 2015, 99, 152–159. [Google Scholar] [CrossRef]
- Oseko, J.; East, A.; Heyes, J. Recent Advances in the Postharvest Technology of Feijoa. Sci. Hortic. 2022, 297, 110969. [Google Scholar] [CrossRef]
- Borsuk, L.J.; Saifert, L.; Villamil, J.; Otalora, M.; Mora, F.D.S.; Nodari, R.O. Phenotypic Variability in Feijoa Fruits [Acca sellowiana (O. Berg.) Burret] on Indigenous Lands, Quilombolas Communities and Protected Areas in the South of Brazil. Rev. Bras. Frutic. 2017, 39, e-699. [Google Scholar] [CrossRef]
- Thorp, T.G. DSIR’s Feijoa Breeding Programme Goes to South America. Orchard. New Zealand 1988, 61, 213–215. [Google Scholar]
- Schotsmans, W.C.; East, A.; Thorp, G.; Woolf, A.B. Feijoa (Acca sellowiana [Berg] Burret). In Postharvest Biology and Technology of Tropical and Subtropical Fruits; Elsevier: Amsterdam, The Netherlands, 2011; pp. 115–135e. [Google Scholar]
- Fischer, G.; Miranda Lasprilla, D.C.; Cayón Salinas, G. Cultivo, Poscosecha y Exportación de La Feijoa (Acca sellowiana Berg); Universidad Nacional de Colombia: Bogota, Colombia, 2003. [Google Scholar]
- Joel, D.; Thiago, S.O.; Juliano, G.B.; Rubens, O.N. Performance of Segregating Populations of Feijoa Cultivated under the Agroforestry Systems in Southern Brazil. Afr. J. Agric. Res. 2014, 9, 3305–3312. [Google Scholar]
- Pasquariello, M.S.; Mastrobuoni, F.; Di Patre, D.; Zampella, L.; Capuano, L.R.; Scortichini, M.; Petriccione, M. Agronomic, Nutraceutical and Molecular Variability of Feijoa (Acca sellowiana (O. Berg) Burret) Germplasm. Sci. Hortic. 2015, 191, 1–9. [Google Scholar] [CrossRef]
- Ramírez, F.; Kallarackal, J. Phenological Growth Stages of Feijoa [Acca sellowiana (O. Berg) Burret] According to the BBCH Scale under Tropical Andean Conditions. Sci. Hortic. 2018, 232, 184–190. [Google Scholar] [CrossRef]
- Agronet—Agricultural Statistics. Report: Area, Production and National Yield per Cultivation. Ministry of Agriculture, Colombia. Available online: https://www.agronet.gov.co/estadistica/Paginas/home.aspx (accessed on 25 March 2024).
- Aitken, A.G.; Hewett, E.W. FreshFacts: New Zealand Horticulture; New Zealand Institute for Plant & Food Research: Auckland, New Zealand, 2016; p. 14. [Google Scholar]
- Guerra, M.P.; Cangahuala-Inocente, G.C.; Vesco, L.L.D.; Pescador, R.; Caprestano, C.A. Micropropagation Systems of Feijoa (Acca sellowiana (O. Berg) Burret). Protoc. Micropropag. Sel. Econ. -Important Hortic. Plants 2013, 11013, 45–62. [Google Scholar]
- Da Silva, J.C.; Da Silva Tognon, L.; Martins, Y.G.; Louise dos Santos, K.; Picolotto, L.; Araújo, L.; Haygert Lencina, K. Enraizamento de Estacas de Feijoa sellowiana Com Uso de Antioxidante. Agropecuária Catarin. 2023, 36, 42–47. [Google Scholar] [CrossRef]
- Vatrano, T.; Amenta, M.; Copetta, A.; Guardo, M.; Nunziata, A.; Strano, M.C.; Petriccione, M. Multifunctional Role of Acca sellowiana from Farm Management to Postharvest Life: A Review. Agronomy 2022, 12, 1802. [Google Scholar] [CrossRef]
- Byrne, D.H. Trends in Fruit Breeding. In Fruit Breeding; Springer: New York, NY, USA, 2012; pp. 3–36. [Google Scholar]
