Garlic (Allium sativum L.) Invertase Genes: Genome-Wide Identification and Expression in Response to Abiotic Stresses and Phytohormones
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification and Structure Analysis of the A. sativum Invertase Gene Family In Silico and In Vitro
2.2. In Silico Profiling of A. sativum Invertase Gene Expression
2.3. Garlic Plants and Abiotic Stress Simulation
2.4. Invertase Gene Expression Analysis
2.5. Soluble Sugar Content
2.6. Statistical Analysis
3. Results
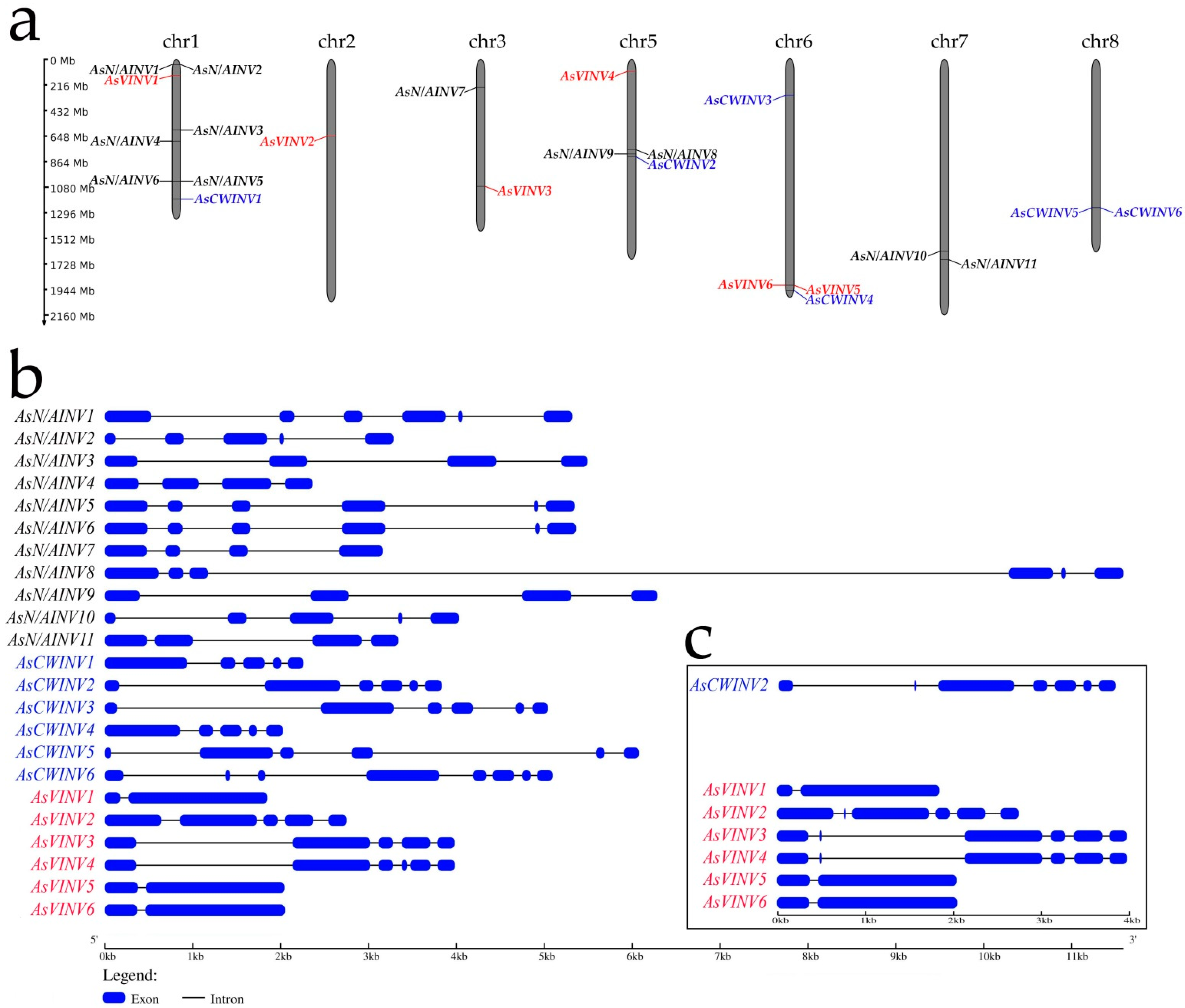
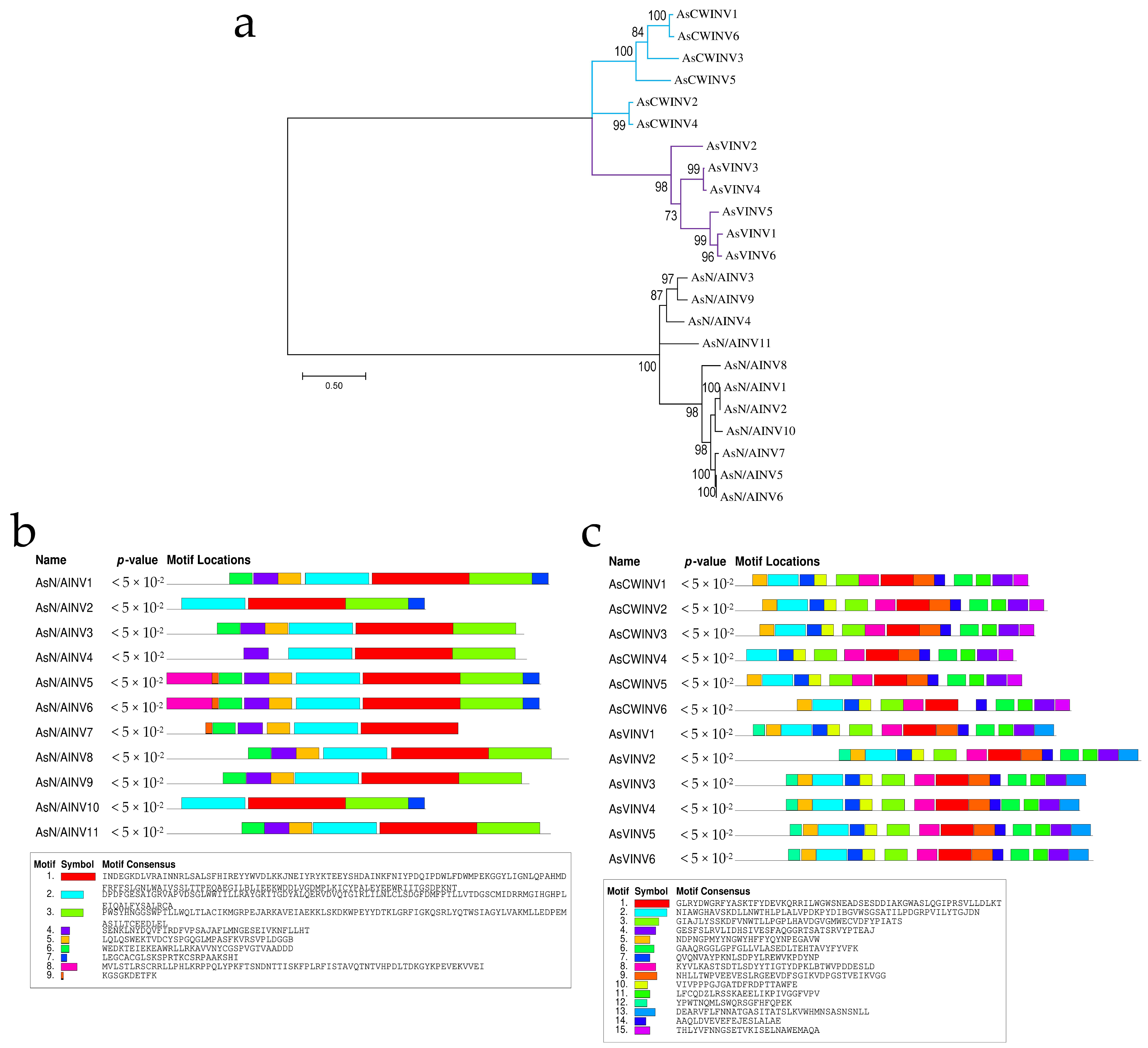
3.1. Invertase Gene Family in A. sativum
3.2. Putative A. sativum Invertase Proteins
3.3. Analysis of Promoter Regions in the A. sativum Invertase Genes
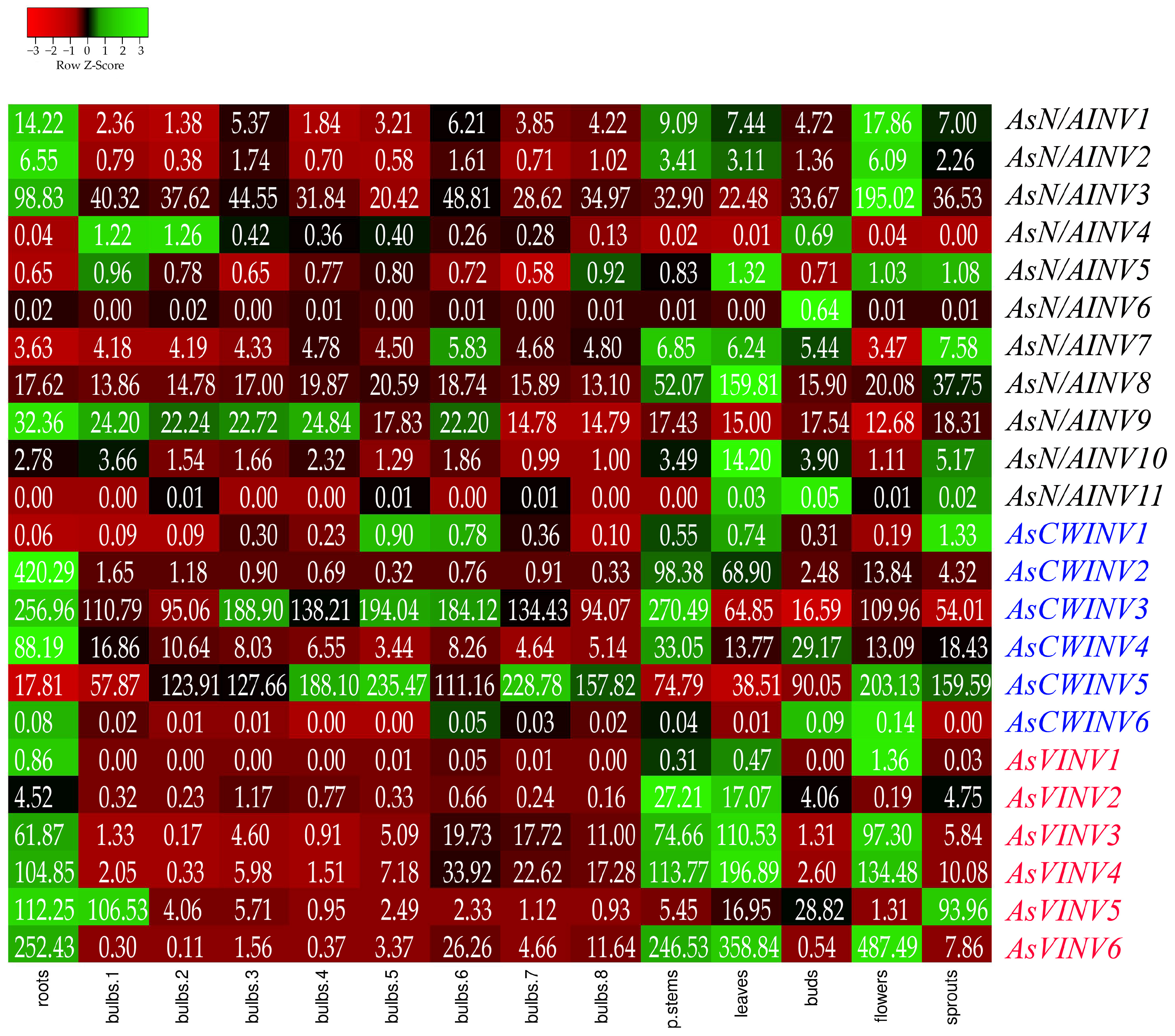
3.4. Organ-Specific Expression Pattern of the Invertase Genes in Garlic
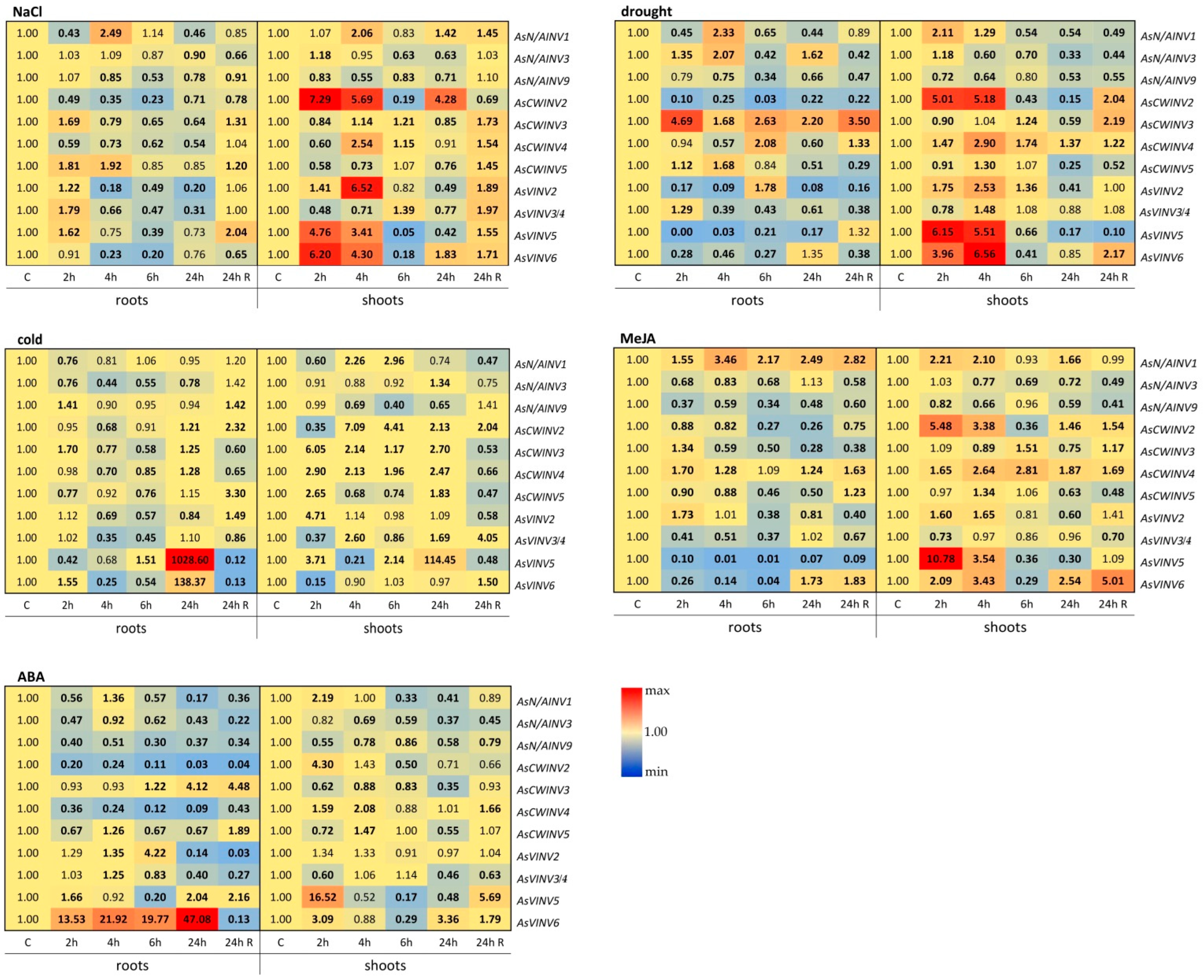
3.5. Expression of Invertase Genes in cv. Sarmat Seedlings in Response to High Salinity, Drought, Cold, and Phytohormones
3.5.1. Response to High Salinity
3.5.2. Response to Drought
3.5.3. Response to Cold
3.5.4. Response to Phytohormones
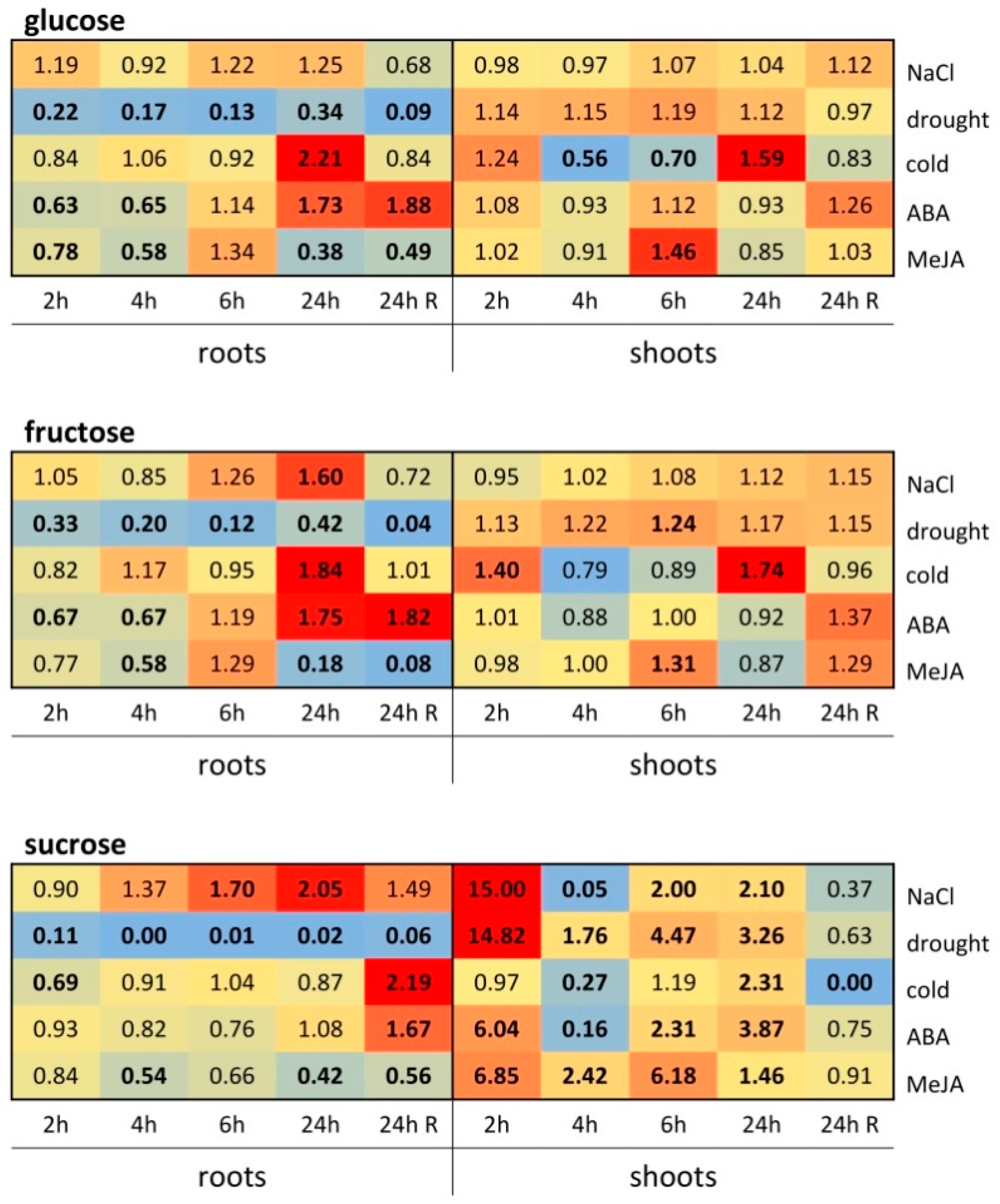
3.6. Sucrose, Glucose, and Fructose Content in cv. Sarmat Seedlings in Response to Cold, Drought, High Salinity, and Phytohormones
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ruan, Y.L. Sucrose metabolism: Gateway to diverse carbon use and sugar signaling. Annu. Rev. Plant Biol. 2014, 65, 33–67. [Google Scholar] [CrossRef]
- Yoon, J.; Cho, L.H.; Tun, W.; Jeon, J.S.; An, G. Sucrose signaling in higher plants. Plant Sci. 2021, 302, 110703. [Google Scholar] [CrossRef]
- Bergareche, D.; Royo, J.; Muñiz, L.M.; Hueros, G. Cell wall invertase activity regulates the expression of the transfer cell-specific transcription factor ZmMRP-1. Planta 2018, 247, 429–442. [Google Scholar] [CrossRef]
