Olive Anthracnose in Portugal Is Still Mostly Caused by Colletotrichum nymphaeae, but C. acutatum Is Spreading and C. alienum and C. cigarro Are Reported for the First Time
Abstract
:1. Introduction
2. Materials and Methods
2.1. Disease Survey and Fungal Isolation
2.2. Molecular Characterization
2.2.1. ISSR Analysis
2.2.2. PCR Amplification and Sequencing
2.2.3. Phylogenetic Analyses
2.3. Morphological Characterization
2.4. Pathogenicity Assays
3. Results
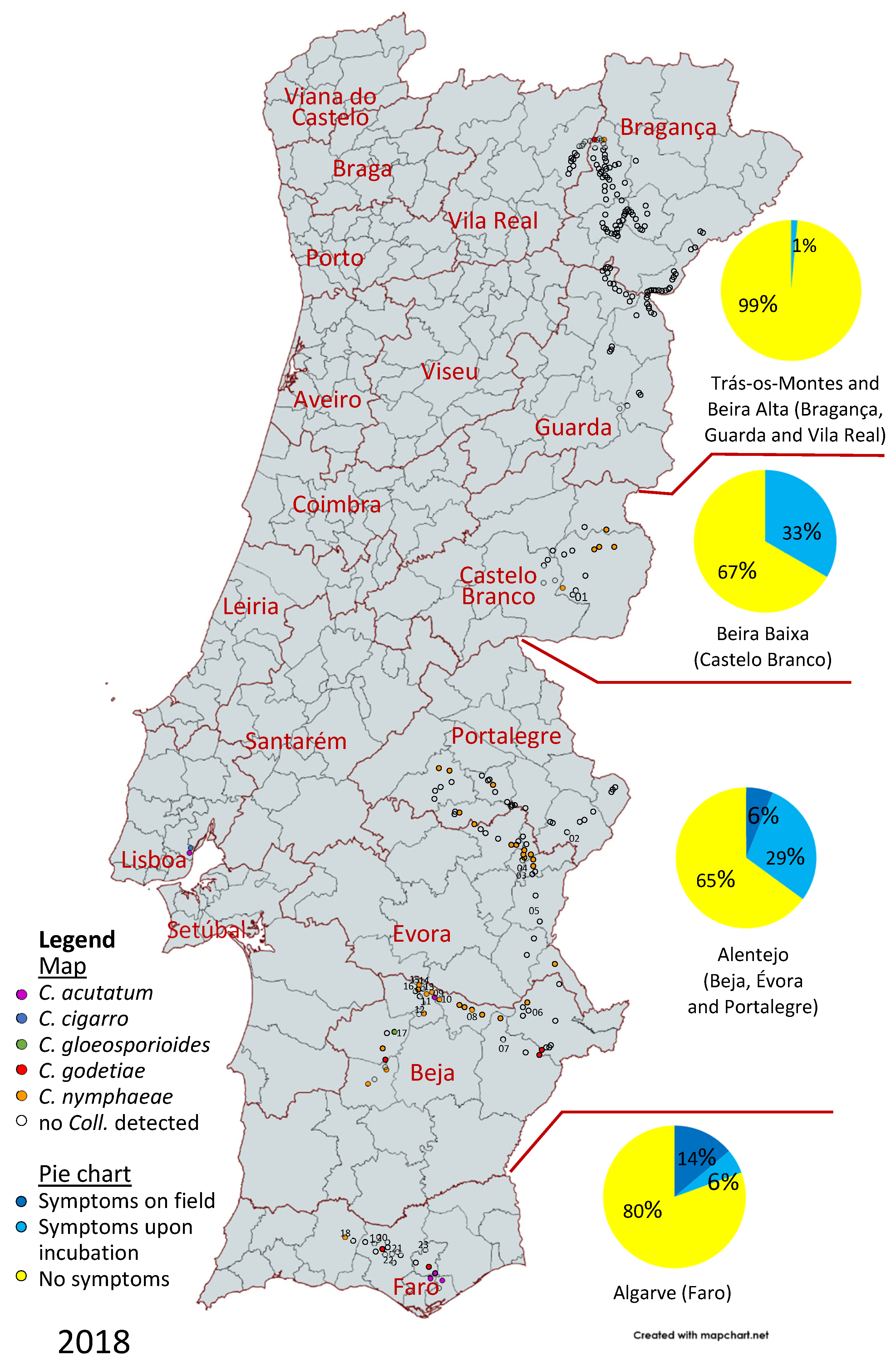
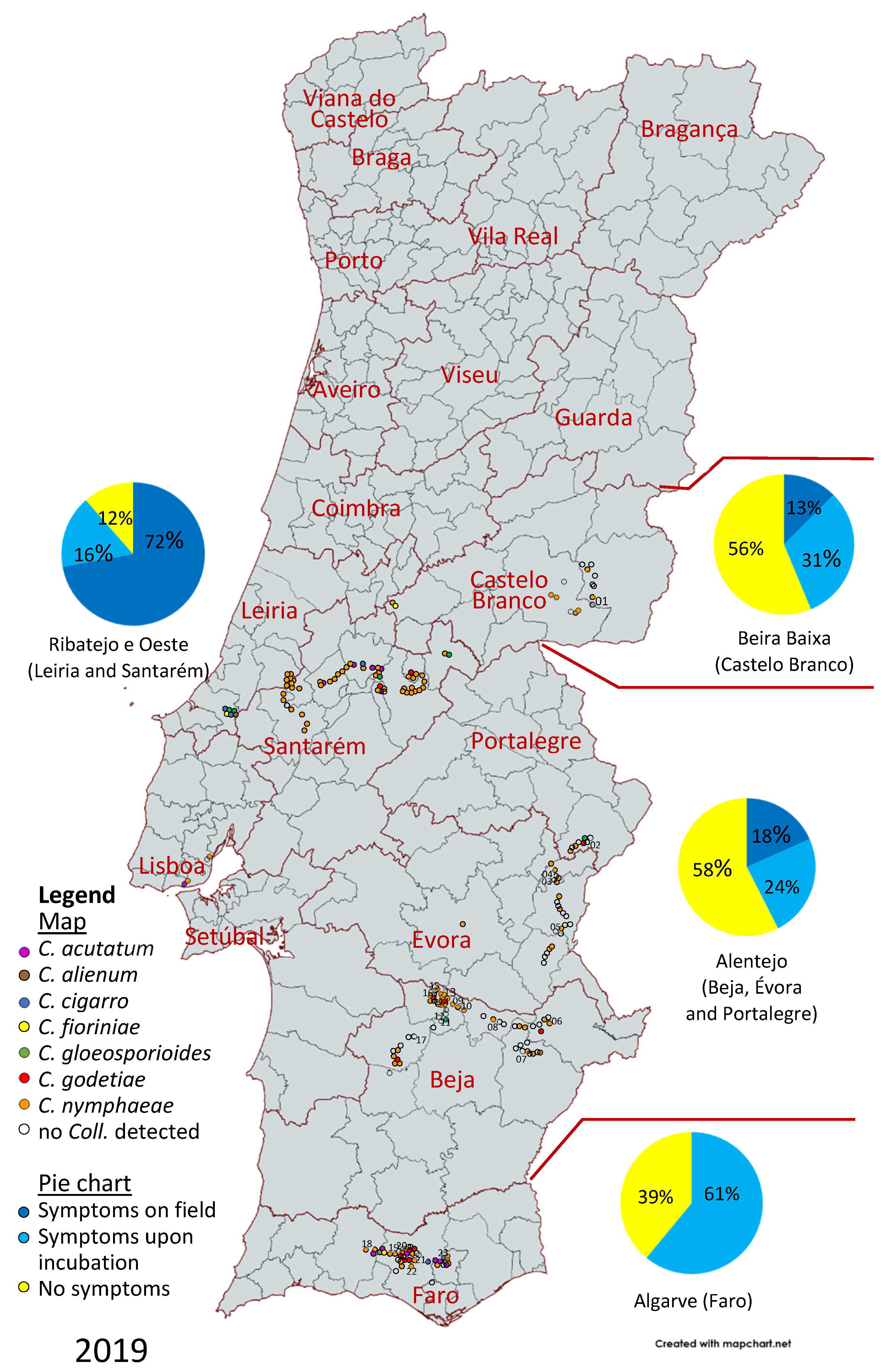
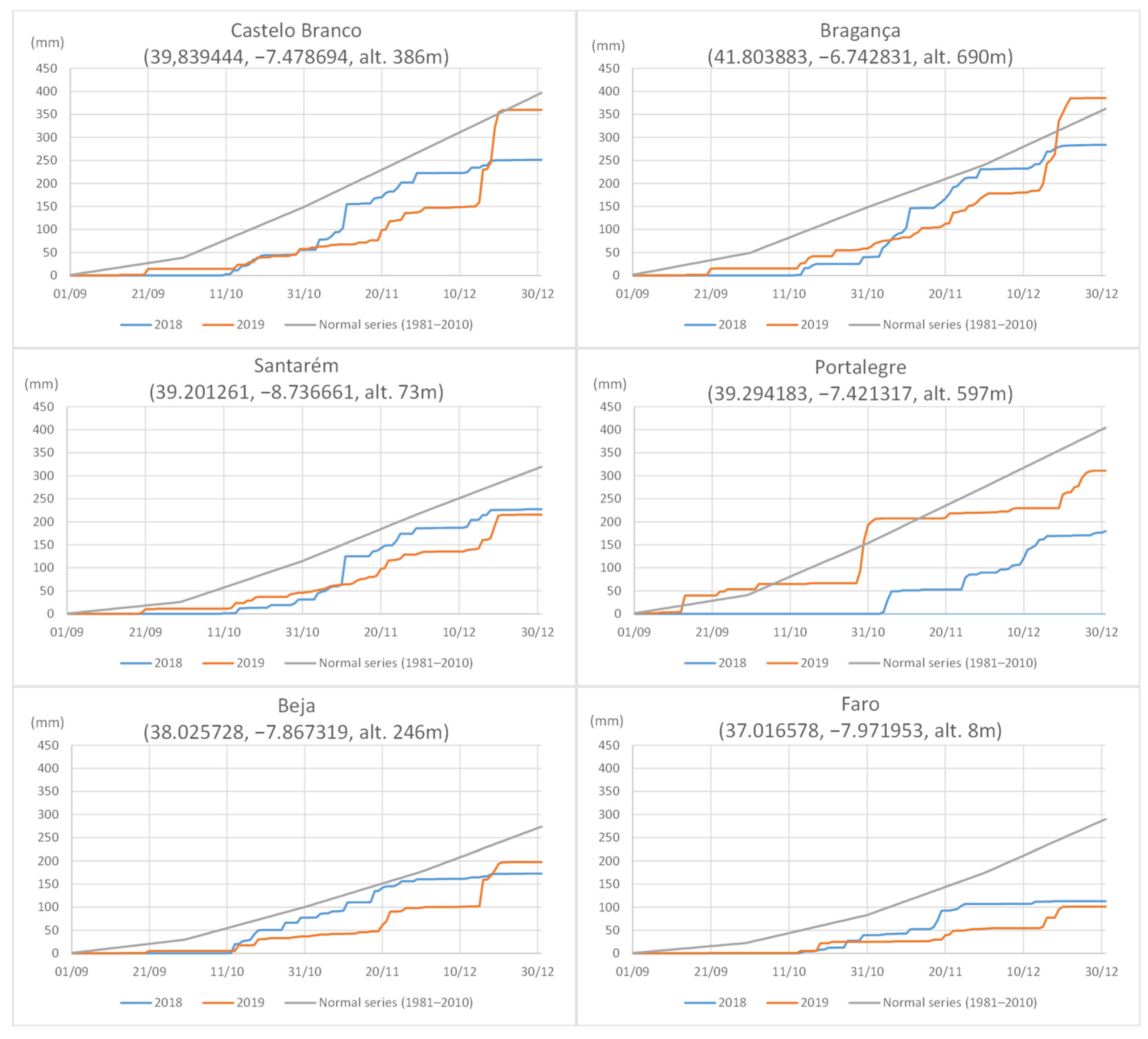