- Amiteye, S. Basic Concepts and Methodologies of DNA Marker Systems in Plant Molecular Breeding. Heliyon 2021, 7, e08093. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Mora, F.D.; Saifert, L.; Ciotta, M.N.; Ribeiro, H.N.; Petry, V.S.; Rojas-Molina, A.M.; Lopes, M.E.; Lombardi, G.G.; dos Santos, K.L.; Ducroquet, J.P.H.J. Characterization of Phenotypic Diversity of Feijoa Fruits of Germplasm Accessions in Brazil. Agrosystems Geosci. Environ. 2019, 2, 1–11. [Google Scholar] [CrossRef]
- Fischer, G.; Parra-Coronado, A. Influence of Some Environmental Factors on the Feijoa (Acca sellowiana [Berg] Burret): A Review. Agron. Colomb. 2020, 38, 388–397. [Google Scholar] [CrossRef]
- Dettori, M.T.; Palombi, M.A. Identification of Feijoa sellowiana Berg Accessions by RAPD Markers. Sci. Hortic. 2000, 86, 279–290. [Google Scholar] [CrossRef]
- dos Santos, K.L.; Welter, L.J.; Dantas, A.C.d.M.; Guerra, M.P.; Ducroquet, J.P.H.J.; Nodari, R.O. Transference of Microsatellite Markers from Eucalyptus spp to Acca sellowiana and the Successful Use of This Technique in Genetic Characterization. Genet. Mol. Biol. 2007, 30, 73–79. [Google Scholar] [CrossRef]
- Santos, K.L.; Santos, M.O.; Laborda, P.R.; Souza, A.P.; Peroni, N.; Nodari, R.O. Isolation and Characterization of Microsatellite Markers in Acca sellowiana (Berg) Burret. Mol. Ecol. Resour. 2008, 8, 1417–1419. [Google Scholar] [CrossRef] [PubMed]
- Donazzolo, J.; Stefenon, V.M.; Guerra, M.P.; Nodari, R.O. On Farm Management of Acca sellowiana (Myrtaceae) as a Strategy for Conservation of Species Genetic Diversity. Sci. Hortic. 2020, 259, 108826. [Google Scholar] [CrossRef]
- Saifert, L.; Sánchez-Mora, F.D.; Borsuk, L.J.; Donazzolo, J.; da Costa, N.C.F.; Ribeiro, H.N.; Nodari, R.O. Evaluation of the Genetic Diversity in the Feijoa Accessions Maintained at Santa Catarina, Brazil. Crop. Sci. 2020, 60, 345–356. [Google Scholar] [CrossRef]
- Quezada, M.; Pastina, M.M.; Ravest, G.; Silva, P.; Vignale, B.; Cabrera, D.; Hinrichsen, P.; Garcia, A.A.F.; Pritsch, C. A First Genetic Map of Acca sellowiana Based on ISSR, AFLP and SSR Markers. Sci. Hortic. 2014, 169, 138–146. [Google Scholar] [CrossRef]
- Quezada, M.; Amadeu, R.R.; Vignale, B.; Cabrera, D.; Pritsch, C.; Garcia, A.A.F. Construction of a High-Density Genetic Map of Acca sellowiana (Berg.) Burret, an Outcrossing Species, Based on Two Connected Mapping Populations. Front. Plant. Sci. 2021, 12, 626811. [Google Scholar] [CrossRef] [PubMed]
- Nybom, H. Comparison of Different Nuclear DNA Markers for Estimating Intraspecific Genetic Diversity in Plants. Mol. Ecol. 2004, 13, 1143–1155. [Google Scholar] [CrossRef]
- Keller, H.A.; Tressens, S.G. New records for Argentina of two multipurpose species: Acca sellowiana (Myrtaceae) and Casearia lasiophylla (Flacourtiaceae). Darwiniana 2007, 45, 204–212. [Google Scholar]
- Nuñez, M.; Keller, H.; Pirondo, A. Etnobotánica de Acca sellowiana (Myrtaceae): Una especie de uso múltiple de Misiones, Argentina. Bonplandia 2013, 32.1, 27–37. [Google Scholar] [CrossRef]
- Doyle, J.J. Isolation of Plant DNA from Fresh Tissue. Focus 1990, 12, 13–15. [Google Scholar]
- López-Sepúlveda, P.; Takayama, K.; Greimler, J.; Peñailillo, P.; Crawford, D.J.; Baeza, M.; Ruiz, E.; Kohl, G.; Tremetsberger, K.; Gatica, A. Genetic Variation (AFLPs and Nuclear Microsatellites) in Two Anagenetically Derived Endemic Species of Myrceugenia (Myrtaceae) on the Juan Fernández Islands, Chile. Am. J. Bot. 2013, 100, 722–734. [Google Scholar] [CrossRef]