- Walker, R.P.; Bonghi, C.; Varotto, S.; Battistelli, A.; Burbidge, C.A.; Castellarin, S.D.; Chen, Z.H.; Darriet, P.; Moscatello, S.; Rienth, M.; et al. Sucrose metabolism and transport in grapevines, with emphasis on berries and leaves, and insights gained from a cross-species comparison. Int. J. Mol. Sci. 2021, 22, 7794. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Li, H.; Zhu, Q.; Liu, D.; Li, Z.; Chen, H.; Luo, J.; Gong, P.; Ismail, A.M.; Zhang, Z. Knockout of the sugar transporter OsSTP15 enhances grain yield by improving tiller number due to increased sugar content in the shoot base of rice (Oryza sativa L.). New Phytol. 2024, 241, 1250–1265. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Zhang, C.X.; Zhu, L.C.; Yang, N.X.; Yang, J.J.; Ma, B.Q.; Ma, F.W.; Li, M.J. MdFRK2-mediated sugar metabolism accelerates cellulose accumulation in apple and poplar. Biotechnol. Biofuels 2021, 14, 137. [Google Scholar] [CrossRef] [PubMed]
- Sturm, A.; Tang, G.Q. The sucrose-cleaving enzymes of plants are crucial for development, growth and carbon partitioning. Trends Plant Sci. 1999, 4, 401–407. [Google Scholar] [CrossRef] [PubMed]
- Schmolzer, K.; Gutmann, A.; Diricks, M.; Desmet, T.; Nidetzky, B. Sucrose synthase: A unique glycosyltransferase for biocatalytic glycosylation process development. Biotechnol. Adv. 2016, 34, 88–111. [Google Scholar] [CrossRef]
- Ruan, Y.-L.; Jin, Y.; Yang, Y.-J.; Li, G.-J.; Boyer, J.S. Sugar input, metabolism, and signaling mediated by invertase: Roles in development, yield potential, and response to drought and heat. Mol. Plant 2010, 3, 942–955. [Google Scholar] [CrossRef]
- Liu, Y.H.; Song, Y.H.; Ruan, Y.L. Sugar conundrum in plant-pathogen interactions: Roles of invertase and sugar transporters depend on pathosystems. J. Exp. Bot. 2022, 73, 1910–1925. [Google Scholar] [CrossRef]
- Tauzin, A.S.; Giardina, T. Sucrose and invertases, a part of the plant defense response to the biotic stresses. Front. Plant Sci. 2014, 5, 293. [Google Scholar] [CrossRef] [PubMed]
