Characteristics and Comparative Analysis of the Complete Plastomes of Apostasia fujianica and Neuwiedia malipoensis (Apostasioideae)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material Collection, DNA Extraction, and Sequencing
2.2. Plastome Assembly and Annotation
2.3. Genome Comparison and Analysis
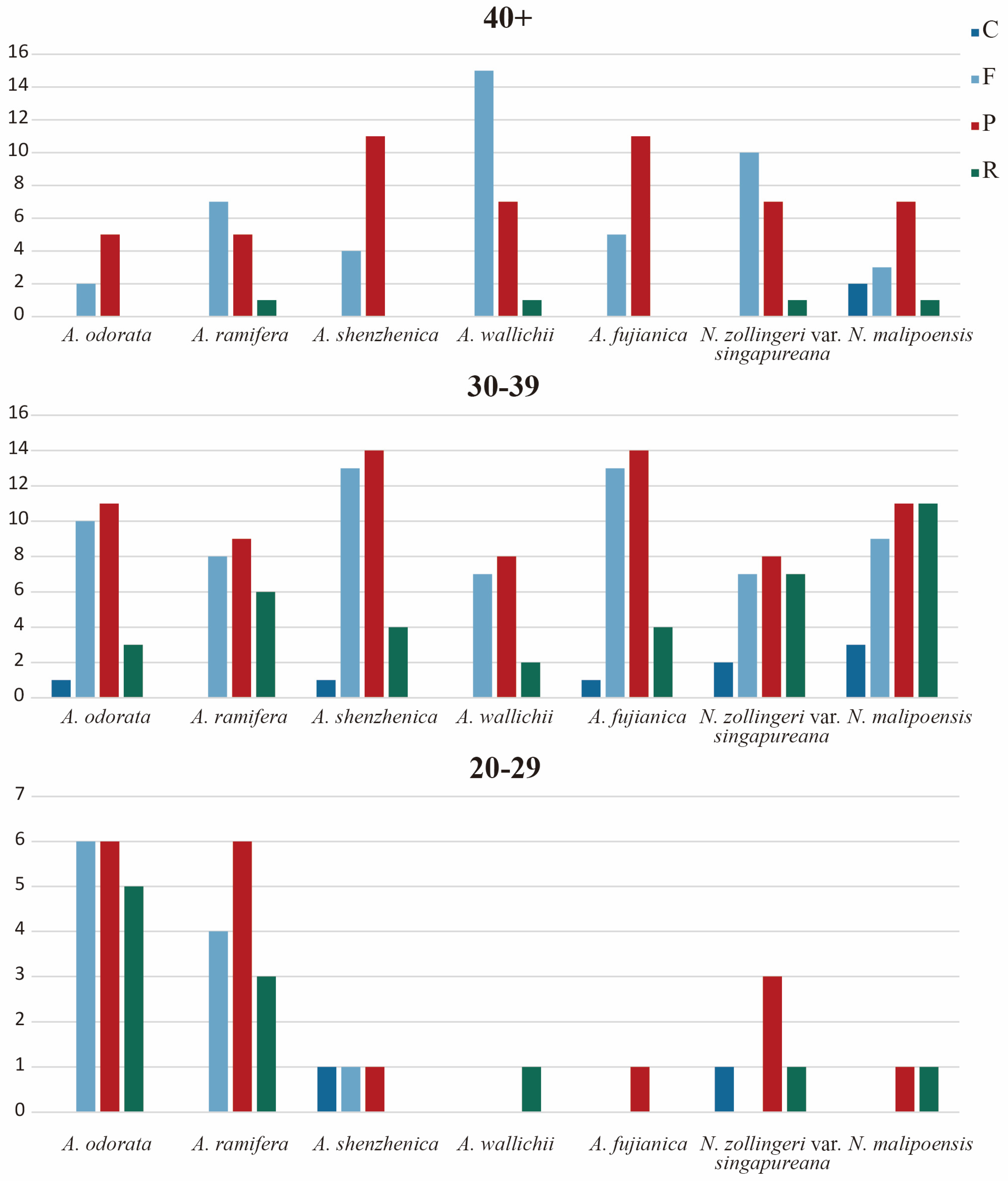
2.4. Repeat Sequences and Codon Usage
2.5. Divergence Analyses and Phylogeny
3. Results
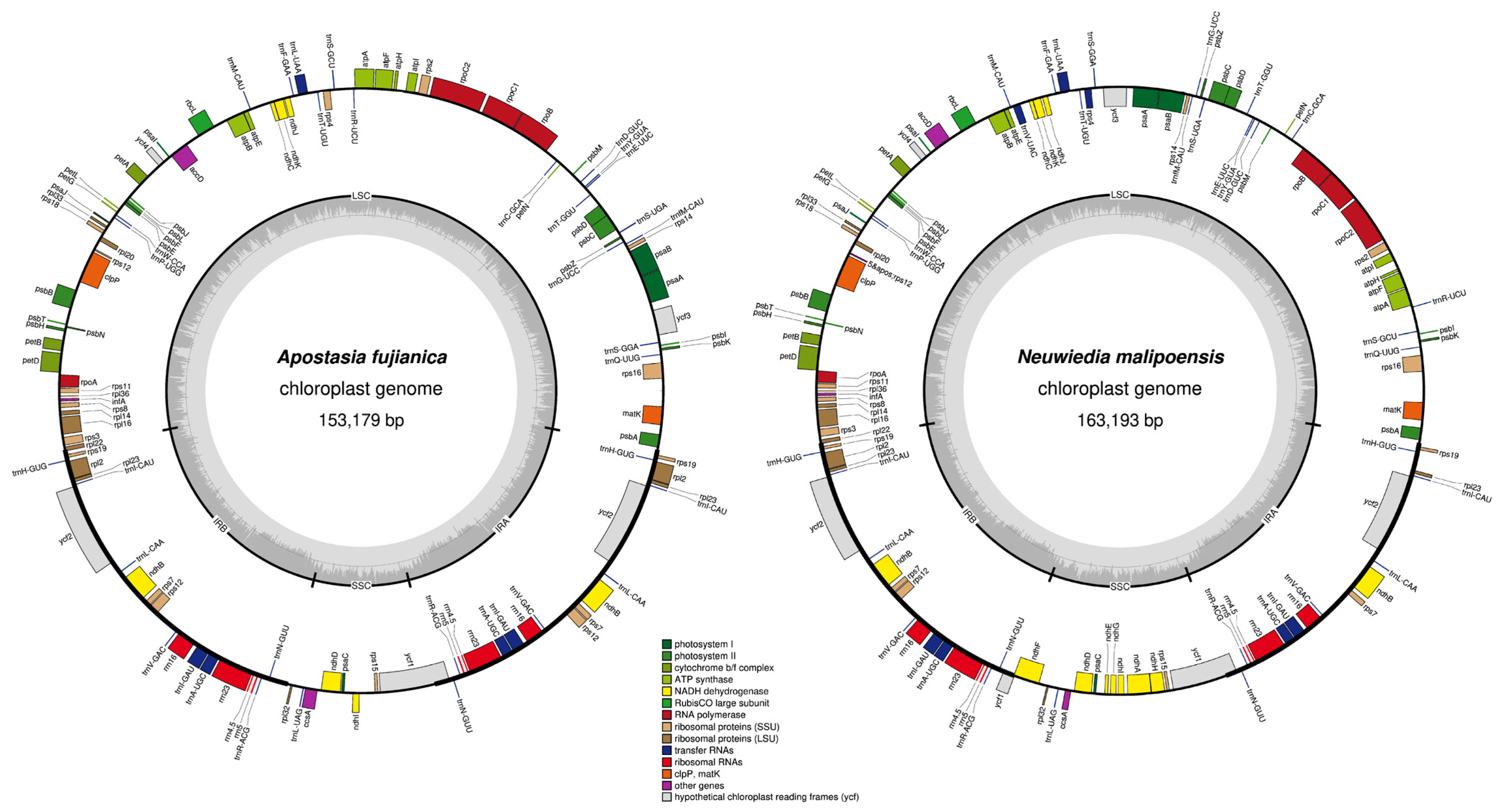
3.1. Plastid Genome Features
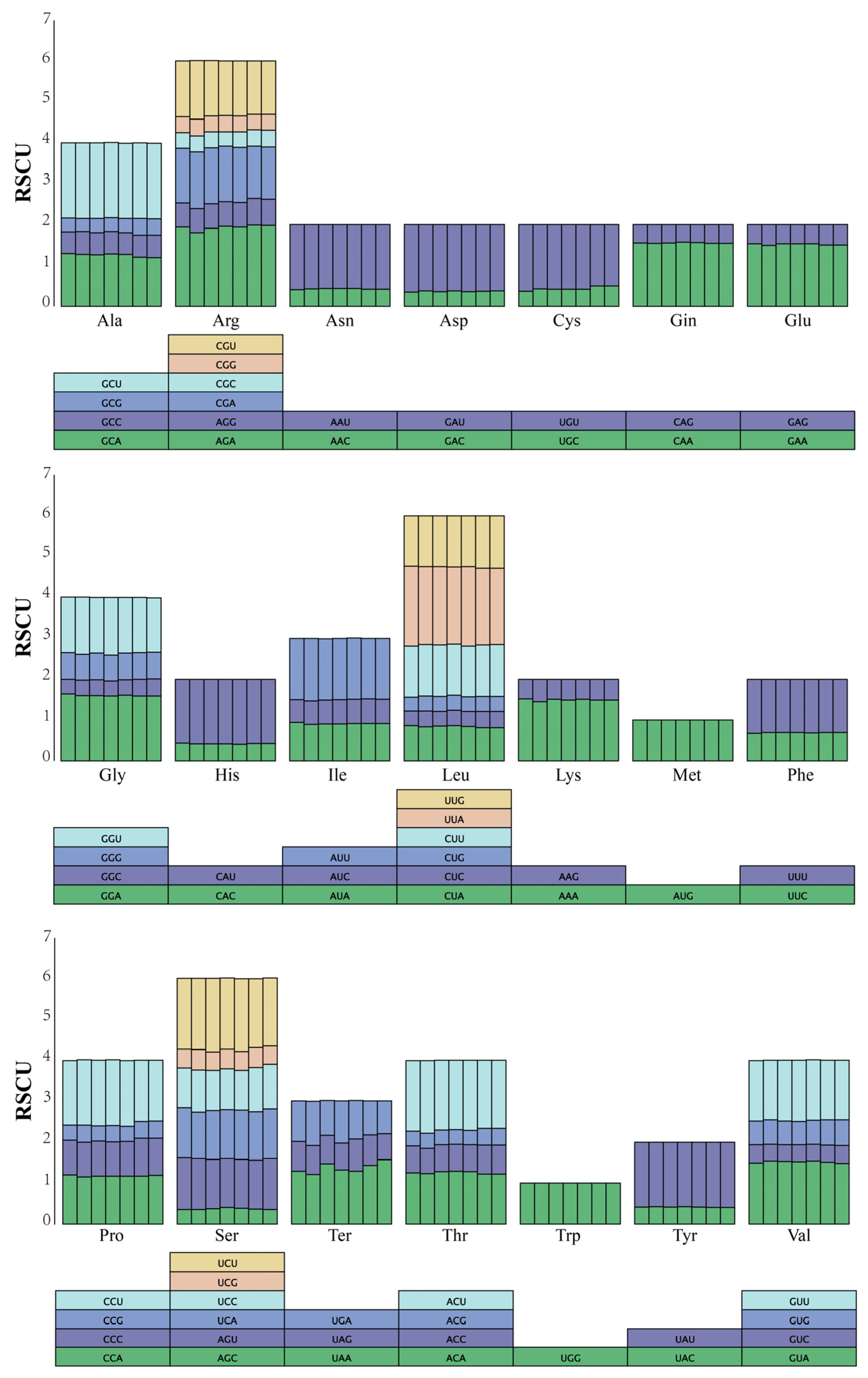
3.2. Codon Preference Analysis
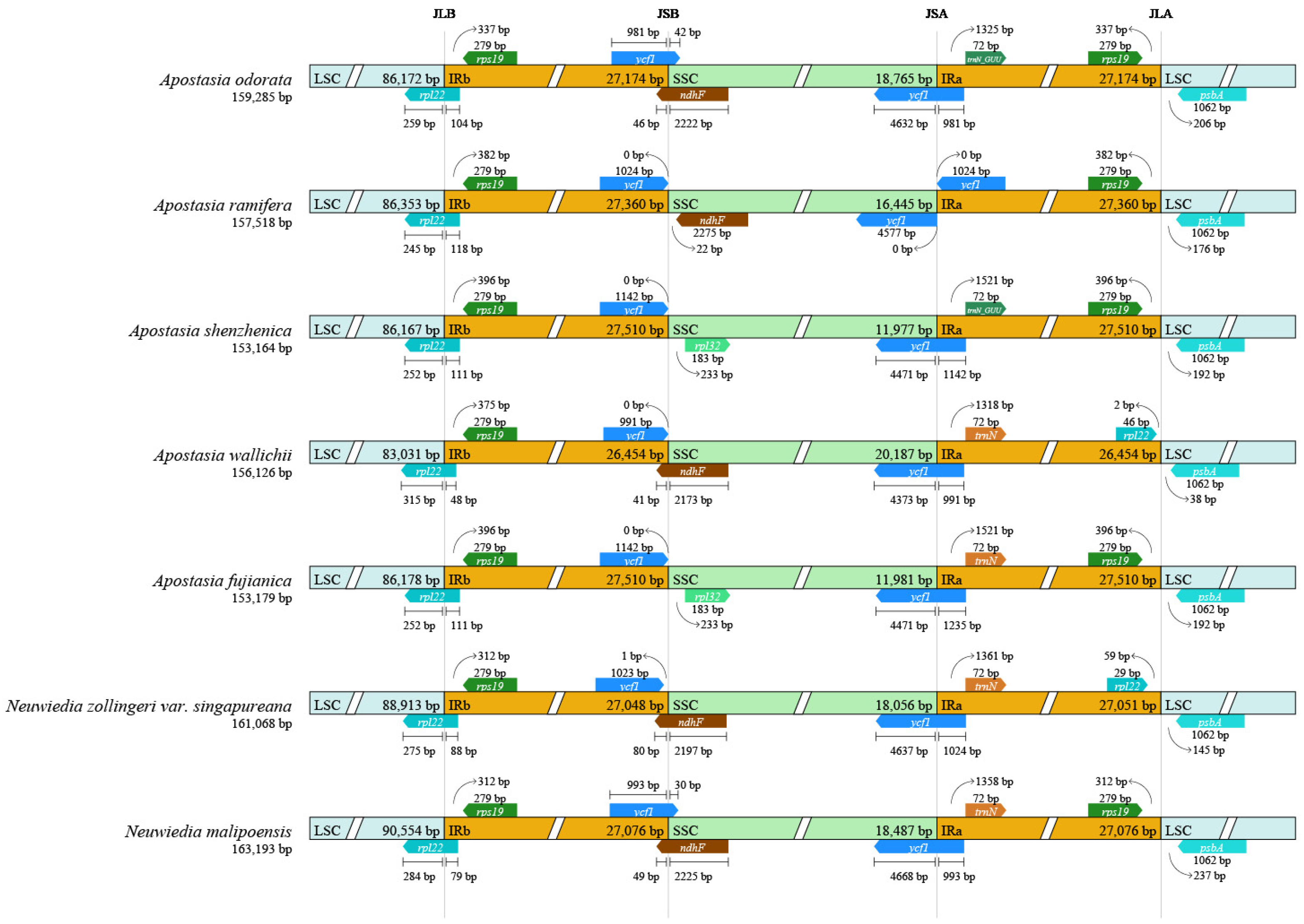
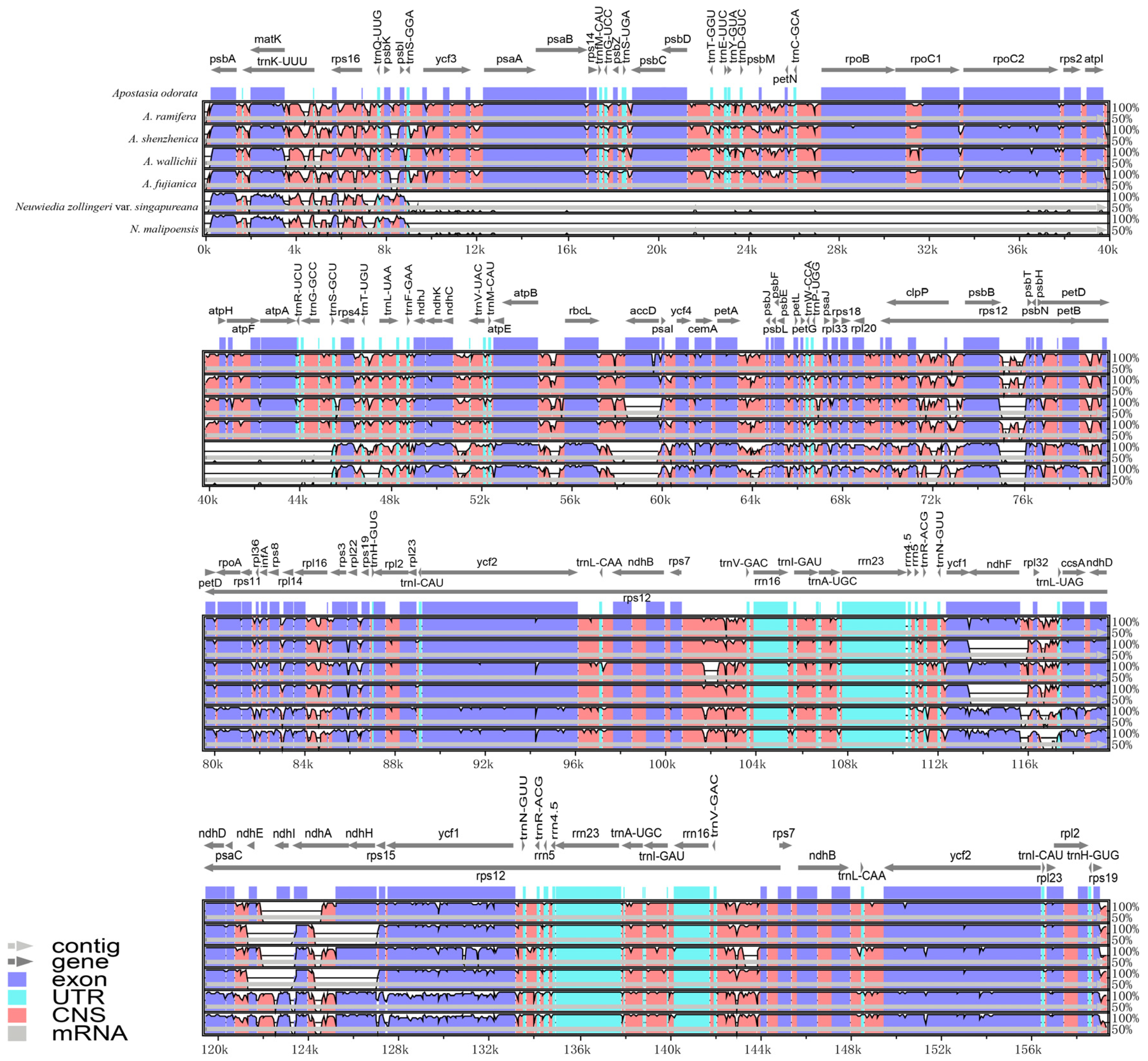
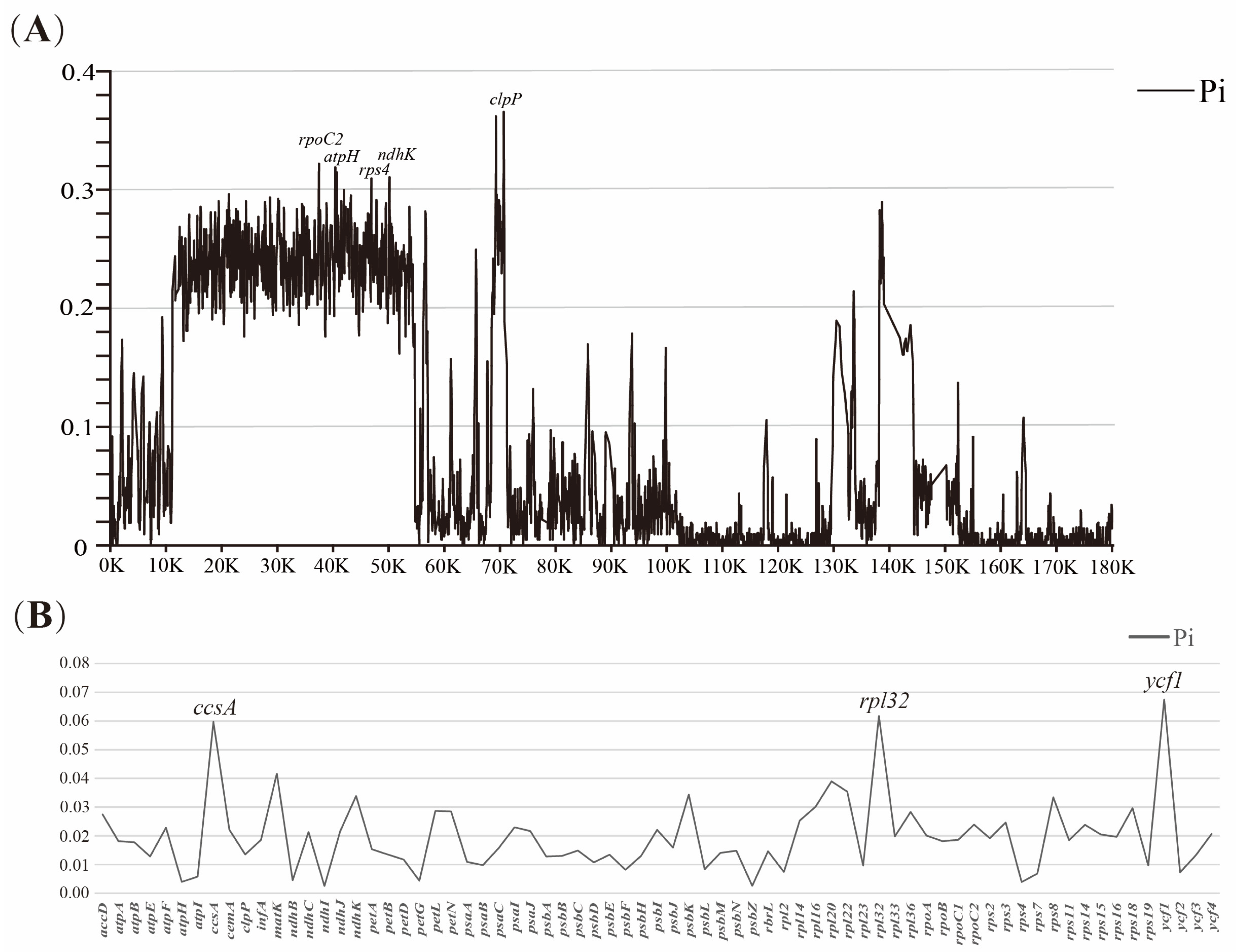
3.3. Plastome Sequence Divergence
3.4. Repeat Analysis and Mutation Hotspot Investigation
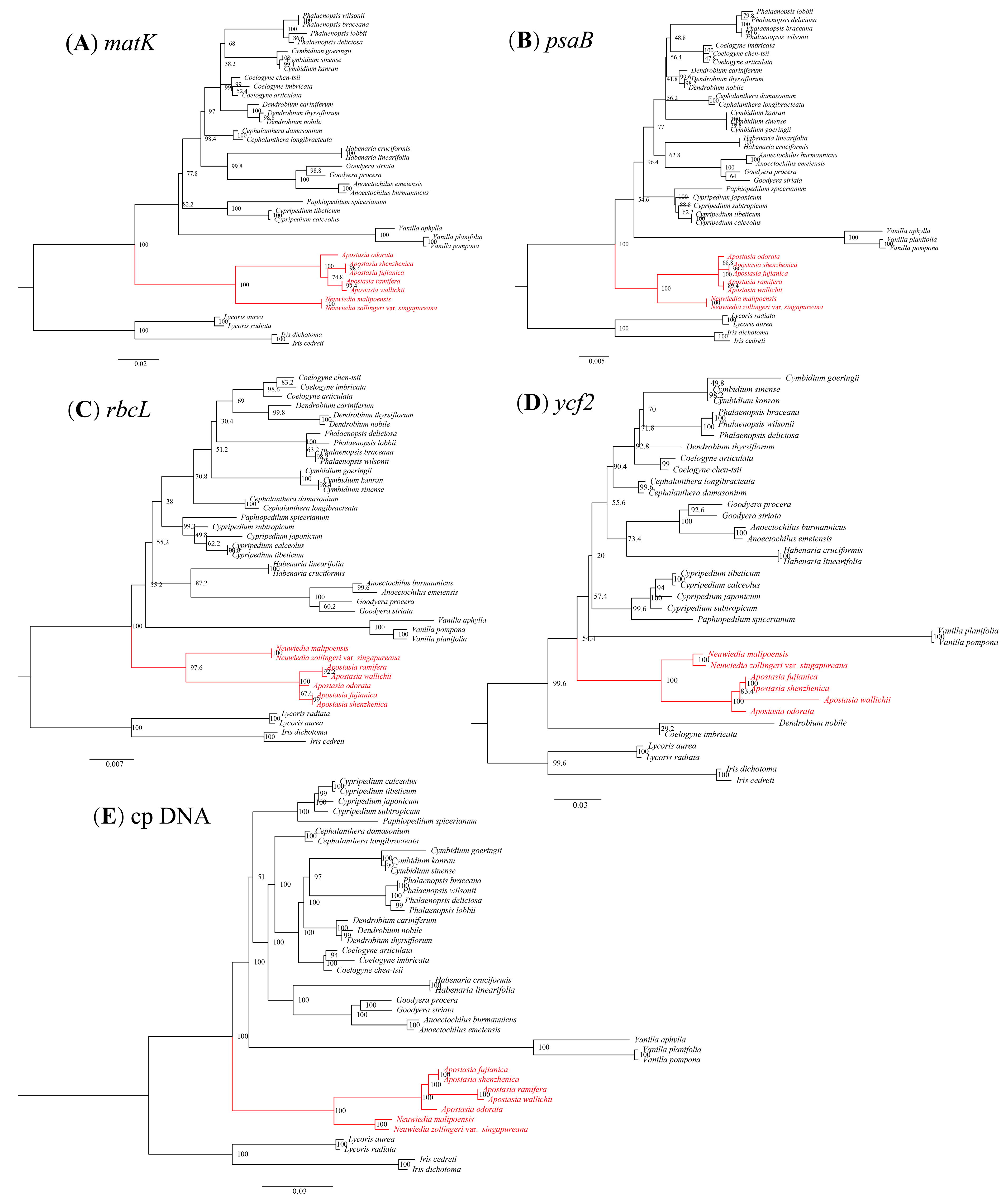
3.5. Phylogenetic Analysis
4. Discussion
4.1. The Plastome Characteristics and Structural Rearrangement
4.2. Polymorphic Loci for Molecular Markers
4.3. Phylogenetic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Scotland, R.W.; Wortley, A.H. How many species of seed plants are there? Taxon 2003, 52, 101–104. [Google Scholar] [CrossRef]
- Christenhusz, M.J.M.; Byng, J.W. The number of known plants species in the world and its annual increase. Phytotaxa 2016, 261, 201–217. [Google Scholar] [CrossRef]
- Givnish, T.J.; Spalink, D.; Ames, M.; Lyon, S.P.; Hunter, S.J.; Zuluaga, A.; Iles, W.J.; Clements, M.A.; Arroyo, M.T.; Leebens-Mack, J.; et al. Orchid phylogenomics and multiple drivers of their extraordinary diversification. Proc. R. Soc. B Biol. Sci. 2015, 282, 20151553. [Google Scholar] [CrossRef]
- Chen, X. Apostasioideae. Flora China 2017, 25, 20. [Google Scholar]