- Coppi, A.; Cecchi, L.; Mengoni, A.; Pustahija, F.; Tomović, G.; Selvi, F. Low Genetic Diversity and Contrasting Patterns of Differentiation in the Two Monotypic Genera Halacsya and Paramoltkia (Boraginaceae) Endemic to the Balkan Serpentines. Flora-Morphol. Distrib. Funct. Ecol. Plants 2014, 209, 5–14. [Google Scholar] [CrossRef]
- Vos, P.; Hogers, R.; Bleeker, M.; Reijans, M.; van de Lee, T.; Hornes, M.; Friters, A.; Pot, J.; Paleman, J.; Kuiper, M. AFLP: A New Technique for DNA Fingerprinting. Nucleic Acids Res. 1995, 23, 4407–4414. [Google Scholar] [CrossRef]
- Coppi, A.; Lastrucci, L.; Cappelletti, D.; Cerri, M.; Ferranti, F.; Ferri, V.; Foggi, B.; Gigante, D.; Venanzoni, R.; Viciani, D. AFLP Approach Reveals Variability in Phragmites australis: Implications for Its Die-Back and Evidence for Genotoxic Effects. Front. Plant Sci. 2018, 9, 386. [Google Scholar] [CrossRef] [PubMed]
- Meudt, H.M.; Clarke, A.C. Almost Forgotten or Latest Practice? AFLP Applications, Analyses and Advances. Trends Plant. Sci. 2007, 12, 106–117. [Google Scholar] [CrossRef] [PubMed]
- Yeh, F.C. POPGENE (Version 1.3. 1). Microsoft Window-Bases Freeware for Population Genetic Analysis. 1999. Available online: http://www.ualberta.ca/~fyeh/ (accessed on 16 January 2024).
- Peakall, R.O.D.; Smouse, P.E. GENALEX 6: Genetic Analysis in Excel. Population Genetic Software for Teaching and Research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of Population Structure Using Multilocus Genotype Data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Earl, D.A.; VonHoldt, B.M. Structure Harvester: A Website and Program for Visualizing Structure Output and Implementing the Evanno Method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the Number of Clusters of Individuals Using the Software Structure: A Simulation Study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Francis, R.M. Pophelper: An R Package and Web App to Analyse and Visualize Population Structure. Mol. Ecol. Resour. 2017, 17, 27–32. [Google Scholar] [CrossRef]
- Jombart, T. Adegenet: A R Package for the Multivariate Analysis of Genetic Markers. Bioinformatics 2008, 24, 1403–1405. [Google Scholar] [CrossRef]
- Kamvar, Z.N.; Tabima, J.F.; Grünwald, N.J. Poppr: An R Package for Genetic Analysis of Populations with Clonal, Partially Clonal, and/or Sexual Reproduction. PeerJ 2014, 2, e281. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013. [Google Scholar]
- Letunic, I.; Bork, P. Interactive Tree of Life (ITOL) v5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Sensi, E.; Vignani, R.; Scali, M.; Masi, E.; Cresti, M. DNA Fingerprinting and Genetic Relatedness among Cultivated Varieties of Olea europaea L. Estimated by AFLP Analysis. Sci. Hortic. 2003, 97, 379–388. [Google Scholar] [CrossRef]
- Gao, J.; Zhang, Y.; Wang, C.; Zhang, S.; Qi, L.; Song, W. AFLP Fingerprinting of Populus deltoides and Populus × canadensis Elite Accessions. New For. 2009, 37, 333–344. [Google Scholar] [CrossRef]
- Christensen, S.; von Bothmer, R.; Poulsen, G.; Maggioni, L.; Phillip, M.; Andersen, B.A.; Jørgensen, R.B. AFLP Analysis of Genetic Diversity in Leafy Kale (Brassica oleracea L. Convar. Acephala (DC.) Alef.) Landraces, Cultivars and Wild Populations in Europe. Genet. Resour. Crop. Evol. 2011, 58, 657–666. [Google Scholar] [CrossRef]
