The AnUFGT1 Is Involved in the Anthurium ‘Alabama’ Anthocyanidin Deficiency
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Sample Preparation
2.2. Metabolomics Analysis
2.3. Extraction and Determination of Anthocyanin Content
2.4. Isolation of the AnUFGT1 Gene
2.5. Bioinformatics Analysis of AnUFGT1
2.6. AnUFGT1 Expression Characteristics and Enzyme Activity Analysis
2.7. Transform Solution of Agrobacterium Containing AnUFGT1
2.8. Transformation of AnUFGT1 into Ufgt and ‘X’
3. Results
3.1. Metabolomics Analysis Further Elucidated the Differences in the Anthocyanin Pathway between ‘Alabama’ and ‘X’
3.1.1. Metabolomic Analysis Revealed the Main Types and Levels of Anthocyanins in ‘Alabama’ and ‘X’
3.1.2. Metabolomics Analysis Revealed the Levels of Intermediate Products of the ASP in ‘Alabama’ and ‘X’
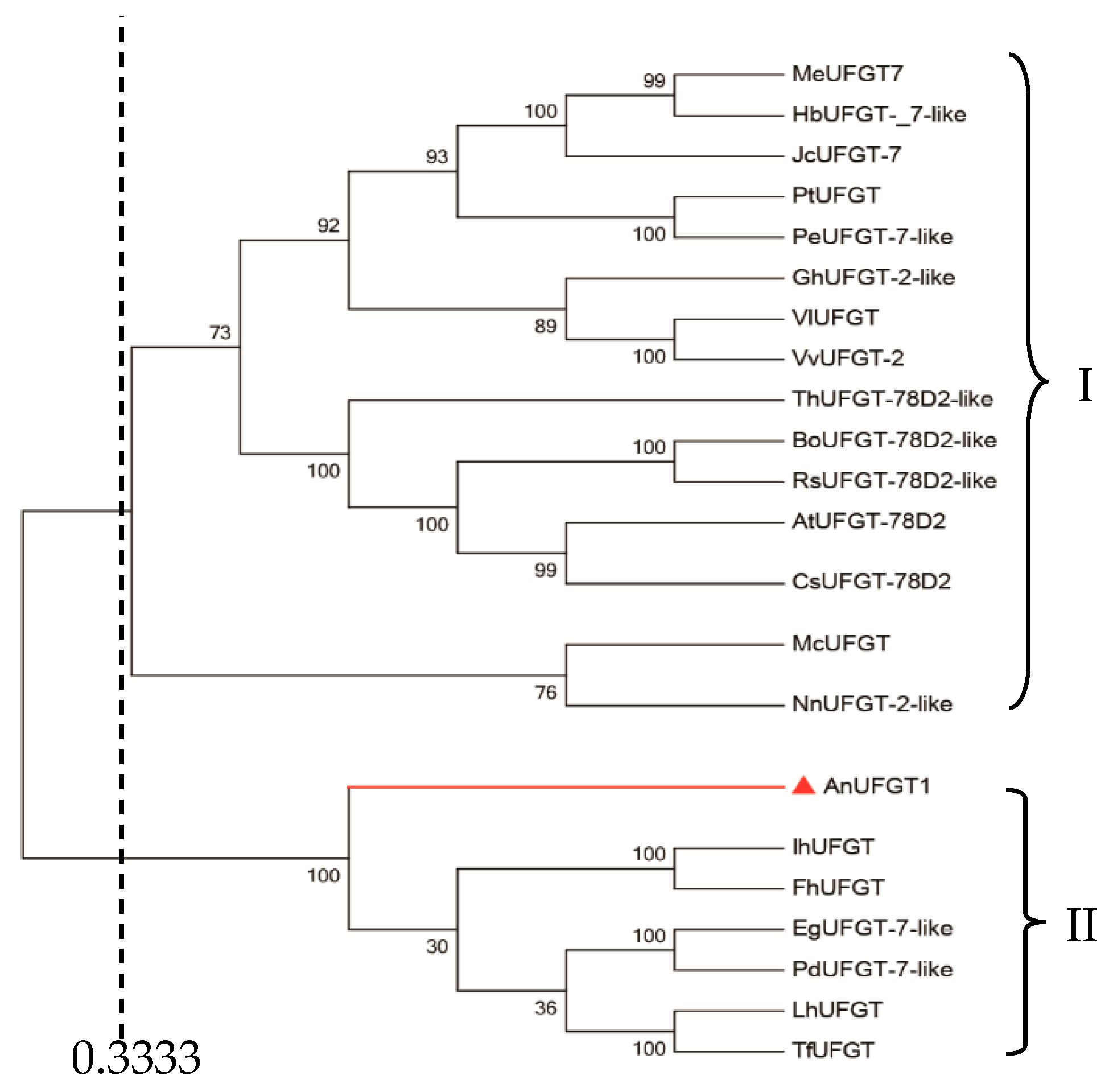
3.2. AnUFGT1 Belongs to the UFGT Genes
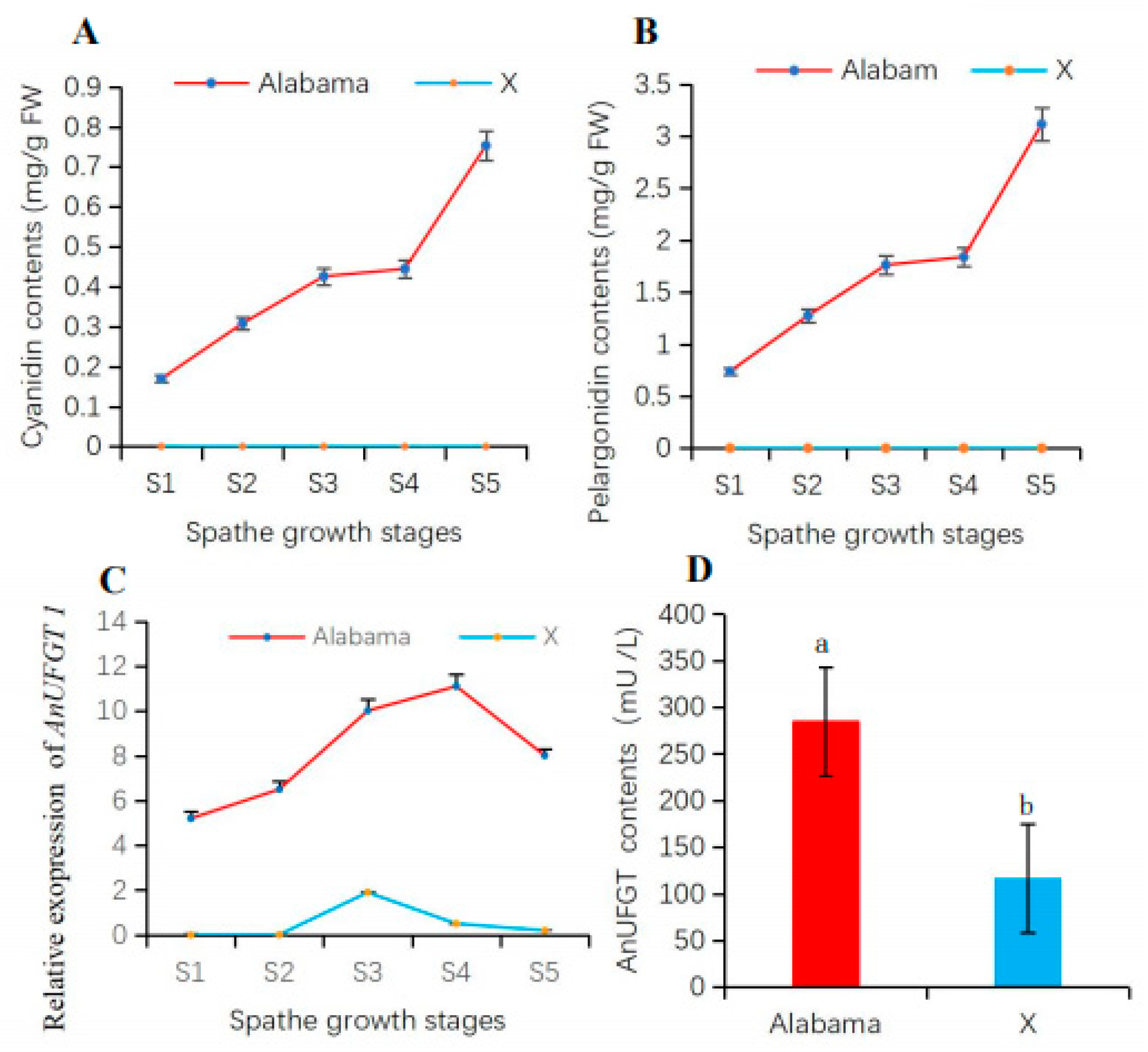
3.3. AnUFGT1 Expression Characteristics Differed between ‘Alabama’ and ‘X’
3.4. The AnUFGT1 Is Related to Anthocyanin Accumulation
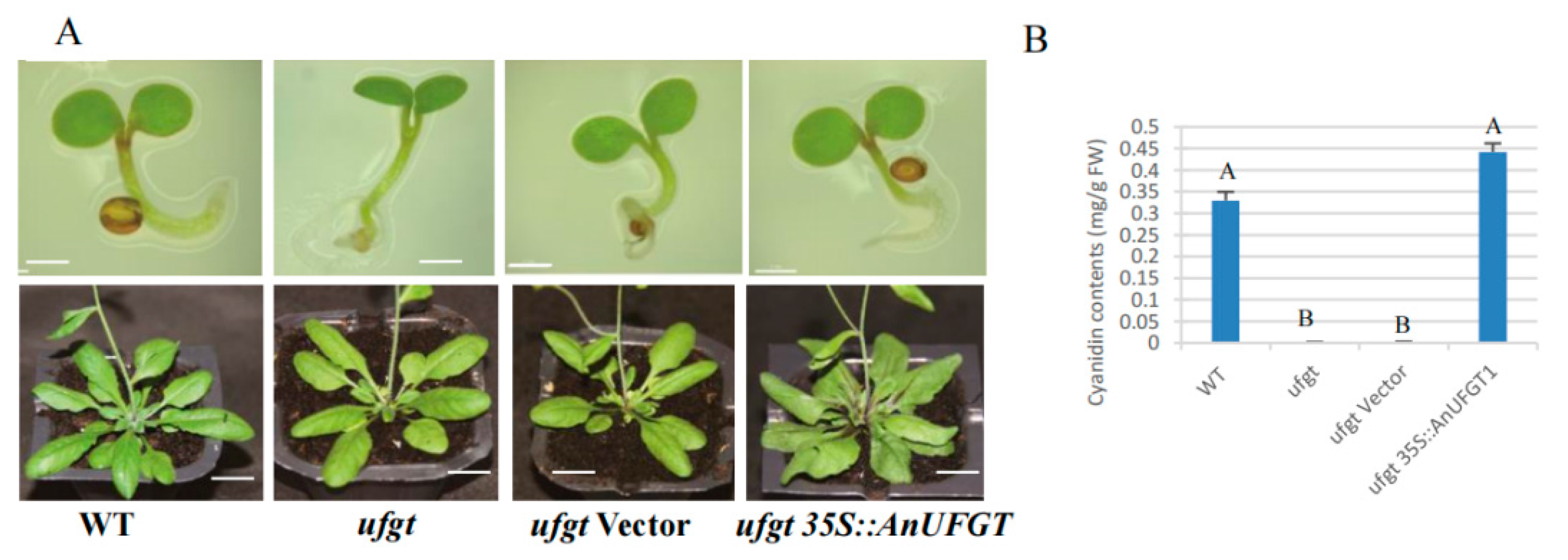
3.5. Overexpression of AnUFGT1 in Arabidopsis Accelerated the Accumulation of Anthocyanins
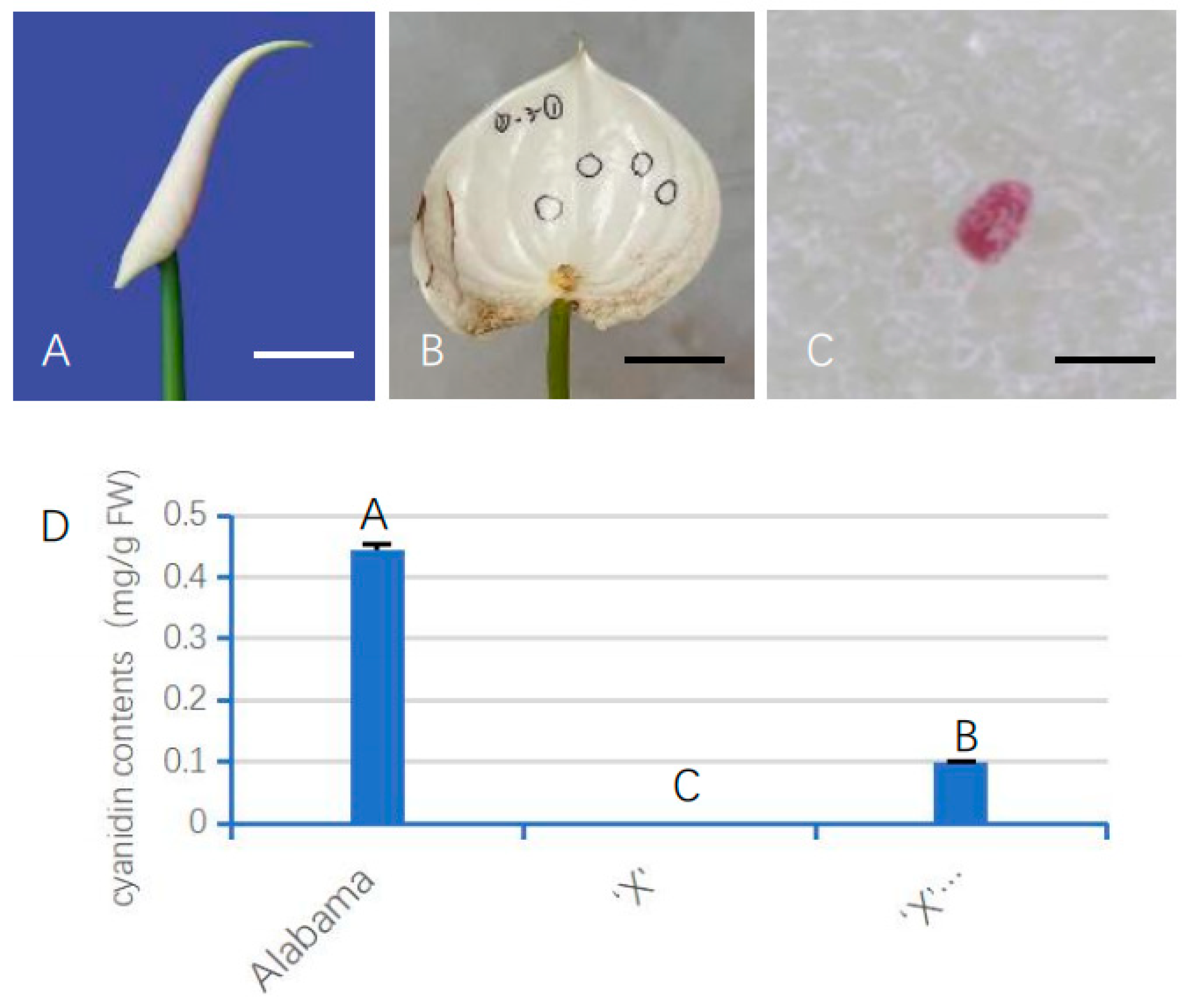
3.6. Transient Expression of AnUFGT1 in ‘X’ Promoted Anthocyanin Synthesis
4. Discussion
4.1. Metabolomics Enables More Accurate Detection of ASP-Related Metabolites in Plants
4.2. AnUFGT1 Gene Is the Key Structural Gene for the Deletion of ‘X’ Anthocyanins
4.3. AnUFGT1 Gene May Act with Transcription Factor in the Deletion of Anthocyanins
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Cong, H.; Xu, L.; Xin, C.; Li, Z. Comparison and Determination of Anthocyanidins in Anthurium Mutant. J. Anhui Agric. Sci. 2009, 37, 7832–7834. [Google Scholar]
- Ben-Simhon, Z.; Judeinstein, S.; Trainin, T.; Harel-Beja, R.; Bar-Ya’Akov, I.; Borochov-Neori, H.; Holland, D. A “White” Anthocyanin-less Pomegranate (Punica granatum L.) Caused by an Insertion in the Coding Region of the Leucoanthocyanidin Dioxygenase (LDOX; ANS) Gene. PLoS ONE 2015, 10, e0142777. [Google Scholar] [CrossRef] [PubMed]