- Rasmussen, H.N.; Rasmussen, F.N. Seedling mycorrhiza: A discussion of origin and evolution in Orchidaceae. Bot. J. Linn. Soc. 2014, 175, 313–327. [Google Scholar] [CrossRef]
- de Vogel, E.F. Monograph of the tribe Apostasieae (Orchidaceae). Blumea Biodivers. Evol. Biogeogr. Plants 1969, 17, 313–350. [Google Scholar]
- Stern, W.L.; Cheadle, V.I.; Thorsch, J. Apostasiads, systematic anatomy, and the origins of Orchidaceae. Bot. J. Linn. Soc. 1993, 111, 411–455. [Google Scholar] [CrossRef]
- Suetsugu, K. A novel seed dispersal mode of Apostasia nipponica could provide some clues to the early evolution of the seed dispersal system in Orchidaceae. Evol. Lett. 2020, 4, 457–464. [Google Scholar] [CrossRef]
- Garay, L.A. On the origin of the Orchidaceae, II. J. Arnold Arbor. 1972, 53, 202–215. [Google Scholar] [CrossRef]
- Burns-Balogh, P.; Funk, V.A. A phylogenetic analysis of the Orchidaceae. Smithson. Contrib. Bot. 1986, 61, 1–79. [Google Scholar] [CrossRef]
- Reveal, J.L.; Hoogland, R.D. Validation of three family names in the Magnoliophyta. Bull. Muséum Natl. D’histoire Nat. Sect. B Adansonia 1991, 13, 91–93. [Google Scholar]
- Rao, V.S. The relationships of the Apostasiaceae on the basis of floral anatomy. Bot. J. Linn. Soc. 1974, 68, 319–327. [Google Scholar] [CrossRef]
- Zheng, F.; Chen, J.B.; Liu, W.R.; Wang, M. The complete chloroplast genome of an endangered species Apostasia ramifera (Orchidaceae). Mitochondrial DNA Part B 2021, 6, 470–471. [Google Scholar] [CrossRef]
- Li, Y.; Li, Z.; Hu, Q.; Zhai, J.; Liu, Z.; Wu, S. Complete plastid genome of Apostasia shenzhenica (Orchidaceae). Mitochondrial DNA Part B 2019, 4, 1388–1389. [Google Scholar] [CrossRef]
- Kocyan, A.; Qiu, Y.L.; Endress, P.K.; Conti, E. A phylogenetic analysis of Apostasioideae (Orchidaceae) based on ITS, trnL–F and matK sequences. Plant Syst. Evol. 2004, 247, 203–213. [Google Scholar] [CrossRef]