- Campbell, D.; Duchesne, P.; Bernatchez, L. AFLP Utility for Population Assignment Studies: Analytical Investigation and Empirical Comparison with Microsatellites. Mol. Ecol. 2003, 12, 1979–1991. [Google Scholar] [CrossRef]
- Mueller, U.G.; Wolfenbarger, L.L. AFLP Genotyping and Fingerprinting. Trends Ecol. Evol. 1999, 14, 389–394. [Google Scholar] [CrossRef] [PubMed]
- Schlötterer, C. The Evolution of Molecular Markers—Just a Matter of Fashion? Nat. Rev. Genet. 2004, 5, 63–69. [Google Scholar] [CrossRef]
- Bensch, S.; Åkesson, M. Ten Years of AFLP in Ecology and Evolution: Why so Few Animals? Mol. Ecol. 2005, 14, 2899–2914. [Google Scholar] [CrossRef]
- Torres, M.M.; Pierantozzi, P.; Cáceres, M.E.; Labombarda, P.; Fontanazza, G.; Maestri, D.M. Genetic and Chemical Assessment of Arbequina Olive Cultivar Grown in Córdoba Province, Argentina. J. Sci. Food Agric. 2009, 89, 523–530. [Google Scholar] [CrossRef]
- Nodari, R.O.; Guerra, M.P.; Meler, K.; Ducroquet, J.P. Genetic Variability of Feijoa sellowiana Germplasm. Acta Hortic. 1997, 452, 41–46. [Google Scholar] [CrossRef]
- Sharpe, R.H.; Sherman, W.B.; Miller, E.P. Feijoa history and improvement. Proc. Fla. State Hort. Soc. 1993, 106, 134–139. [Google Scholar]
- Besnard, G.; Terral, J.-F.; Cornille, A. On the Origins and Domestication of the Olive: A Review and Perspectives. Ann. Bot. 2018, 121, 385–403. [Google Scholar] [CrossRef] [PubMed]
- Sanz-Cortés, F.; Parfitt, D.E.; Romero, C.; Struss, D.; Llácer, G.; Badenes, M.L. Intraspecific Olive Diversity Assessed with AFLP. Plant Breed. 2003, 122, 173–177. [Google Scholar] [CrossRef]
- Bandelj, D.; Jakše, J.; Javornik, B. Assessment of Genetic Variability of Olive Varieties by Microsatellite and AFLP Markers. Euphytica 2004, 136, 93–102. [Google Scholar] [CrossRef]
- Grati-Kamoun, N.; Mahmoud, F.L.; Rebaï, A.; Gargouri, A.; Panaud, O.; Saar, A. Genetic Diversity of Tunisian Olive Tree (Olea europaea L.) Cultivars Assessed by AFLP Markers. Genet. Resour. Crop. Evol. 2006, 53, 265–275. [Google Scholar] [CrossRef]
- Corrado, G.; la Mura, M.; Ambrosino, O.; Pugliano, G.; Varricchio, P.; Rao, R. Relationships of Campanian Olive Cultivars: Comparative Analysis of Molecular and Phenotypic Data. Genome 2009, 52, 692–700. [Google Scholar] [CrossRef]
- Hutchison, D.W.; Templeton, A.R. Correlation of Pairwise Genetic and Geographic Distance Measures: Inferring the Relative Influences of Gene Flow and Drift on the Distribution of Genetic Variability. Evolution 1999, 53, 1898–1914. [Google Scholar] [CrossRef]
| Accession of Genotype | Collection Site | Country of Origin |
|---|---|---|
| WT1 | Misiones (AR) | Argentina |
| WT2 | Misiones (AR) | Argentina |
| WT3 | Misiones (AR) | Argentina |
| WT4 | Misiones (AR) | Argentina |
| WT5 | Misiones (AR) | Argentina |
| WT6 | Misiones (AR) | Argentina |
| Moore | ALSIA (IT) | Unknown |
| Nazemetz | CREA-OFA-Caserta (IT) | USA |
| Roundjon | CREA-OFA-Caserta (IT) | New Zealand |
| Pagliaia | CREA-OFA-Caserta (IT) | Italy |
| Coolidge | CREA-OFA-Caserta (IT) | USA |
| Mammouth | CREA-OFA-Caserta (IT) | New Zealand |