- Qian, W.; Yue, C.; Wang, Y.; Cao, H.; Li, N.; Wang, L.; Hao, X.; Wang, X.; Xiao, B.; Yang, Y. Identification of the invertase gene family (INVs) in tea plant and their expression analysis under abiotic stress. Plant Cell Rep. 2016, 35, 2269–2283. [Google Scholar] [CrossRef] [PubMed]
- Coculo, D.; Lionetti, V. The plant invertase/pectin methylesterase inhibitor superfamily. Front. Plant Sci. 2022, 13, 863892. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Gao, K.; Su, X.; Rao, P.; An, X. Genome-wide identification of the invertase gene family in populus. PLoS ONE 2015, 10, e0138540. [Google Scholar] [CrossRef] [PubMed]
- Abbas, A.; Shah, A.N.; Shah, A.A.; Nadeem, M.A.; Alsaleh, A.; Javed, T.; Alotaibi, S.S.; Abdelsalam, N.R. genome-wide analysis of invertase gene family, and expression profiling under abiotic stress conditions in potato. Biology 2022, 11, 539. [Google Scholar] [CrossRef] [PubMed]
- Juárez-Colunga, S.; López-González, C.; Morales-Elías, N.C.; Massange-Sánchez, J.A.; Trachsel, S.; Tiessen, A. Genome-wide analysis of the invertase gene family from maize. Plant Mol. Biol. 2018, 97, 385–406. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Jin, J.; He, X.; Luo, Z.; Wang, Z.; Yang, J.; Xu, X. Genome-wide identification and analysis of the invertase gene family in tobacco (Nicotiana tabacum) reveals NtNINV10 participating the sugar metabolism. Front. Plant Sci. 2023, 14, 1164296. [Google Scholar] [CrossRef]
- Wan, H.; Zhang, Y.; Wu, L.; Zhou, G.; Pan, L.; Fernie, A.R.; Ruan, Y.L. Evolution of cytosolic and organellar invertases empowered the colonization and thriving of land plants. Plant Physiol. 2023, 193, 1227–1243. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Li, Y.; Du, Y.; Pan, L.; Huang, Y.; Liu, H.; Zhao, Y.; Shi, Y.; Ruan, Y.L.; Dong, Z.; et al. Maize cytosolic invertase INVAN6 ensures faithful meiotic progression under heat stress. New Phytol. 2022, 236, 2172–2188. [Google Scholar] [CrossRef]