- Castillejo, C.; Waurich, V.; Wagner, H.; Ramos, R.; Oiza, N.; Muñoz, P.; Triviño, J.C.; Caruana, J.; Liu, Z.; Cobo, N.; et al. Allelic Variation of MYB10 Is the Major Force Controlling Natural Variation in Skin and Flesh Color in Strawberry (Fragaria spp.) Fruit. Plant Cell 2020, 32, 3723–3749. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Cheng, J.; Xiang, J.; Zheng, T.; Wu, J. Transcriptome and proteome analysis of the fig (Ficus carica L.) cultivar Orphan and its mutant Hongyan based on the fruit peel colour in South China. Czech J. Genet. Plant Breed. 2022, 59, 33–42. [Google Scholar] [CrossRef]
- Yuan, K.; Zhao, X.; Sun, W.; Yang, L.; Zhang, Y.; Wang, Y.; Ji, J.; Han, F.; Fang, Z.; Lv, H. Map-based cloning and CRISPR/Cas9-based editing uncover BoNA1 as the causal gene for the no-anthocyanin-accumulation phenotype in curly kale (Brassica oleracea var. sabellica). Hortic. Res. 2023, 10, uhad133. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Luo, X.; Liu, Z.; Wang, C.; Lin, A.; Xiao, K.; Cao, M.; Fan, J.; Lian, H.; Xu, P. FvDFR2 rather than FvDFR1 play key roles for anthocyanin synthesis in strawberry petioles. Plant Sci. 2024, 340, 111960. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Olsen, O.; Knudsen, S. Expression of the Dihydroflavonol Reductase Gene in an Anthocyanin-Free Barley Mutant. Hereditas 1993, 119, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Toda, K.; Yang, D.; Yamanaka, N.; Watanabe, S.; Harada, K.; Takahashi, R. A single-base deletion in soybean flavonoid 3′-hydroxylase gene is associated with gray pubescence color. Plant Mol. Biol. 2002, 50, 187–196. [Google Scholar] [CrossRef]
- Li, J.; Tan, Q.; Yi, M.; Yu, Z.; Xia, Q.; Zheng, L.; Chen, J.; Zhou, X.; Zhang, X.-Q.; Guo, H.-R. Identification of key genes responsible for green and white colored spathes in Anthurium andraeanum (Hort.). Front. Plant Sci. 2023, 14, 1208226. [Google Scholar] [CrossRef]