- Yin, Y.Y.; Zhong, P.S.; Zhang, G.Q.; Chen, L.J.; Zeng, S.J.; Li, M.H.; Liu, Z.J. Morphological, genome-size and molecular analyses of Apostasia fogangica (Apostasioideae, Orchidaceae), a new species from China. Phytotaxa 2016, 277, 59–67. [Google Scholar] [CrossRef]
- Jersáková, J.; Trávníček, P.; Kubátová, B.; Krejčíková, J.; Urfus, T.; Liu, Z.J.; Lamb, A.; Ponert, J.; Schulte, K.; Čurn, V.; et al. Genome size variation in Orchidaceae subfamily Apostasioideae: Filling the phylogenetic gap. Bot. J. Linn. Soc. 2013, 172, 95–105. [Google Scholar] [CrossRef]
- Li, Y.; Ma, L.; Liu, D.; Zhao, X.; Zhang, D.; Ke, S.; Chen, G.Z.; Zheng, Q.; Liu, Z.J.; Lan, S. Apostasia fujianica (Apostasioideae, Orchidaceae), a new Chinese species: Evidence from morphological, genome size and molecular analyses. Phytotaxa 2023, 583, 277–284. [Google Scholar] [CrossRef]
- Kim, Y.K.; Jo, S.; Cheon, S.H.; Joo, M.J.; Hong, J.R.; Kwak, M. Plastome evolution and phylogeny of Orchidaceae, with 24 new sequences. Front. Plant Sci. 2020, 11, 22. [Google Scholar] [CrossRef]
- Kim, H.T.; Kim, J.S.; Moore, M.J.; Neubig, K.M.; Williams, N.H.; Whitten, W.M.; Kim, J.H. Seven new complete plastome sequences reveal rampant independent loss of the ndh gene family across orchids and associated instability of the inverted repeat/small single-copy region boundaries. PLoS ONE 2015, 10, e0142215. [Google Scholar] [CrossRef]
- Niu, Z.; Pan, J.; Zhu, S.; Li, L.; Xue, Q.; Liu, W.; Ding, X. Comparative analysis of the complete plastomes of Apostasia wallichii and Neuwiedia singapureana (Apostasioideae) reveals different evolutionary dynamics of IR/SSC boundary among photosynthetic orchids. Front. Plant Sci. 2017, 8, 1713. [Google Scholar] [CrossRef]