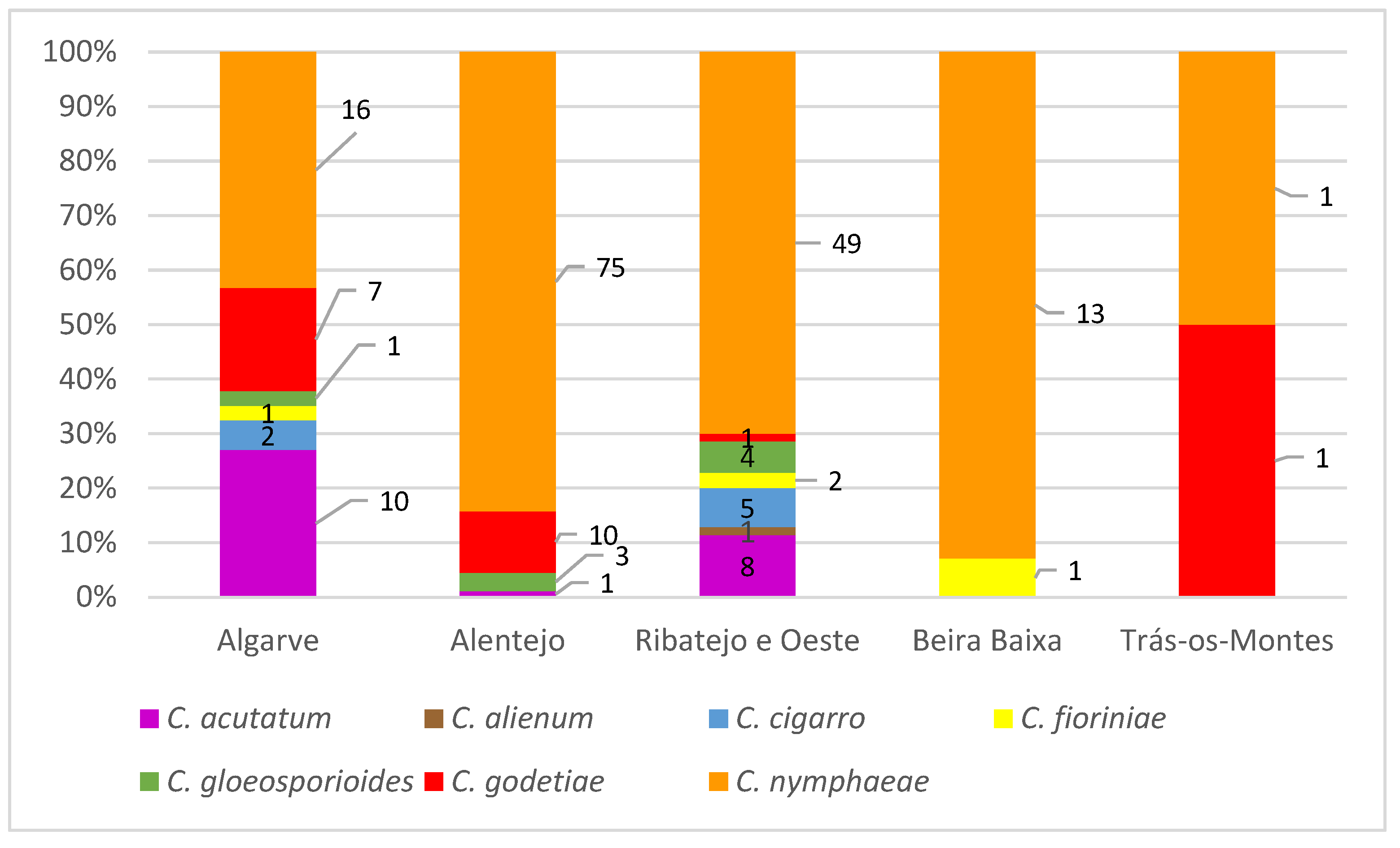
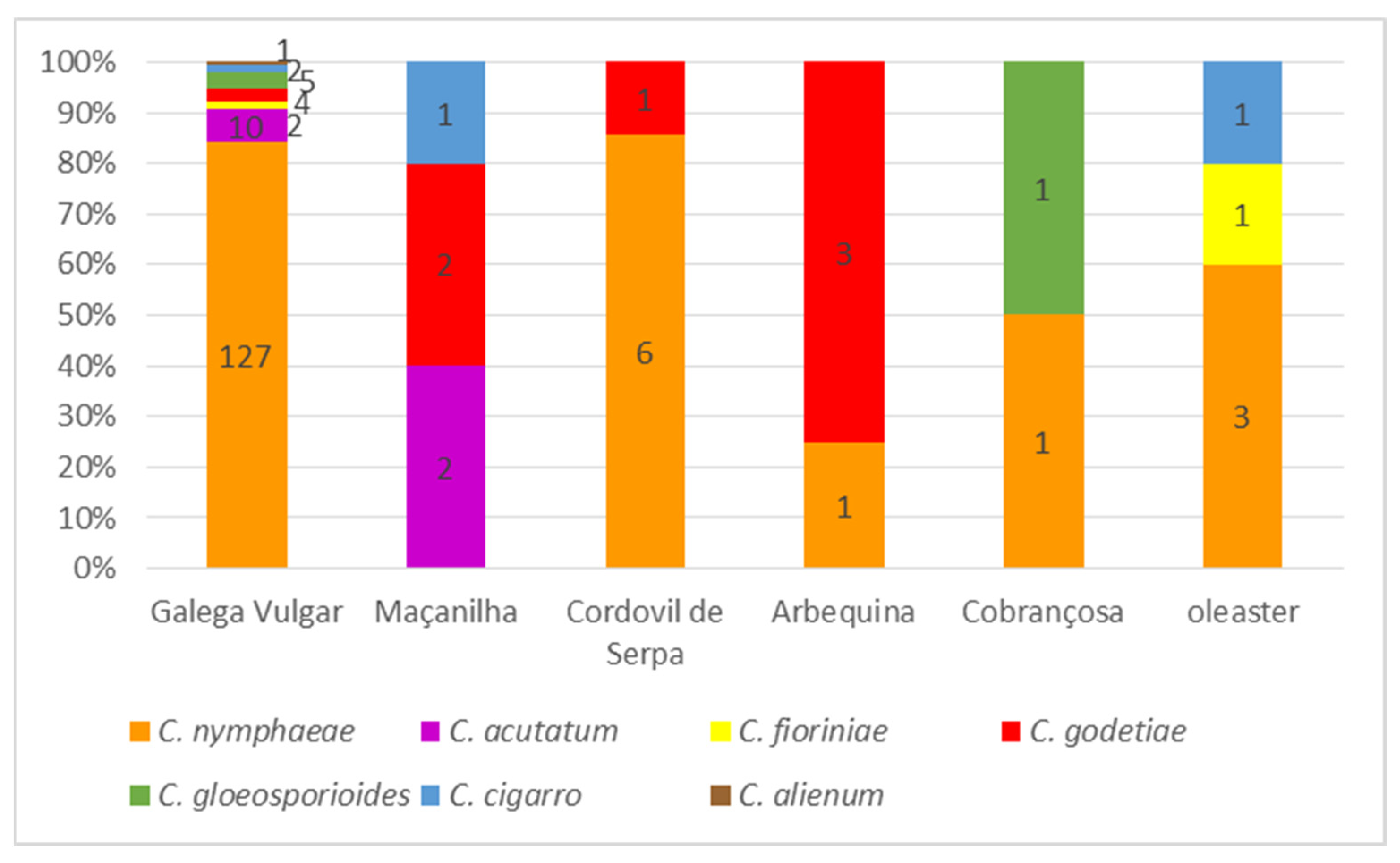
3.1. Disease Survey
3.2. Molecular Characterization
3.2.1. ISSR Analysis
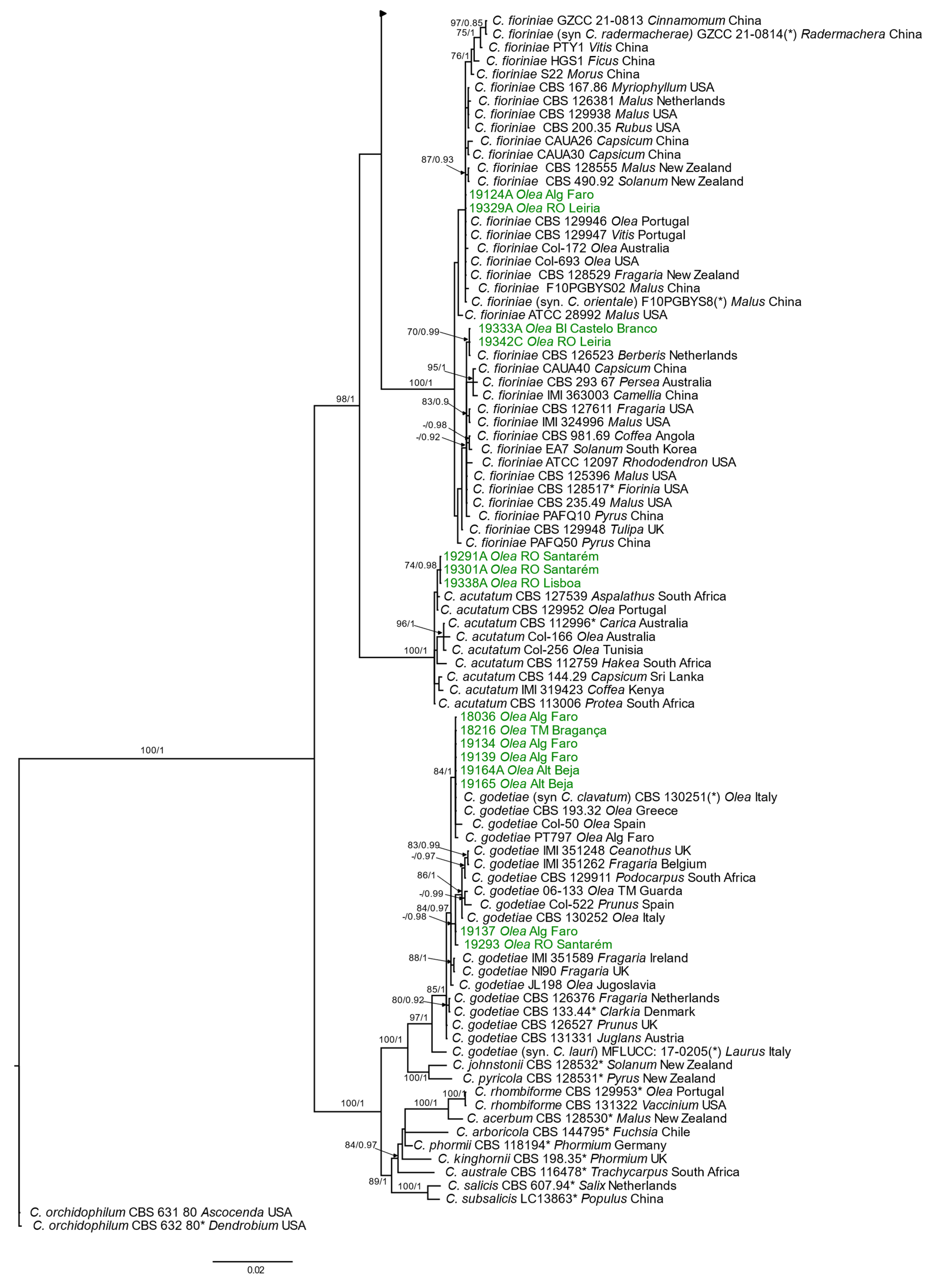
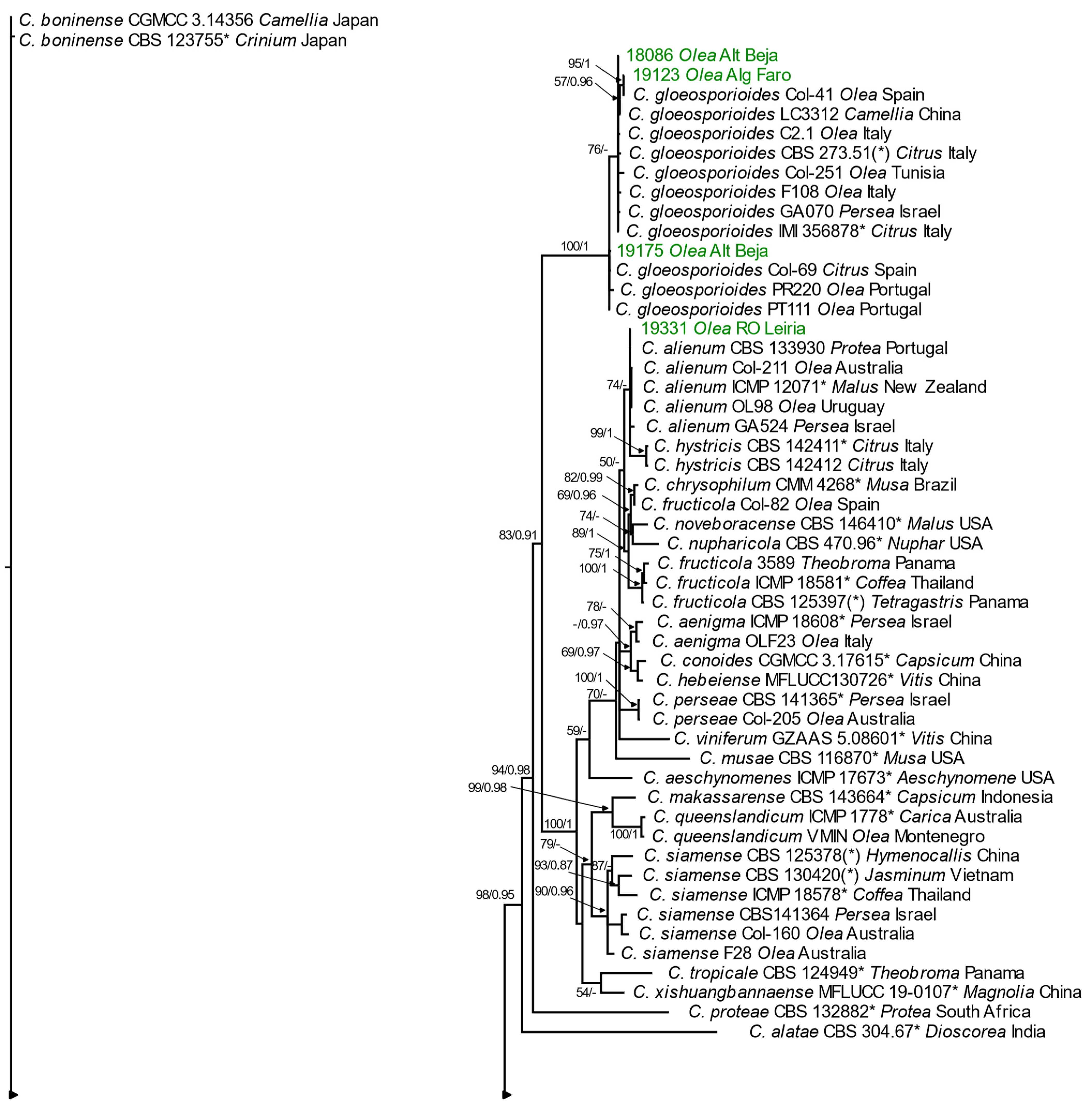
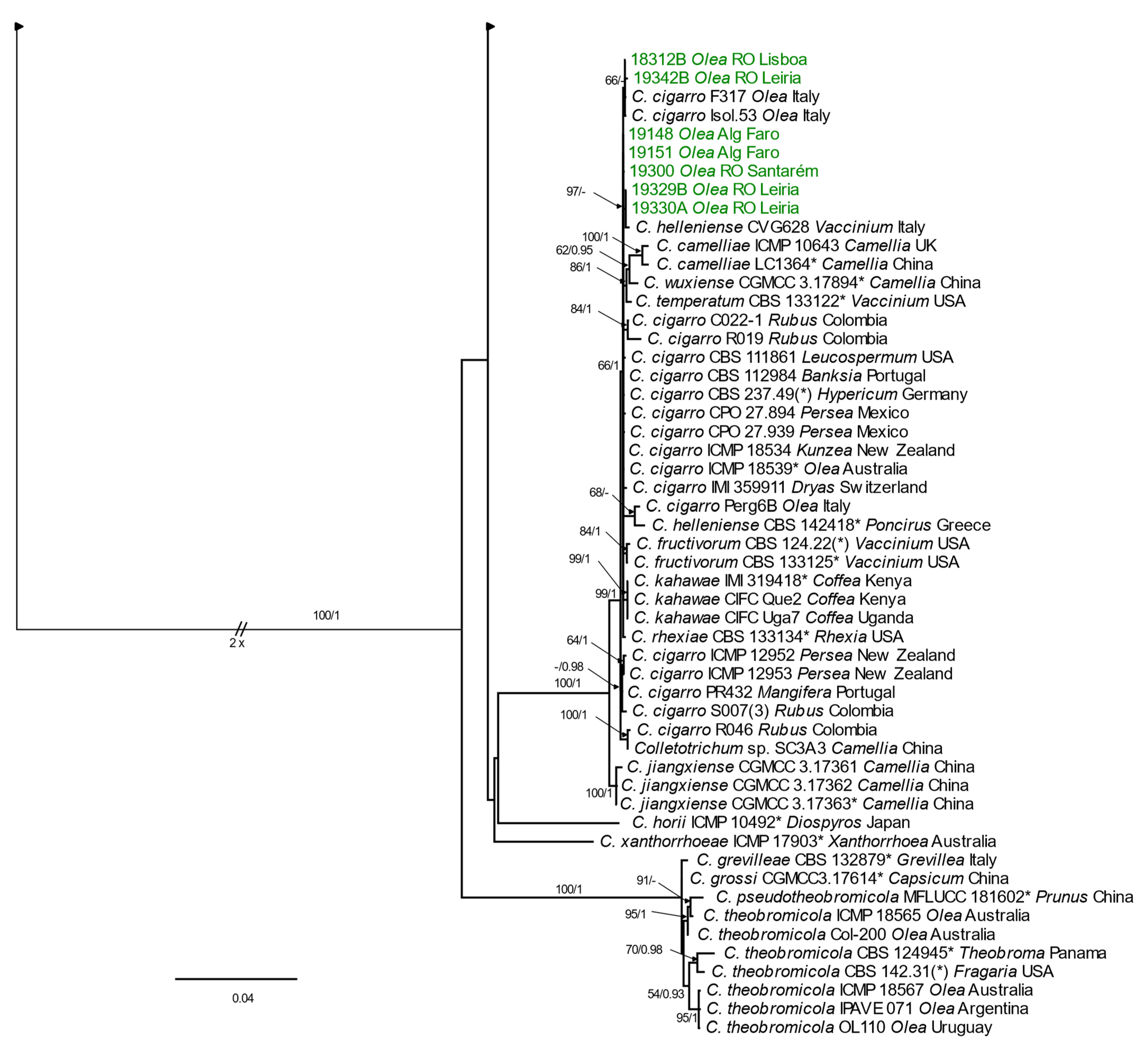
3.2.2. Phylogenetic Analyses