- Liu, Y.; Zhou, B.; Qi, Y.; Liu, C.; Liu, Z.; Ren, X. Biochemical and functional characterization of AcUFGT3a, a galactosyltransferase involved in anthocyanin biosynthesis in the red-fleshed kiwifruit (Actinidia chinensis). Physiol. Plant. 2018, 162, 409–426. [Google Scholar] [CrossRef]
- Huang, B.-H.; Chen, Y.-W.; Huang, C.-L.; Gao, J.; Liao, P.-C. Diversifying selection of the anthocyanin biosynthetic downstream gene UFGT accelerates floral diversity of island Scutellaria species. BMC Evol. Biol. 2016, 16, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Ni, X.; Ni, Z.; Ouma, K.O.; Gao, Z. Mutations in PmUFGT3 contribute to color variation of fruit skin in Japanese apricot (Prunus mume Sieb. et Zucc.). BMC Plant Biol. 2022, 22, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Burklew, C.E.; Yang, Y.; Liu, M.; Xiao, P.; Zhang, B.; Qiu, D. De novo sequencing and a comprehensive analysis of purple sweet potato (Impomoea batatas L.) transcriptome. Planta 2012, 236, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Lu, Z.; Guo, J.; Luo, Y.; Li, H.; Li, L.; Gao, F. Cloning and characterization of the cDNA and promoter of UDP-glucose: Flavonoid 3-O-glucosyltransferase gene from a purple-fleshed sweet potato. S. Afr. J. Bot. 2016, 106, 211–220. [Google Scholar] [CrossRef]
- Morita, Y.; Ishiguro, K.; Tanaka, Y.; Iida, S.; Hoshino, A. Spontaneous mutations of the UDP-glucose:flavonoid 3-O-glucosyltransferase gene confers pale- and dull-colored flowers in the Japanese and common morning glories. Planta 2015, 242, 575–587. [Google Scholar] [CrossRef] [PubMed]
- Liao, L.; Li, Y.; Lan, X.; Yang, Y.; Wei, W.; Ai, J.; Feng, X.; Chen, H.; Tang, Y.; Xi, L.; et al. Integrative Analysis of Fruit Quality and Anthocyanin Accumulation of Plum cv. ‘Cuihongli’ (Prunus salicina Lindl.) and Its Bud Mutation. Plants 2023, 12, 1357. [Google Scholar] [CrossRef] [PubMed]