- Jin, J.J.; Yu, W.B.; Yang, J.B.; Song, Y.; DePamphilis, C.W.; Yi, T.S.; Li, D.Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef]
- Qu, X.J.; Moore, M.J.; Li, D.Z.; Yi, T.S. PGA: A software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods 2019, 15, 50. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef]
- Darling, A.C.E.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef]
- Li, H.; Guo, Q.; Xu, L.; Gao, H.; Liu, L.; Zhou, X. CPJSdraw: Analysis and visualization of junction sites of chloroplast genomes. PeerJ 2023, 11, e15326. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Li, Z.H.; Jiang, Y.; Ma, X.; Li, J.W.; Yang, J.B.; Wu, J.Y.; Jin, X.H. Plastid genome evolution in the subtribe Calypsoinae (Epidendroideae, Orchidaceae). Genome Biol. Evol. 2020, 12, 867–870. [Google Scholar] [CrossRef]
- Garay, L.A. On the origin of the Orchidaceae. Bot. Mus. Leafl. Harv. Univ. 1960, 19, 57–96. [Google Scholar] [CrossRef]
- Dahlgren, R.M.T.; Clifford, H.T.; Yeo, P.F. The Families of the Monocotyledons: Structure, Evolution, and Taxonomy; Springer Science & Business Media: Berlin/Heidelberg, Germany, 1984. [Google Scholar]
- Suetsugu, K.; Matsubayashi, J. Evidence for mycorrhizal cheating in Apostasia nipponica, an early-diverging member of the Orchidaceae. New Phytol. 2021, 229, 2302–2310. [Google Scholar] [CrossRef]
- Burrows, P.A.; Sazanov, L.A.; Svab, Z.; Maliga, P.; Nixon, P.J. Identification of a functional respiratory complex in chloroplasts through analysis of tobacco mutants containing disrupted plastid ndh genes. EMBO J. 1998, 17, 868–876. [Google Scholar] [CrossRef]