- Coluccio Leskow, C.; Conte, M.; Del Pozo, T.; Bermúdez, L.; Lira, B.S.; Gramegna, G.; Baroli, I.; Burgos, E.; Zavallo, D.; Kamenetzky, L.; et al. The cytosolic invertase NI6 affects vegetative growth, flowering, fruit set, and yield in tomato. J. Exp. Bot. 2021, 72, 2525–2543. [Google Scholar] [CrossRef]
- Ansary, J.; Forbes-Hernández, T.Y.; Gil, E.; Cianciosi, D.; Zhang, J.; Elexpuru-Zabaleta, M.; Simal-Gandara, J.; Giampieri, F.; Battino, M. Potential health benefit of garlic based on human intervention studies: A brief overview. Antioxidants 2020, 9, 619. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Wu, C.; Cheng, Z.; Meng, H. Growth and physiological changes in continuously cropped eggplant (Solanum melongena L.) upon relay intercropping with garlic (Allium sativum L.). Front. Plant Sci. 2015, 6, 262. [Google Scholar] [CrossRef] [PubMed]
- Shemesh, E.; Scholten, O.; Rabinowitch, H.D.; Kamenetsky, R. Unlocking variability: Inherent variation and developmental traits of garlic plants originated from sexual reproduction. Planta 2008, 227, 1013–1024. [Google Scholar] [CrossRef] [PubMed]
- Egea, L.A.; Mérida-García, R.; Kilian, A.; Hernandez, P.; Dorado, G. Assessment of genetic diversity and structure of large garlic (Allium sativum L.) germplasm bank, by diversity arrays technology “Genotyping-by-Sequencing” platform (DArTseq). Front. Genet. 2017, 8, 98. [Google Scholar] [CrossRef] [PubMed]
- Casals, J.; Rivera, A.; Campo, S.; Aymerich, E.; Isern, H.; Fenero, D.; Garriga, A.; Palou, A.; Monfort, A.; Howad, W.; et al. Phenotypic diversity and distinctiveness of the Belltall garlic landrace. Front. Plant Sci. 2023, 13, 1004069. [Google Scholar] [CrossRef] [PubMed]