- Medda, S.; Sanchez-Ballesta, M.T.; Romero, I.; Dessena, L.; Mulas, M. Expression of Structural Flavonoid Biosynthesis Genes in Dark-Blue and White Myrtle Berries (Myrtus communis L.). Plants 2021, 10, 316. [Google Scholar] [CrossRef]
- Kapoor, P.; Sharma, S.; Tiwari, A.; Kaur, S.; Kumari, A.; Sonah, H.; Goyal, A.; Krishania, M.; Garg, M. Genome–Transcriptome Transition Approaches to Characterize Anthocyanin Biosynthesis Pathway Genes in Blue, Black and Purple Wheat. Genes 2023, 14, 809. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wang, J.; Zhang, X.; Xu, L. Comparative Transcriptome Analysis of Anthurium “Albama” and Its Anthocyanin-Loss Mutant. PLoS ONE 2015, 10, e0119027. [Google Scholar] [CrossRef]
- Tel-Zur, N.; Abbo, S.; Myslabodski, D.; Mizrahi, Y. Modified CTAB Procedure for DNA Isolation from Epiphytic Cacti of the Genera Hylocereus and Selenicereus (Cactaceae). Plant Mol. Biol. Rep. 1999, 17, 249–254. [Google Scholar] [CrossRef]
- Kenneth, J.L.; Thomas, D.S. Analysis of relative gene expression data using real-time quantitative PCR and the 2-△△CT method. Methods 2001, 25, 402–408. [Google Scholar]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Lister, C.E.; Lancaster, J.E.; Sutton, K.H.; Walker, J.R. Developmental changes in the concentration and composition of flavonoids in skin of a red and a green apple cultivar. J. Sci. Food Agric. 1994, 64, 155–161. [Google Scholar] [CrossRef]
- Bonaccorsi, P.; Caristi, C.; Gargiulli, C.; Leuzzi, U. Flavonol glucosides in Allium species: A comparative study by means of HPLC–DAD–ESI-MS–MS. Food Chem. 2007, 107, 1668–1673. [Google Scholar] [CrossRef]
- Termentzi, A.; Kefalas, P.; Kokkalou, E. LC–DAD–MS (ESI+) analysis of the phenolic content of Sorbus domestica fruits in relation to their maturity stage. Food Chem. 2007, 106, 1234–1245. [Google Scholar] [CrossRef]