| David | CREA-OFA-Caserta (IT) | New Zealand |
| Smith | CREA-OFA-Caserta (IT) | USA |
| Russo | CREA-OFA-Caserta (IT) | Italy |
| Robert | CREA-OFA-Caserta (IT) | New Zealand |
| Apollo | CREA-OFA-Caserta (IT) | New Zealand |
| Gemini | CREA-OFA-Caserta (IT) | New Zealand |
| Triumph | CREA-OFA-Caserta (IT) | New Zealand |
| Edenvale Improved | CREA-OFA-Caserta (IT) | USA |
| Edenvale Supreme | CREA-OFA-Caserta (IT) | USA |
| Edenvale Late | CREA-OFA-Caserta (IT) | USA |
| Marion | CREA-OFA-Caserta (IT) | New Zealand |
| Kakariki | Uzein (FR) | New Zealand |
| Unique | Uzein (FR) | New Zealand |
| Anatoki | Uzein (FR) | New Zealand |
| Oligo Name | Sequence 5′–3′ | Labeling | Refs. |
|---|---|---|---|
| Adapt. EcoRI_2 | AATTGGTACGCAGTCTAC | ||
| Adapt. MseI_1 | GACGATGAGTCCTGAG | ||
| Adapt. MseI_2 | TACTCAGGACTCAT | ||
| Primer pair 1 | EcoRI–ACG/MseI–CTC | HEX | [37] |
| Primer pair 2 | EcoRI–ACT/MseI–CTT | FAM | [36] |
| Primer pair 3 | EcoRI–ATG/MseI–CTT | FAM | [36] |
| Primer pair 4 | EcoRI–ATG/MseI–CTG | FAM | [36] |
| Primers | N. Total Loci | N. Polymorphic Loci | PPB | ne | h | I |
|---|---|---|---|---|---|---|
| EcoRI–ACG/MseI–CTC | 40.00 | 27.00 | 67.50% | 1.41 | 0.26 | 0.41 |
| EcoRI–ACT/MseI–CTT | 53.00 | 36.00 | 67.92% | 1.41 | 0.26 | 0.42 |
| EcoRI–ATG/MseI–CTT | 69.00 | 51.00 | 73.91% | 1.42 | 0.26 | 0.42 |
| EcoRI–ATG/MseI–CTG | 47.00 | 32.00 | 68.09% | 1.49 | 0.30 | 0.46 |
| Group | Sample Size | N. Polymorphic Loci | ne | h | I |
|---|---|---|---|---|---|
| Cultivars | 20.00 | 115 | 1.43 | 0.27 | 0.43 |
| WT | 6.00 | 75 | 1.62 | 0.37 | 0.55 |
| Source | df | SS | MS | Est. Var. | % | PhiPT | Nm |
|---|---|---|---|---|---|---|---|
| Among Pops | 1 | 114.985 | 114.985 | 10.665 | 39% | ||
| Within Pops | 24 | 396.900 | 16.538 | 16.538 | 61% | ||
| Total | 25 | 511.885 | 27.203 | 100% | 0.392 *** | 0.775 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bini, L.; Gori, M.; Novello, M.A.; Biricolti, S.; Giordani, E.; Lara, M.V.; Niella, F.; Nunziata, A.; Rocha, P.; Filippi, J.M.; et al. Assessing the Genetic Diversity of Wild and Commercial Feijoa sellowiana Accessions Using AFLPs. Horticulturae 2024, 10, 366. https://doi.org/10.3390/horticulturae10040366
Bini L, Gori M, Novello MA, Biricolti S, Giordani E, Lara MV, Niella F, Nunziata A, Rocha P, Filippi JM, et al. Assessing the Genetic Diversity of Wild and Commercial Feijoa sellowiana Accessions Using AFLPs. Horticulturae. 2024; 10(4):366. https://doi.org/10.3390/horticulturae10040366
Chicago/Turabian StyleBini, Lorenzo, Massimo Gori, Maria Angelina Novello, Stefano Biricolti, Edgardo Giordani, María Valeria Lara, Fernando Niella, Angelina Nunziata, Patricia Rocha, Jacopo Mattia Filippi, and et al. 2024. "Assessing the Genetic Diversity of Wild and Commercial Feijoa sellowiana Accessions Using AFLPs" Horticulturae 10, no. 4: 366. https://doi.org/10.3390/horticulturae10040366
APA StyleBini, L., Gori, M., Novello, M. A., Biricolti, S., Giordani, E., Lara, M. V., Niella, F., Nunziata, A., Rocha, P., Filippi, J. M., & Natale, R. (2024). Assessing the Genetic Diversity of Wild and Commercial Feijoa sellowiana Accessions Using AFLPs. Horticulturae, 10(4), 366. https://doi.org/10.3390/horticulturae10040366