- Matsubayashi, T.; Wakasugi, T.; Shinozaki, K.; Yamaguchi-Shinozaki, K.; Zaita, N.; Hidaka, T.; Meng, B.Y.; Ohto, C.; Tanaka, M.; Kato, A.; et al. Six chloroplast genes (ndhA-F) homologous to human mitochondrial genes encoding components of the respiratory chain NADH dehydrogenase are actively expressed: Determination of the splice sites in ndhA and ndhB pre-mRNAs. Mol. Gen. Genet. MGG 1987, 210, 385–393. [Google Scholar] [CrossRef]
- Martín, M.; Sabater, B. Plastid ndh genes in plant evolution. Plant Physiol. Biochem. 2010, 48, 636–645. [Google Scholar] [CrossRef]
- Niu, Z.; Zhu, S.; Pan, J.; Li, L.; Jing, S.; Ding, X. Comparative analysis of Dendrobium plastomes and utility of plastomic mutational hotspots. Sci. Rep. 2017, 7, 2073. [Google Scholar]
- Perini, V.R.; Leles, B.; Furtado, C.; Prosdocimi, F. Complete chloroplast genome of the orchid Cattleya crispata (Orchidaceae: Laeliinae), a Neotropical rupiculous species. Mitochondrial DNA Part A 2016, 27, 4075–4077. [Google Scholar] [CrossRef]
- He, S.; Yang, Y.; Li, Z.; Wang, X.; Guo, Y.; Wu, H. Comparative analysis of four Zantedeschia chloroplast genomes: Expansion and contraction of the IR region, phylogenetic analyses and SSR genetic diversity assessment. PeerJ 2020, 8, e9132. [Google Scholar] [CrossRef]
- Dugas, D.V.; Hernandez, D.; Koenen, E.J.M.; Schwarz, E.; Straub, S.; Hughes, C.E.; Jansen, R.K.; Nageswara-Rao, M.; Staats, M.; Trujillo, J.T.; et al. Mimosoid legume plastome evolution: IR expansion, tandem repeat expansions and accelerated rate of evolution in clpP. Sci. Rep. 2015, 5, 16958. [Google Scholar] [CrossRef]
- Lin, C.S.; Chen, J.J.; Huang, Y.T.; Chan, M.T.; Daniell, H.; Chang, W.J.; Hsu, C.T.; Liao, D.C.; Wu, F.H.; Lin, S.Y.; et al. The location and translocation of ndh genes of chloroplast origin in the Orchidaceae family. Sci. Rep. 2015, 5, 9040. [Google Scholar] [CrossRef]