- Yamazaki, M.; Nakajima, J.-I.; Yamanashi, M.; Sugiyama, M.; Makita, Y.; Springob, K.; Awazuhara, M.; Saito, K. Metabolomics and differential gene expression in anthocyanin chemo-varietal forms of Perilla frutescens. Phytochemistry 2003, 62, 987–995. [Google Scholar] [CrossRef] [PubMed]
- Tweeddale, H.; Notley-McRobb, L.; Ferenci, T. Effect of Slow Growth on Metabolism of Escherichia coli, as Revealed by Global Metabolite Pool (“Metabolome”) Analysis. J. Bacteriol. 1998, 180, 5109–5116. [Google Scholar] [CrossRef] [PubMed]
- Iwata, R.Y.; Tang, C.S.; Kamemoto, H. Concentration of anthocyanins affecting spathe colour in Anthuriums. J. Am. Soc. Hortic. Sci. 1985, 110, 383–385. [Google Scholar] [CrossRef]
- Iwata, R.Y.; Tang, C.S.; Kamemoto, H. Anthocyanins of Anthurium andraeanum Lind. J. Am. Soc. Hortic. 1979, 104, 464–466. [Google Scholar] [CrossRef]
- Marutani, M.; Tang, C.S.; Paull, R.; Kamemoto, H. Anthocyanins in the Lavender Anthurium. HortScience 1987, 22, 620–622. [Google Scholar] [CrossRef]
- Meng, X.; Li, G.; Gu, L.; Sun, Y.; Li, Z.; Liu, J.; Wu, X.; Dong, T.; Zhu, M. Comparative metabolomic and transcriptome analysis reveal distinct flavonoid biosynthesis regulation between petals of white and purple Phalaenopsis amabilis. J. Plant Growth Regul. 2020, 39, 823–840. [Google Scholar] [CrossRef]
- Wang, Z.; Li, X.; Chen, M.; Yang, L.; Zhang, Y. Molecular and Metabolic Insights into Anthocyanin Biosynthesis for Spot Formation on Lilium leichtlinii var. maximowiczii Flower Petals. Int. J. Mol. Sci. 2023, 24, 1844. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.-Z.; Hu, F.-C.; Hu, G.-B.; Li, X.-J.; Huang, X.-M.; Wang, H.-C. Differential Expression of Anthocyanin Biosynthetic Genes in Relation to Anthocyanin Accumulation in the Pericarp of Litchi chinensis Sonn. PLoS ONE 2011, 6, e19455. [Google Scholar] [CrossRef] [PubMed]
- Meinke, D.W.; Cherry, J.M.; Dean, C.; Rounsley, S.D.; Koornneef, M. Arabidopsis thaliana: A Model Plant for Genome Analysis. Science 1998, 282, 662–682. [Google Scholar] [CrossRef] [PubMed]