- Bian, H.; Zhou, Q.; Du, Z.; Zhang, G.; Han, R.; Chen, L.; Tian, J.; Li, Y. Integrated transcriptomics and metabolomics analysis of the fructan metabolism response to low-temperature stress in garlic. Genes 2023, 14, 1290. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.-L.; Ren, X.-Q.; Liu, J.-X.; Yang, F.; Wang, Y.-P.; Xiong, A.-S. Transcript profiling reveals an important role of cell wall remodeling and hormone signaling under salt stress in garlic. Plant Physiol. Biochem. 2019, 135, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Kong, Q.; Mostafa, H.H.A.; Yang, W.; Wang, J.; Nuerawuti, M.; Wang, Y.; Song, J.; Zhang, X.; Ma, L.; Wang, H.; et al. Comparative transcriptome profiling reveals that brassinosteroid-mediated lignification plays an important role in garlic adaption to salt stress. Plant Physiol. Biochem. 2021, 158, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Sun, H.; Zhang, G.; Wang, J.; Tian, J. Gene co-expression analysis reveals the transcriptome changes and hub genes of fructan metabolism in garlic under drought stress. Plants 2023, 12, 3357. [Google Scholar] [CrossRef]
- Valluru, R.; Ende, W. Plant fructans in stress environments: Emerging concepts and future prospects. J. Exp. Bot. 2008, 59, 2905–2916. [Google Scholar] [CrossRef]
- Van den Ende, W. Multifunctional fructans and raffinose family oligosaccharides. Front. Plant Sci. 2013, 4, 247. [Google Scholar] [PubMed]
- Van den Ende, W. Different evolutionary pathways to generate plant fructan exohydrolases. J. Exp. Bot. 2022, 73, 4620–4623. [Google Scholar] [CrossRef] [PubMed]
- Le Roy, K.; Lammens, W.; Verhaest, M.; De Coninck, B.; Rabijns, A.; Van Laere, A.; Van den Ende, W. Unraveling the difference between invertases and fructan exohydrolases: A single amino acid (Asp-239) substitution transforms Arabidopsis cell wall invertase1 into a fructan 1-exohydrolase. Plant Physiol. 2007, 145, 616–625. [Google Scholar] [CrossRef]
- Sun, X.; Zhu, S.; Li, N.; Cheng, Y.; Zhao, J.; Qiao, X.; Lu, L.; Liu, S.; Wang, Y.; Liu, C.; et al. A Chromosome-level genome assembly of garlic (Allium sativum) provides insights into genome evolution and allicin biosynthesis. Mol. Plant 2020, 13, 1328–1339. [Google Scholar] [CrossRef]