- Kim, Y.K.; Cheon, S.H.; Hong, J.R.; Kim, K.J. Evolutionary Patterns of the Chloroplast Genome in Vanilloid Orchids (Vanilloideae, Orchidaceae). Int. J. Mol. Sci. 2023, 24, 3808. [Google Scholar] [CrossRef]
- Tu, X.D.; Liu, D.K.; Xu, S.W.; Zhou, C.Y.; Gao, X.Y.; Zeng, M.Y.; Zhang, S.; Chen, J.L.; Ma, L.; Zhou, Z.; et al. Plastid phylogenomics improves resolution of phylogenetic relationship in the Cheirostylis and Goodyera clades of Goodyerinae (Orchidoideae, Orchidaceae). Mol. Phylogenetics Evol. 2021, 164, 107269. [Google Scholar] [CrossRef]
- Liu, D.K.; Zhou, C.Y.; Tu, X.D.; Zhao, Z.; Chen, J.L.; Gao, X.Y.; Xu, S.W.; Zeng, M.Y.; Ma, L.; Ahmad, S.; et al. Comparative and phylogenetic analysis of Chiloschista (Orchidaceae) species and DNA barcoding investigation based on plastid genomes. BMC Genom. 2023, 24, 749. [Google Scholar] [CrossRef]
- Dong, W.; Liu, J.; Yu, J.; Wang, L.; Zhou, S. Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. PLoS ONE 2012, 7, e35071. [Google Scholar] [CrossRef]
- Jiao, L.; Lu, Y.; He, T.; Li, J.; Yin, Y. A strategy for developing high-resolution DNA barcodes for species discrimination of wood specimens using the complete chloroplast genome of three Pterocarpus species. Planta 2019, 250, 95–104. [Google Scholar] [CrossRef]
- Li, Y.; Tong, Y.; Xing, F. DNA barcoding evaluation and its taxonomic implications in the recently evolved genus Oberonia Lindl. (Orchidaceae) in China. Front. Plant Sci. 2016, 7, 1791. [Google Scholar] [CrossRef]
- Smidt, E.C.; Páez, M.Z.; Vieira, L.N.; Viruel, J.; Baura, V.A.; Balsanelli, E.; de Souza, E.M.; Chase, M.W. Characterization of sequence variability hotspots in Cranichideae plastomes (Orchidaceae, Orchidoideae). PLoS ONE 2020, 15, e0227991. [Google Scholar] [CrossRef]