- Walker, A.R.; Lee, E.; Bogs, J.; McDavid, D.A.; Thomas, M.R.; Robinson, S.P. White grapes arose through the mutation of two similar and adjacent regulatory genes. Plant J. 2007, 49, 772–785. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, S.; Goto-Yamamoto, N.; Hirochika, H. Association of VvmybAl Gene Expression with Anthocyanin Production in Grape (Vitis vinifera) Skin-color Mutants. J. Jpn. Soc. Hortic. Sci. 2005, 74, 196–203. [Google Scholar] [CrossRef]
- Kobayashi, S.; Ishimaru, M.; Ding, C.; Yakushiji, H.; Goto, N. Comparison of UDP-glucose:flavonoid 3-O-glucosyltransferase (UFGT) gene sequences between white grapes (Vitis vinifera) and their sports with red skin. Plant Sci. 2001, 160, 543–550. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kim, D.-H.; Lee, J.-Y.; Lim, S.-H. The R3-Type MYB Transcription Factor BrMYBL2.1 Negatively Regulates Anthocyanin Biosynthesis in Chinese Cabbage (Brassica rapa L.) by Repressing MYB–bHLH–WD40 Complex Activity. Int. J. Mol. Sci. 2022, 23, 3382. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Xu, Y.; Zhu, G.-F.; Tan, J.; Lin, J.; Huang, L.; Ye, Y.; Liu, J. Pigment Diversity in Leaves of Caladium × hortulanum Birdsey and Transcriptomic and Metabolic Comparisons between Red and White Leaves. Int. J. Mol. Sci. 2024, 25, 605. [Google Scholar] [CrossRef]
- Muhammad, N.; Uddin, N.; Khan, M.K.U.; Ali, N.; Ali, K.; Jones, D.A. Diverse role of basic Helix-Loop-Helix (bHLH) transcription factor superfamily genes in the fleshy fruit-bearing plant species. Czech J. Genet. Plant Breed. 2022, 59, 1–13. [Google Scholar] [CrossRef]
- Yuan, H.; Cai, W.; Chen, X.; Pang, F.; Wang, J.; Zhao, M. Heterozygous frameshift mutation in FaMYB10 is responsible for the natural formation of red and white-fleshed strawberry (Fragaria x ananassa Duch). Front. Plant Sci. 2022, 13, 1027567. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Li, Z.; Cong, H.; Li, X. Cloning of Anthurium MYB Transcription Factor AN2-Like and Construction of Expression Vector. Chinese Invention Patent ZL201310140892.0, 19 November 2014. [Google Scholar]