Acutatum Species Complex
Gloeosporioides Species Complex
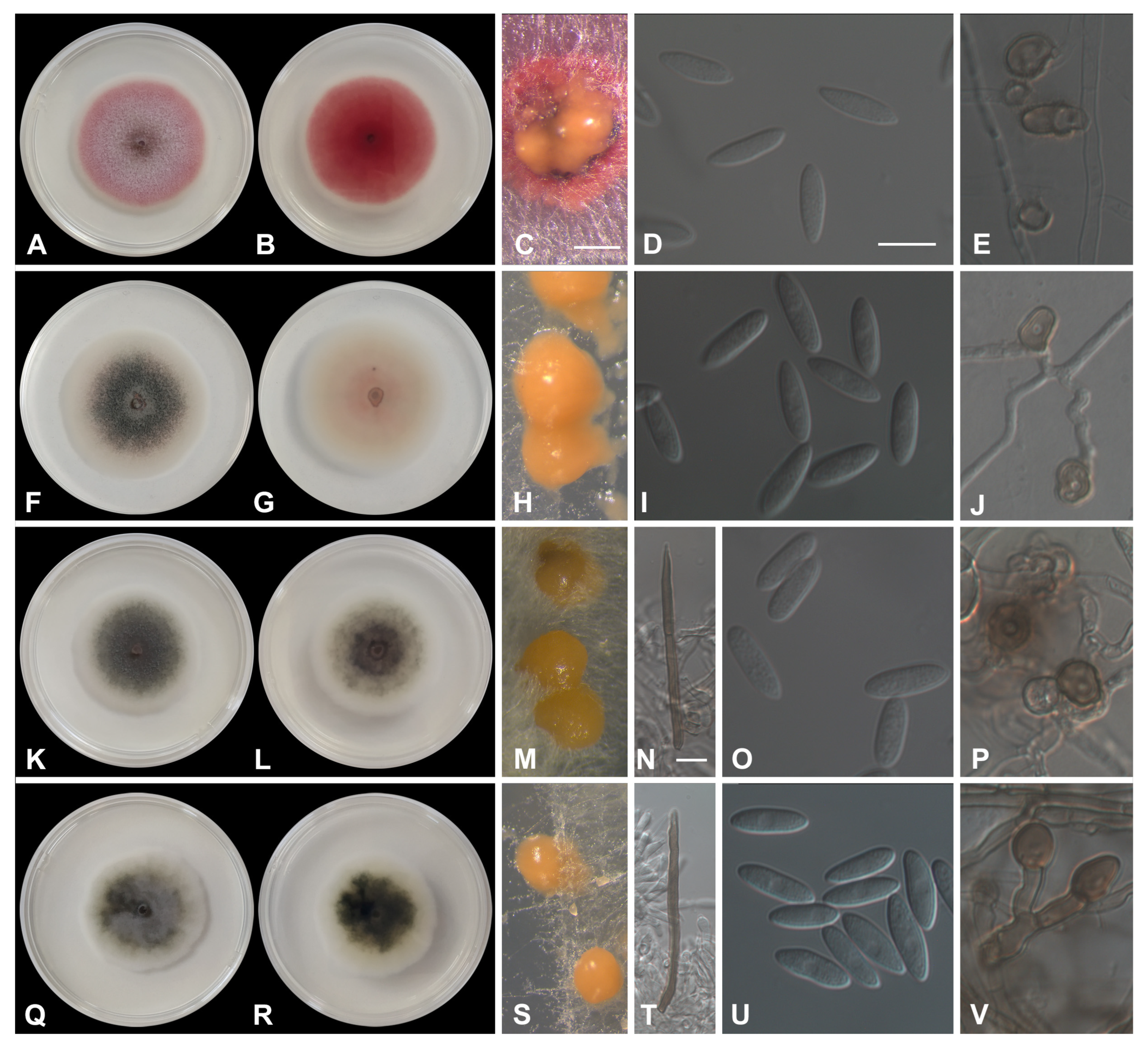
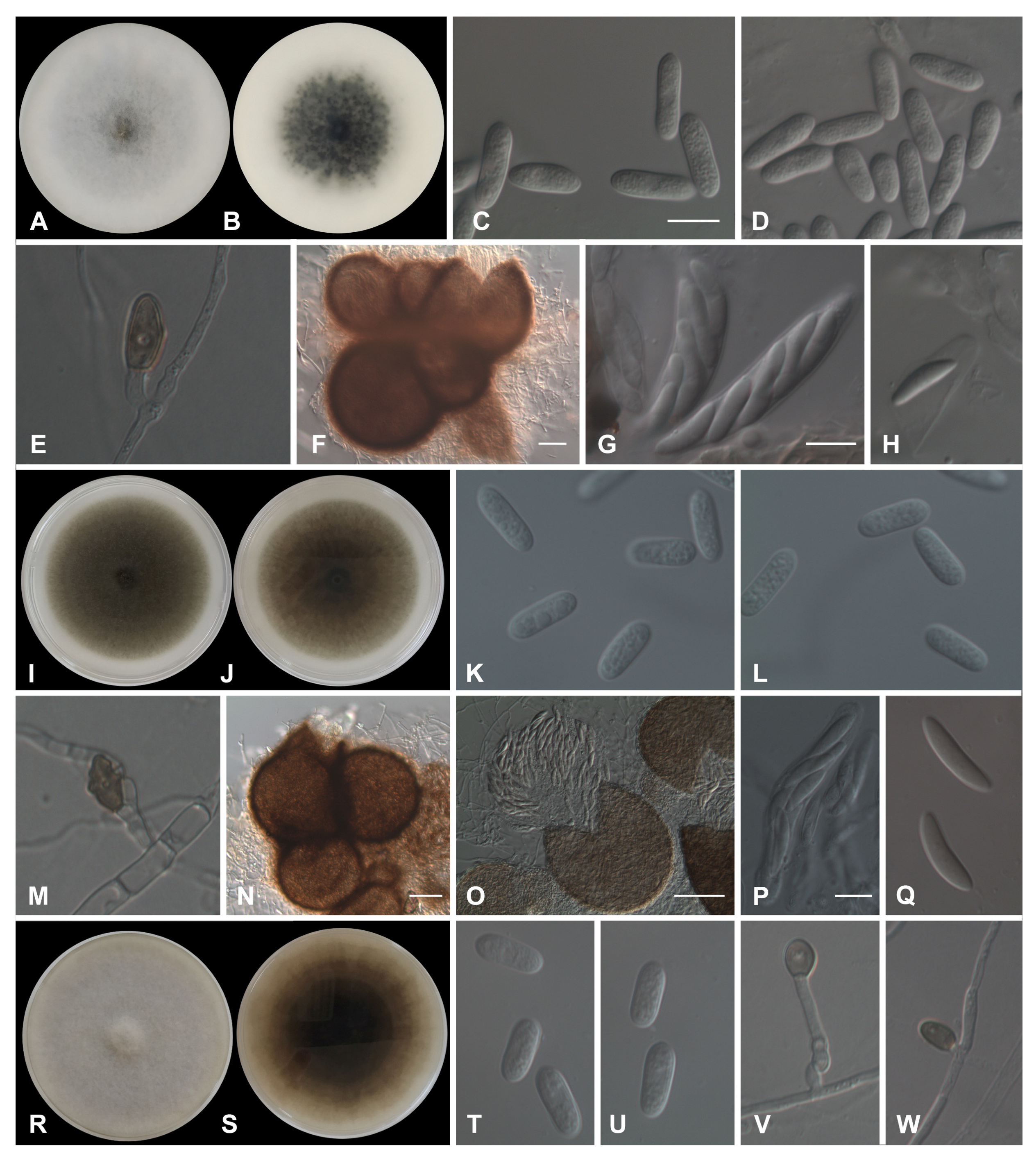
3.3. Morphological Characterization
3.3.1. Acutatum Species Complex
3.3.2. Gloeosporioides Species Complex
3.4. Population Structure
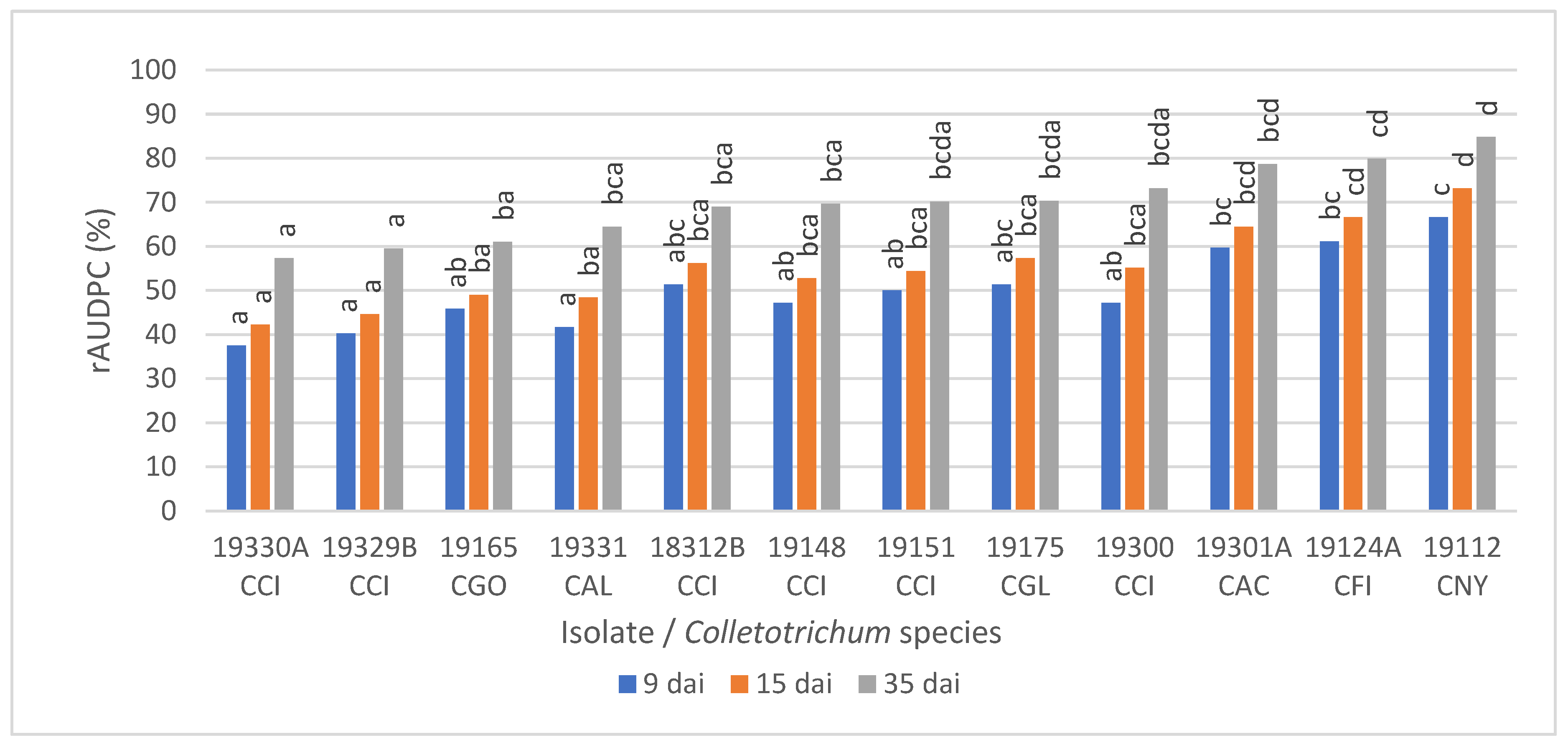
3.5. Pathogenicity on Detached Olive Fruit
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Food and Agricultural Organisation of the United Nations, FAOSTAT-Crops and Livestock Products. Available online: https://www.fao.org/faostat/en/#data/QCL/visualize (accessed on 6 February 2024).