- Cavalier-Smith, T. Skeletal DNA and the evolution of genome size. Annu. Rev. Biophys. Bioeng. 1982, 11, 273–302. [Google Scholar] [CrossRef]
- Britten, R.J. Transposable element insertions have strongly affected human evolution. Proc. Natl. Acad. Sci. 2010, 107, 19945–19948. [Google Scholar] [CrossRef]
- Chen, J.; Wang, F.; Zhou, C.; Ahamd, S.; Zhou, Y.; Li, M.; Liu, Z.; Peng, D. Comparative Phylogenetic Analysis for Aerides (Aeridinae, Orchidaceae) Based on Six Complete Plastid Genomes. Int. J. Mol. Sci. 2023, 24, 12473. [Google Scholar] [CrossRef]
- Zhou, C.Y.; Zeng, M.Y.; Gao, X.; Zhao, Z.; Li, R.; Wu, Y.; Liu, Z.J.; Zhang, D.; Li, M.H. Characteristics and Comparative Analysis of Seven Complete Plastomes of Trichoglottis sl (Aeridinae, Orchidaceae). Int. J. Mol. Sci. 2023, 24, 14544. [Google Scholar] [CrossRef]
- Zhao, Z.; Zeng, M.Y.; Wu, Y.W.; Li, J.W.; Zhou, Z.; Liu, Z.J.; Li, M.H. Characterization and Comparative Analysis of the Complete Plastomes of Five Epidendrum (Epidendreae, Orchidaceae) Species. Int. J. Mol. Sci. 2023, 24, 14437. [Google Scholar] [CrossRef]
- Kocyan, A.; Endress, P.K. Floral structure and development of Apostasia and Neuwiedia (Apostasioideae) and their relationships to other Orchidaceae. Int. J. Plant Sci. 2001, 162, 847–867. [Google Scholar] [CrossRef]
- Chase, M.W. Classification of Orchidaceae in the age of DNA data. Curtis’s Bot. Mag. 2005, 22, 2–7. [Google Scholar] [CrossRef]
- Cameron, K.M.; Chase, M.W.; Whitten, W.M.; Kores, P.J.; Jarrell, D.C.; Albert, V.A.; Yukawa, T.; Hills, H.G.; Goldman, D.H. A phylogenetic analysis of the Orchidaceae: Evidence from rbcL nucleotide sequences. Am. J. Bot. 1999, 86, 208–224. [Google Scholar] [CrossRef]


| Species Name | Size (bp) | GC Content (%) | LSC Size in bp (%) | IR Size in bp (%) | SSC Size in bp (%) | Total Number of Genes | Protein-Encoding Genes | tRNA | rRNA | Number of ndh Fragments |