- Sun, C.; Wang, C.; Zhang, W.; Liu, S.; Wang, W.; Yu, X.; Song, T.; Yu, M.; Yu, W.; Qu, S. The R2R3-type MYB transcription factor MdMYB90-like is responsible for the enhanced skin color of an apple bud sport mutant. Hortic. Res. 2021, 8, 156. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Su, J.; Zhu, Y.; Yao, G.; Allan, A.C.; Ampomah-Dwamena, C.; Shu, Q.; Lin-Wang, K.; Zhang, S.; Wu, J. The involvement of PybZIPa in light-induced anthocyanin accumulation via the activation of PyUFGT through binding to tandem G-boxes in its promoter. Hortic. Res. 2019, 6, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Wang, J.; Zhang, Z.; Fan, Y.; Qin, H.; Yin, Y.; Dai, G.; Cao, Y.; Tang, L. Anthocyanin biosynthesis in goji berry is inactivated by deletion in a bHLH transcription factor LrLAN1b promoter. Plant Physiol. 2024, kiae122. [Google Scholar] [CrossRef] [PubMed]

| Compound | Alabama | X | Comparative Result |
|---|---|---|---|
| Cyanidin-3-O-glucoside | 1.37 × 108 | 3.08 × 105 | Down |
| Pelargonidin-3-O-glucoside | 2.19 × 107 | 9.00 × 100 | Down |
| Pelargonidin-3-O-glucoside-5-O-arabinoside | 1.02 × 106 | 9.00 × 100 | Down |
| Cyanidin-3-O-(2″-O-glucosyl) glucoside | 1.24 × 106 | 9.00 × 100 | Down |
| Cyanidin-3-O-rutinoside | 1.59 × 107 | 9.00 × 100 | Down |
| Compound | Alabama | X | Type | ||||
|---|---|---|---|---|---|---|---|
| A1 | A2 | A3 | X1 | X2 | X3 | ||
| Dihydroquercetin (Taxifolin) | 5.58 × 104 | 7.72 × 104 | 5.62 × 104 | 3.73 × 104 | 4.37 × 104 | 4.03 × 104 | insig |
| Aromadendrin (Dihydrokaempferol) | 1.03 × 105 | 1.03 × 105 | 1.16 × 105 | 1.21 × 105 | 1.09 × 105 | 1.08 × 105 | insig |
| Naringenin (5,7,4′-Trihydroxyflavanone) | 1.25 × 106 | 1.15 × 106 | 1.08 × 106 | 1.76 × 106 | 1.67 × 106 | 1.90 × 106 | insig |
| Eriodictyol (5,7,3′,4′-Tetrahydroxyflavanone) | 9.08 × 103 | 7.91 × 103 | 1.36 × 104 | 7.89 × 103 | 7.35 × 103 | 8.15 × 103 | insig |