- Anisimova, O.K.; Shchennikova, A.V.; Kochieva, E.Z.; Filyushin, M.A. Pathogenesis-related genes of PR1, PR2, PR4, and PR5 families are involved in the response to Fusarium infection in garlic (Allium sativum L.). Int. J. Mol. Sci. 2021, 22, 6688. [Google Scholar] [CrossRef]
- Anisimova, O.K.; Kochieva, E.Z.; Shchennikova, A.V.; Filyushin, M.A. Thaumatin-like protein (TLP) genes in garlic (Allium sativum L.): Genome-wide identification, characterization, and expression in response to Fusarium proliferatum infection. Plants 2022, 11, 748. [Google Scholar] [CrossRef]
- Filyushin, M.A.; Anisimova, O.K.; Kochieva, E.Z.; Shchennikova, A.V. Genome-wide identification and expression of chitinase class i genes in garlic (Allium sativum L.) cultivars resistant and susceptible to Fusarium proliferatum. Plants 2021, 10, 720. [Google Scholar] [CrossRef] [PubMed]
- Filyushin, M.A.; Anisimova, O.K.; Shchennikova, A.V.; Kochieva, E.Z. Genome-wide identification, expression, and response to Fusarium infection of the SWEET gene family in garlic (Allium sativum L.). Int. J. Mol. Sci. 2023, 24, 7533. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An Upgraded Gene Feature Visualization Server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Babicki, S.; Arndt, D.; Marcu, A.; Liang, Y.; Grant, J.R.; Maciejewski, A.; Wishart, D.S. Heatmapper: Web-enabled heat mapping for all. Nucl. Acids Res. 2016, 44, W147–W153. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Wang, Q.; Li, Y.; Gao, L.; Lv, F.; Yang, R.; Wang, P. Candidate reference genes for quantitative gene expression analysis in Lagerstroemia indica. Mol. Biol. Rep. 2021, 48, 1677–1685. [Google Scholar] [CrossRef]
- Sturm, A. Invertases. Primary structures, functions, and roles in plant development and sucrose partitioning. Plant Physiol. 1999, 121, 1–8. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- Waadt, R.; Seller, C.A.; Hsu, P.K.; Takahashi, Y.; Munemasa, S.; Schroeder, J.I. Plant hormone regulation of abiotic stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 680–694. [Google Scholar] [CrossRef] [PubMed]