- Superfície das Principais Culturas Agrícolas (ha) por Localização Geográfica (Região Agrária) e Espécie; Anual-INE, Estatísticas da Produção Vegetal. Available online: https://www.ine.pt/xportal/xmain?xpid=INE&xpgid=ine_indicadores&indOcorrCod=0000019 (accessed on 6 February 2024).
- Talhinhas, P.; Loureiro, A.; Oliveira, H. Olive Anthracnose: A Yield- and Oil Quality-degrading Disease Caused by Several Species of Colletotrichum that Differ in Virulence, Host Preference and Geographical Distribution. Mol. Plant Pathol. 2018, 19, 1797–1807. [Google Scholar] [CrossRef]
- Moral, J.; Agustí-Brisach, C.; Raya, M.C.; Jurado-Bello, J.; López-Moral, A.; Roca, L.F.; Chattaoui, M.; Rhouma, A.; Nigro, F.; Sergeeva, V.; et al. Diversity of Colletotrichum Species Associated with Olive Anthracnose Worldwide. J. Fungi 2021, 7, 741. [Google Scholar] [CrossRef] [PubMed]
- Peres, F.; Talhinhas, P.; Afonso, H.; Alegre, H.; Oliveira, H.; Ferreira-Dias, S. Olive Oils from Fruits Infected with Different Anthracnose Pathogens Show Sensory Defects Earlier Than Chemical Degradation. Agronomy 2021, 11, 1041. [Google Scholar] [CrossRef]
- Moral, J.; Xaviér, C.; Roca, L.F.; Romero, J.; Moreda, W.; Trapero, A. La Antracnosis Del Olivo y Su Efecto En La Calidad Del Aceite. Grasas Aceites 2014, 65, e028. [Google Scholar] [CrossRef]
- Damm, U.; Cannon, P.F.; Woudenberg, J.H.C.; Johnston, P.R.; Weir, B.S.; Tan, Y.P.; Shivas, R.G.; Crous, P.W. The Colletotrichum boninense Species Complex. Stud. Mycol. 2012, 73, 1–36. [Google Scholar] [CrossRef] [PubMed]
- Damm, U.; Cannon, P.F.; Woudenberg, J.H.C.; Crous, P.W. The Colletotrichum acutatum Species Complex. Stud. Mycol. 2012, 73, 37–113. [Google Scholar] [CrossRef] [PubMed]
- Weir, B.S.; Johnston, P.R.; Damm, U. The Colletotrichum gloeosporioides Species Complex. Stud. Mycol. 2012, 73, 115–180. [Google Scholar] [CrossRef] [PubMed]
- Talhinhas, P.; Neves-Martins, J.; Oliveira, H.; Sreenivasaprasad, S. The Distinctive Population Structure of Colletotrichum Species Associated with Olive Anthracnose in the Algarve Region of Portugal Reflects a Host-Pathogen Diversity Hot Spot. FEMS Microbiol. Lett. 2009, 296, 31–38. [Google Scholar] [CrossRef]
- Talhinhas, P.; Mota-Capitão, C.; Martins, S.; Ramos, A.P.; Neves-Martins, J.; Guerra-Guimarães, L.; Várzea, V.; Silva, M.C.; Sreenivasaprasad, S.; Oliveira, H. Epidemiology, Histopathology and Aetiology of Olive Anthracnose Caused by Colletotrichum acutatum and C. gloeosporioides in Portugal. Plant Pathol. 2011, 60, 483–495. [Google Scholar] [CrossRef]
- Cacciola, S.O.; Faedda, R.; Fulvia, S.; Agosteo, G.E.; Schena, L.; Frisullo, S.; Magnano di San Lio, G. Olive Anthracnose. J. Plant Pathol. 2012, 94, 29–44. [Google Scholar]
- Mosca, S.; Li Destri Nicosia, M.G.; Cacciola, S.O.; Schena, L. Molecular Analysis of Colletotrichum Species in the Carposphere and Phyllosphere of Olive. PLoS ONE 2014, 9, e114031. [Google Scholar] [CrossRef] [PubMed]
- Talhinhas, P.; Sreenivasaprasad, S.; Neves-Martins, J.; Oliveira, H. Molecular and Phenotypic Analyses Reveal Association of Diverse Colletotrichum acutatum Groups and a Low Level of C. gloeosporioides with Olive Anthracnose. Appl. Environ. Microbiol. 2005, 71, 2987–2998. [Google Scholar] [CrossRef] [PubMed]
- Talhinhas, P.; Gonçalves, E.; Sreenivasaprasad, S.; Oliveira, H. Virulence Diversity of Anthracnose Pathogens (Colletotrichum acutatum and C. gloeosporioides Species Complexes) on Eight Olive Cultivars Commonly Grown in Portugal. Eur. J. Plant Pathol. 2015, 142, 73–83. [Google Scholar] [CrossRef]
- Riolo, M.; Pane, A.; Santilli, E.; Moricca, S.; Cacciola, S.O. Susceptibility of Italian Olive Cultivars to Various Colletotrichum Species Associated with Fruit Anthracnose. Plant Pathol. 2023, 72, 255–267. [Google Scholar] [CrossRef]
- Garcia-Lopez, M.T.; Serrano, M.S.; Camiletti, B.X.; Gordon, A.; Estudillo, C.; Trapero, A.; Diez, C.M.; Moral, J. Study of the Competition between Colletotrichum godetiae and C. nymphaeae, Two Pathogenic Species in Olive. Sci. Rep. 2023, 13, 5344. [Google Scholar] [CrossRef] [PubMed]
- Velásquez, A.C.; Castroverde, C.D.M.; He, S.Y. Plant–Pathogen Warfare under Changing Climate Conditions. Curr. Biol. 2018, 28, R619–R634. [Google Scholar] [CrossRef] [PubMed]
- Pangallo, S.; Li Destri Nicosia, M.G.; Agosteo, G.E.; Schena, L. Control of Olive Anthracnose and Leaf Spot Disease by Bloom Treatments with a Pomegranate Peel Extract. J. Saudi Soc. Agric. Sci. 2022, 21, 248–254. [Google Scholar] [CrossRef]
- Schena, L.; Abdelfattah, A.; Mosca, S.; Li Destri Nicosia, M.G.; Agosteo, G.E.; Cacciola, S.O. Quantitative Detection of Colletotrichum godetiae and C. acutatum sensu stricto in the Phyllosphere and Carposphere of Olive during Four Phenological Phases. Eur. J. Plant Pathol. 2017, 149, 337–347. [Google Scholar] [CrossRef]
- Antelmi, I.; Sion, V.; Nigro, F. First Report of Colletotrichum nymphaeae on Olive in Italy. Plant Dis. 2019, 103, 765. [Google Scholar] [CrossRef]
- Lima, N.B.; Pastor, S.E.; Maza, C.E.; Conforto, C.; Vargas-Gil, S.; Roca, M. First Report of Anthracnose of Olive Fruit Caused by Colletotrichum theobromicola in Argentina. Plant Dis. 2020, 104, 589. [Google Scholar] [CrossRef]
- Moreira, V.; Mondino, P.; Alaniz, S. Olive Anthracnose Caused by Colletotrichum in Uruguay: Symptoms, Species Diversity and Pathogenicity on Flowers and Fruits. Eur. J. Plant Pathol. 2021, 160, 663–681. [Google Scholar] [CrossRef]
- Cenis, J.L. Rapid Extraction of Fungal DNA for PCR Amplification. Nucleic Acids Res. 1992, 20, 2380. [Google Scholar] [CrossRef] [PubMed]
- Zietkiewicz, E.; Rafalski, A.; Labuda, D. Genome Fingerprinting by Simple Sequence Repeat (SSR)-Anchored Polymerase Chain Reaction Amplification. Genomics 1994, 20, 176–183. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, R.J.; Yoder, O.C. A Family of Conserved Repetitive DNA Elements from the Fungal Plant Pathogen Glomerella cingulata (Colletotrichum lindemuthianum). Exp. Mycol. 1991, 15, 232–242. [Google Scholar] [CrossRef]
- Reis, P.; Cabral, A.; Nascimento, T.; Oliveira, H. Diversity of Ilyonectria Species in a Young Vineyard Affected by Black Foot Disease. Phytopathol. Mediterr. 2013, 52, 335–346. [Google Scholar]