| Naringenin chalcone | 1.34 × 106 | 1.13 × 106 | 1.21 × 106 | 1.81 × 106 | 1.75 × 106 | 1.97 × 106 | insig |
| Epicatechin | 5.38 × 106 | 5.34 × 106 | 4.64 × 106 | 4.49 × 106 | 5.78 × 106 | 5.05 × 106 | insig |
| Procyanidine B4 | 2.51 × 107 | 2.63 × 107 | 2.59 × 107 | 2.82 × 107 | 2.24 × 107 | 2.24 × 107 | insig |
| Procyanidine C1 | 5.63 × 106 | 5.00 × 106 | 4.80 × 106 | 6.71 × 106 | 6.78 × 106 | 6.89 × 106 | insig |
| Procyanidine B1 | 1.28 × 106 | 1.62 × 106 | 1.12 × 106 | 4.19 × 106 | 3.72 × 106 | 3.39 × 106 | up |
| Procyanidine C2 | 8.60 × 105 | 9.04 × 105 | 6.96 × 105 | 2.04 × 106 | 2.09 × 106 | 2.04 × 106 | up |
| Procyanidine A2 | 8.18 × 105 | 9.94 × 105 | 1.35 × 106 | 1.24 × 106 | 1.23 × 106 | 1.06 × 106 | insig |
| Procyanidine A1 | 3.25 × 105 | 3.55 × 105 | 5.80 × 105 | 4.66 × 105 | 4.53 × 105 | 4.12 × 105 | insig |
| Procyanidine A6 | 1.22 × 105 | 4.39 × 104 | 1.43 × 105 | 8.61 × 104 | 1.04 × 105 | 1.04 × 105 | insig |
| Procyanidine B3 | 1.50 × 104 | 1.82 × 104 | 1.45 × 104 | 4.43 × 104 | 4.26 × 104 | 4.10 × 104 | up |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Wang, J.; Gao, Y.; Jing, Y.; Li, J.; Xu, L. The AnUFGT1 Is Involved in the Anthurium ‘Alabama’ Anthocyanidin Deficiency. Horticulturae 2024, 10, 369. https://doi.org/10.3390/horticulturae10040369
Li Z, Wang J, Gao Y, Jing Y, Li J, Xu L. The AnUFGT1 Is Involved in the Anthurium ‘Alabama’ Anthocyanidin Deficiency. Horticulturae. 2024; 10(4):369. https://doi.org/10.3390/horticulturae10040369
Chicago/Turabian StyleLi, Zhiying, Jiabin Wang, Yu Gao, Yonglin Jing, Junguo Li, and Li Xu. 2024. "The AnUFGT1 Is Involved in the Anthurium ‘Alabama’ Anthocyanidin Deficiency" Horticulturae 10, no. 4: 369. https://doi.org/10.3390/horticulturae10040369
APA StyleLi, Z., Wang, J., Gao, Y., Jing, Y., Li, J., & Xu, L. (2024). The AnUFGT1 Is Involved in the Anthurium ‘Alabama’ Anthocyanidin Deficiency. Horticulturae, 10(4), 369. https://doi.org/10.3390/horticulturae10040369