- Baillo, E.H.; Kimotho, R.N.; Zhang, Z.; Xu, P. Transcription factors associated with abiotic and biotic stress tolerance and their potential for crops improvement. Genes 2019, 10, 771. [Google Scholar] [CrossRef] [PubMed]
- Ouwerkerk, P.B.; Trimborn, T.O.; Hilliou, F.; Memelink, J. Nuclear factors GT-1 and 3AF1 interact with multiple sequences within the promoter of the Tdc gene from Madagascar periwinkle: GT-1 is involved in UV light-induced expression. Mol. Gen. Genet. 1999, 261, 610–622. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wang, G.; Wen, X.; Qu, X.; Zhang, Y.; Zhang, X.; Deng, P.; Chen, C.; Ji, W.; Zhang, H. Characteristics and expression analysis of invertase gene family in common wheat (Triticum aestivum L.). Genes 2022, 14, 41. [Google Scholar] [CrossRef]
- Saddhe, A.A.; Manuka, R.; Penna, S. Plant sugars: Homeostasis and transport under abiotic stress in plants. Physiol. Plant. 2021, 171, 739–755. [Google Scholar] [CrossRef]
- Peng, Y.; Zhu, L.; Tian, R.; Wang, L.; Su, J.; Yuan, Y.; Ma, F.; Li, M.; Ma, B. Genome-wide identification, characterization and evolutionary dynamic of invertase gene family in apple, and revealing its roles in cold tolerance. Int. J. Biol. Macromol. 2023, 229, 766–777. [Google Scholar] [CrossRef]



| Gene | Gene ID/Transcript ID [34] | Genomic Localization | Gene, bp | CDS, bp | Protein, aa | MW, kDa | pI | GRAVY |
|---|---|---|---|---|---|---|---|---|
| AsN/AINV1 | Asa1G00182.1/Asa7G01205.1 | ch1: 40208894-40214212 | 5319 | 1776 | 591 | 66.78 | 5.88 | −0.313 |
| AsN/AINV2 | Asa1G00196.1/Asa7G01204.1 | ch1: 45988527-45991812 | 3286 | 1203 | 400 | 45.55 | 5.53 | −0.303 |
| AsN/AINV3 | Asa1G02231.1/Asa2G02607.1 | ch1: 597271753-597277244 | 5492 | 1659 | 552 | 62.96 | 6.11 | −0.188 |