- Talhinhas, P.; Sreenivasaprasad, S.; Neves-Martins, J.; Oliveira, H. Genetic and Morphological Characterization of Colletotrichum acutatum Causing Anthracnose of Lupins. Phytopathology 2002, 92, 986–996. [Google Scholar] [CrossRef] [PubMed]
- Carbone, I.; Kohn, L.M. A Method for Designing Primer Sets for Speciation Studies in Filamentous Ascomycetes. Mycologia 1999, 91, 553–556. [Google Scholar] [CrossRef]
- O’Donnell, K.; Cigelnik, E. Two Divergent Intragenomic rDNA ITS2 Types within a Monophyletic Lineage of the Fungus Fusarium Are Nonorthologous. Mol. Phylogenet Evol. 1997, 7, 103–116. [Google Scholar] [CrossRef] [PubMed]
- Glass, N.L.; Donaldson, G.C. Development of Primer Sets Designed for Use with the PCR to Amplify Conserved Genes from Filamentous Ascomycetes. Appl. Environ. Microbiol. 1995, 61, 1323–1330. [Google Scholar] [CrossRef]
- Crous, P.W.; Groenewald, J.Z.; Risède, J.-M.; Simoneau, P.; Hywel-Jones, N.L. Calonectria Species and Their Cylindrocladium Anamorphs: Species with Sphaeropedunculate Vesicles. Stud. Mycol. 2004, 50, 415–430. [Google Scholar]
- de Hoog, G.S.; Gerrits van den Ende, A.H. Molecular Diagnostics of Clinical Strains of Filamentous Basidiomycetes. Mycoses 1998, 41, 183–189. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. PCR Protocols: A Guide to Methods and Applications. In PCR Protocols; Academic Press, Inc.: New York, NY, USA, 1990. [Google Scholar]
- Guerber, J.C.; Liu, B.; Correll, J.C.; Johnston, P.R. Characterization of Diversity in Colletotrichum acutatum sensu lato by Sequence Analysis of Two Gene Introns, mtDNA and Intron RFLPs, and Mating Compatibility. Mycologia 2003, 95, 872–895. [Google Scholar] [CrossRef] [PubMed]
- Stephenson, S.A.; Green, J.R.; Manners, J.M.; Maclean, D.J. Cloning and Characterisation of Glutamine Synthetase from Colletotrichum gloeosporioides and Demonstration of Elevated Expression during Pathogenesis on Stylosanthes guianensis. Curr. Genet. 1997, 31, 447–454. [Google Scholar] [CrossRef] [PubMed]
- Silva, D.N.; Talhinhas, P.; Várzea, V.; Cai, L.; Paulo, O.S.; Batista, D. Application of the Apn2/MAT Locus to Improve the Systematics of the Colletotrichum gloeosporioides Complex: An Example from Coffee (Coffea spp.) Hosts. Mycologia 2012, 104, 396–409. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT Online Service: Multiple Sequence Alignment, Interactive Sequence Choice and Visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Vaidya, G.; Lohman, D.J.; Meier, R. SequenceMatrix: Concatenation Software for the Fast Assembly of Multi-Gene Datasets with Character Set and Codon Information. Cladistics 2011, 27, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Nylander, J.A.A. MrModelTest v2 Program Distributed by the Author; Evolutionary Biology Centre, Uppsala University: Uppsala, Sweden, 2004. [Google Scholar]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for Inference of Large Phylogenetic Trees. In Proceedings of the 2010 Gateway Computing Environments Workshop, GCE, New Orleans, LA, USA, 14 November 2010. [Google Scholar]
- Nirenberg, H. Untersuchungen Über Die Morphologische Und Biologische Differenzierung in Der Fusarium-Sektion Liseola. Mitt. Biol. Bundesanst. Land. Forstwirtsch. Berl. Dahl. 1976, 169, 1–167. [Google Scholar] [CrossRef]
- Rayner, R.W. A Mycological Colour Chart; Commonwealth Mycological Institute: Kew, UK, 1970; pp. 1–34. [Google Scholar]
- Guerber, J.C.; Correll, J.C. Characterization of Glomerella acutata, the Teleomorph of Colletotrichum acutatum. Mycologia 2001, 93, 216–229. [Google Scholar] [CrossRef]
- International Olive Council. Guide for the Determination of the Characteristics of Oil-Olives. COI/OH/Doc. No 1. 2011. Available online: https://www.internationaloliveoil.org/wp-content/uploads/2019/11/COI-OH-Doc.-1-2011-Eng.pdf (accessed on 20 March 2024).
- Madden, L.V.; Hughes, G.; van den Bosch, F. Chapter 4: Temporal Analysis I: Quantifying and Comparing Epidemics. In The Study of Plant Disease Epidemics; Madden, L.V., Hughes, G., van den Bosch, F., Eds.; The American Phytopathological Society: St. Paul, MN, USA, 2017; pp. 63–116. [Google Scholar]
- Yu, Z.; Jiang, X.; Zheng, H.; Zhang, H.; Qiao, M. Fourteen New Species of Foliar Colletotrichum Associated with the Invasive Plant Ageratina adenophora and Surrounding Crops. J. Fungi 2022, 8, 185. [Google Scholar] [CrossRef]
- Damm, U.; Sun, Y.C.; Huang, C.J. Colletotrichum eriobotryae sp. nov. and C. nymphaeae, the Anthracnose Pathogens of Loquat Fruit in Central Taiwan, and Their Sensitivity to Azoxystrobin. Mycol. Prog. 2020, 19, 367–380. [Google Scholar] [CrossRef]
- Crouch, J.A.; Clarke, B.B.; Hillman, B.I. What Is the Value of ITS Sequence Data in Colletotrichum Systematics and Species Diagnosis? A Case Study Using the Falcate-Spored Graminicolous Colletotrichum Group. Mycologia 2009, 101, 648–656. [Google Scholar] [CrossRef]
- Liu, F.; Weir, B.S.; Damm, U.; Crous, P.W.; Wang, Y.; Liu, B.; Wang, M.; Zhang, M.; Cai, L. Unravelling Colletotrichum Species Associated with Camellia Employing ApMat and GS Loci to Resolve Species in the C. gloeosporioides Complex. Persoonia 2015, 35, 63–86. [Google Scholar] [CrossRef] [PubMed]
- Jayawardena, R.S.; Hyde, K.D.; de Farias, A.R.G.; Bhunjun, C.S.; Ferdinandez, H.S.; Manamgoda, D.S.; Udayanga, D.; Herath, I.S.; Thambugala, K.M.; Manawasinghe, I.S.; et al. What Is a Species in Fungal Plant Pathogens? Fungal Divers. 2021, 109, 239–266. [Google Scholar] [CrossRef]
- Chen, Y.; Fu, D.; Wang, W.; Gleason, M.L.; Zhang, R.; Liang, X.; Sun, G. Diversity of Colletotrichum Species Causing Apple Bitter Rot and Glomerella Leaf Spot in China. J. Fungi 2022, 8, 740. [Google Scholar] [CrossRef]
- Zhang, Q.; Nizamani, M.M.; Feng, Y.; Yang, Y.Q.; Jayawardena, R.S.; Hyde, K.D.; Wang, Y.; Li, C. Genome-Scale and Multi-Gene Phylogenetic Analyses of Colletotrichum spp. Host Preference and Associated with Medicinal Plants. Mycosphere 2023, 14, 1–106. [Google Scholar] [CrossRef]