|---|---|---|---|---|---|---|---|---|---|---|
| A. odorata | 159,285 | 35.7 | 86,288 (54.17) | 27,116 (17.02) | 18,765 (11.79) | 133 | 86 | 38 | 8 | 11 |
| A. ramifera | 157,518 | 35.8 | 86,353 (54.82) | 27,360 (17.37) | 16,445 (10.44) | 134 | 78 | 38 | 8 | 9 |
| A. shenzhenica | 153,164 | 35.9 | 86,167 (56.26) | 27,510 (17.96) | 11,977 (7.82) | 130 | 77 | 40 | 8 | 6 |
| A. wallichii | 156,126 | 36.1 | 83,035 (53.18) | 26,452 (16.94) | 20,187 (12.94) | 135 | 71 | 39 | 8 | 11 |
| A. fujianica | 153,179 | 35.9 | 86,178 (56.26) | 27,510 (17.96) | 11,981 (7.82) | 123 | 74 | 35 | 8 | 6 |
| N. zollingeri var. singapureana | 161,068 | 36.0 | 89,060 (55.29) | 26,959 (16.75) | 18,058 (11.21) | 135 | 71 | 39 | 8 | 11 |
| N. malipoensis | 163,193 | 35.6 | 90,554 (55.49) | 27,076 (16.59) | 18,487 (11.33) | 129 | 74 | 37 | 8 | 11 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, Q.; Wu, Y.; Ke, S.-J.; Liu, D.-K.; Liu, Z.-J. Characteristics and Comparative Analysis of the Complete Plastomes of Apostasia fujianica and Neuwiedia malipoensis (Apostasioideae). Horticulturae 2024, 10, 383. https://doi.org/10.3390/horticulturae10040383
Zheng Q, Wu Y, Ke S-J, Liu D-K, Liu Z-J. Characteristics and Comparative Analysis of the Complete Plastomes of Apostasia fujianica and Neuwiedia malipoensis (Apostasioideae). Horticulturae. 2024; 10(4):383. https://doi.org/10.3390/horticulturae10040383
Chicago/Turabian StyleZheng, Qinyao, Yuwei Wu, Shi-Jie Ke, Ding-Kun Liu, and Zhong-Jian Liu. 2024. "Characteristics and Comparative Analysis of the Complete Plastomes of Apostasia fujianica and Neuwiedia malipoensis (Apostasioideae)" Horticulturae 10, no. 4: 383. https://doi.org/10.3390/horticulturae10040383