- Zhang, Y.X.; Chen, J.W.; Manawasinghe, I.S.; Lin, Y.H.; Jayawardena, R.S.; Mckenzie, E.H.C.; Hyde, K.D.; Xiang, M.M. Identification and Characterization of Colletotrichum Species Associated with Ornamental Plants in Southern China. Mycosphere 2023, 14, 262–302. [Google Scholar] [CrossRef]
- Huang, F.; Chen, G.Q.; Hou, X.; Fu, Y.S.; Cai, L.; Hyde, K.D.; Li, H.Y. Colletotrichum Species Associated with Cultivated Citrus in China. Fungal Divers. 2013, 61, 61–74. [Google Scholar] [CrossRef]
- Zapata, M.; Rodríguez-Serrano, E.; Castro, J.F.; Santelices, C.; Carrasco-Fernández, J.; Damm, U.; Palfner, G. Novel Species and Records of Colletotrichum Associated with Native Woody Plants in South-Central Chile. Mycol. Prog. 2024, 23, 18. [Google Scholar] [CrossRef]
- Cabral, A.; Azinheira, H.G.; Talhinhas, P.; Batista, D.; Ramos, A.P.; Silva, M.D.C.; Oliveira, H.; Várzea, V. Pathological, Morphological, Cytogenomic, Biochemical and Molecular Data Support the Distinction between Colletotrichum cigarro comb. et stat. nov. and Colletotrichum kahawae. Plants 2020, 9, 502. [Google Scholar] [CrossRef]
- Liu, F.; Ma, Z.Y.; Hou, L.W.; Diao, Y.Z.; Wu, W.P.; Damm, U.; Song, S.; Cai, L. Updating Species Diversity of Colletotrichum, with a Phylogenomic Overview. Stud. Mycol. 2022, 101, 1–56. [Google Scholar] [CrossRef]
- Licciardello, G.; Moral, J.; Strano, M.C.; Caruso, P.; Sciara, M.; Bella, P.; Sorrentino, G.; Di Silvestro, S. Characterization of Colletotrichum Strains Associated with Olive Anthracnose in Sicily. Phytopathol. Mediterr. 2022, 61, 139–151. [Google Scholar] [CrossRef]
- López-Moral, A.; Raya-Ortega, M.C.; Agustí-Brisach, C.; Roca, L.F.; Lovera, M.; Luque, F.; Arquero, O.; Trapero, A. Morphological, Pathogenic, and Molecular Characterization of Colletotrichum acutatum Isolates Causing Almond Anthracnose in Spain. Plant Dis. 2017, 101, 2034–2045. [Google Scholar] [CrossRef]
- Materatski, P.; Varanda, C.; Carvalho, T.; Dias, A.B.; Campos, M.D.; Rei, F.; Félix, M.R. Diversity of Colletotrichum Species Associated with Olive Anthracnose and New Perspectives on Controlling the Disease in Portugal. Agronomy 2018, 8, 301. [Google Scholar] [CrossRef]
- Filoda, P.F.; Arellano, A.D.V.; Dallagnol, L.J.; Chaves, F.C. Colletotrichum acutatum and Colletotrichum nymphaeae Causing Blossom Blight and Fruit Anthracnose on Olives in Southern Brazil. Eur. J. Plant Pathol. 2021, 161, 993–998. [Google Scholar] [CrossRef]
- Talhinhas, P.; Baroncelli, R. Colletotrichum Species and Complexes: Geographic Distribution, Host Range and Conservation Status. Fungal Divers. 2021, 110, 109–198. [Google Scholar] [CrossRef]
- López-Moral, A.; Agustí-Brisach, C.; Lovera, M.; Arquero, O.; Trapero, A. Almond Anthracnose: Current Knowledge and Future Perspectives. Plants 2020, 9, 945. [Google Scholar] [CrossRef]
- Shivas, R.G.; Tan, Y.P.; Edwards, J.; Dinh, Q.; Maxwell, A.; Andjic, V.; Liberato, J.R.; Anderson, C.; Beasley, D.R.; Bransgrove, K.; et al. Colletotrichum Species in Australia. Australas. Plant Path 2016, 45, 447–464. [Google Scholar] [CrossRef]
- Chattaoui, M.; Raya, M.C.; Bouri, M.; Moral, J.; Perez-Rodriguez, M.; Trapero, A.; Msallem, M.; Rhouma, A. Characterization of a Colletotrichum Population Causing Anthracnose Disease on Olive in Northern Tunisia. J. Appl. Microbiol. 2016, 120, 1368–1381. [Google Scholar] [CrossRef] [PubMed]
- Cara, M.; Iliadi, M.K.; Lagogianni, C.S.; Paplomatas, E.J.; Merkuri, J.; Tsitsigiannis, D.I. First Report of Colletotrichum acutatum Causing Anthracnose on Olives in Albania. Plant Dis. 2021, 105, 495. [Google Scholar] [CrossRef] [PubMed]
- Iliadi, M.K.; Tjamos, E.C.; Antoniou, P.P.; Tsitsigiannis, D.I. First Report of Colletotrichum acutatum Causing Anthracnose on Olives in Greece. Plant Dis. 2018, 102, 820. [Google Scholar] [CrossRef]
- Nawaz, H.H.; Manzoor, A.; Iqbal, M.Z.; Ansar, M.R.; Ali, M.; Muhammad Kakar, K.; Ali Awan, A.; Weiguo, M. Colletotrichum acutatum: Causal Agent of Olive Anthracnose Isolation, Characterization, and Fungicide Susceptibility Screening in Punjab, Pakistan. Plant Dis. 2023, 107, 1329–1342. [Google Scholar] [CrossRef] [PubMed]
- Riolo, M.; Cacciola, S.O. First Report of Colletotrichum fioriniae Associated with Olive Anthracnose in Italy. J. Plant Pathol. 2022, 105, 363. [Google Scholar] [CrossRef]
- Liu, F.; Damm, U.; Cai, L.; Crous, P.W. Species of the Colletotrichum gloeosporioides Complex Associated with Anthracnose Diseases of Proteaceae. Fungal Divers. 2013, 61, 89–105. [Google Scholar] [CrossRef]
- EFSA PLH Panel (EFSA Panel on Plant Health); Bragard, C.; Baptista, P.; Chatzivassiliou, E.; Di Serio, F.; Gonthier, P.; Jaques Miret, J.A.; Justesen, A.F.; MacLeod, A.; Magnusson, C.S.; et al. Pest Categorisation of Colletotrichum aenigma, C. alienum, C. perseae, C. siamense and C. theobromicola. EFSA J. 2022, 20, e07529. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cabral, A.; Nascimento, T.; Azinheira, H.G.; Loureiro, A.; Talhinhas, P.; Oliveira, H. Olive Anthracnose in Portugal Is Still Mostly Caused by Colletotrichum nymphaeae, but C. acutatum Is Spreading and C. alienum and C. cigarro Are Reported for the First Time. Horticulturae 2024, 10, 434. https://doi.org/10.3390/horticulturae10050434
Cabral A, Nascimento T, Azinheira HG, Loureiro A, Talhinhas P, Oliveira H. Olive Anthracnose in Portugal Is Still Mostly Caused by Colletotrichum nymphaeae, but C. acutatum Is Spreading and C. alienum and C. cigarro Are Reported for the First Time. Horticulturae. 2024; 10(5):434. https://doi.org/10.3390/horticulturae10050434
Chicago/Turabian StyleCabral, Ana, Teresa Nascimento, Helena G. Azinheira, Andreia Loureiro, Pedro Talhinhas, and Helena Oliveira. 2024. "Olive Anthracnose in Portugal Is Still Mostly Caused by Colletotrichum nymphaeae, but C. acutatum Is Spreading and C. alienum and C. cigarro Are Reported for the First Time" Horticulturae 10, no. 5: 434. https://doi.org/10.3390/horticulturae10050434
APA StyleCabral, A., Nascimento, T., Azinheira, H. G., Loureiro, A., Talhinhas, P., & Oliveira, H. (2024). Olive Anthracnose in Portugal Is Still Mostly Caused by Colletotrichum nymphaeae, but C. acutatum Is Spreading and C. alienum and C. cigarro Are Reported for the First Time. Horticulturae, 10(5), 434. https://doi.org/10.3390/horticulturae10050434