| AsN/AINV4 | Asa1G02550.1/Asa2G02247.1 | ch1: 692918625-692920987 | 2363 | 1671 | 556 | 62.74 | 6.32 | −0.147 |
| AsN/AINV5 | Asa1G03777.1/Asa2G00901.1 | ch1: 1030642218-1030647564 | 5347 | 1737 | 578 | 66.01 | 6.39 | −0.277 |
| AsN/AINV6 | Asa1G03784.1/Asa2G00903.1 | ch1: 1031491320-1031496681 | 5362 | 1737 | 578 | 66.01 | 6.39 | −0.277 |
| AsN/AINV7 | Asa3G00854.1/Asa8G04269.1 | ch3: 235042209-235045372 | 3164 | 1353 | 450 | 50.99 | 6.11 | −0.212 |
| AsN/AINV8 | Asa5G02977.1/Asa6G02773.1 | ch5: 760809575-760821162 | 11,588 | 1866 | 621 | 69.83 | 6.61 | −0.286 |
| AsN/AINV9 | Asa5G03124.1/Asa3G04501.1 | ch5: 803147316-803153602 | 6287 | 1683 | 560 | 63.46 | 6.51 | −0.189 |
| AsN/AINV10 | Asa7G05846.1/Asa5G05408.1 | ch7: 1623050841-1623054871 | 4031 | 1203 | 400 | 45.81 | 5.76 | −0.285 |
| AsN/AINV11 | Asa7G06162.1/Asa5G05675.1 | ch7: 1691491430-1691494767 | 3338 | 1782 | 593 | 67.39 | 8.73 | −0.378 |
| AsCWINV1 | Asa1G04355.1/Asa2G00250.1 | ch1: 1178706763-1178709021 | 2259 | 1608 | 535 | 60.55 | 5.86 | −0.321 |
| AsCWINV2 | Asa5G03190.1/Asa6G03410.1 | ch5: 820997521-821001354 | 3834 | 1707 | 568 | 64.43 | 7.72 | −0.407 |
| AsCWINV3 | Asa6G01167.1/Asa6G07032.1 | ch6: 308416872-308421914 | 5043 | 1641 | 546 | 62.42 | 5.72 | −0.315 |
| AsCWINV4 | Asa6G07063.1/Asa1G04317.1 | ch6: 1957444508-1957446535 | 2028 | 1539 | 512 | 58.27 | 8.48 | −0.505 |
| AsCWINV5 | Asa8G04697.1/Asa7G04854.1 | ch8: 1251531812-1251537888 | 6077 | 1569 | 522 | 58.96 | 5.86 | −0.319 |
| AsCWINV6 | Asa8G04714.1/Asa7G04858.1 | ch8: 1255046760-1255051854 | 5095 | 1836 | 611 | 69.34 | 6.35 | −0.391 |
| AsVINV1 | Asa1G00540.1/Asa0G04097.1 | ch1: 133485285-133487133 | 1849 | 1755 | 584 | 64.87 | 4.9 | −0.279 |
| AsVINV2 | Asa2G02355.1/Asa3G02390.1 | ch2: 650863353-650866104 | 2752 | 2217 | 738 | 81.18 | 5.16 | −0.253 |
| AsVINV3 | Asa3G03921.1/Asa5G01574.1 | ch3: 1070971636-1070975615 | 3980 | 1920 | 639 | 70.28 | 4.94 | −0.135 |
| AsVINV4 | Asa5G00414.1/Asa6G00588.1 | ch5: 101522664-101526645 | 3982 | 1884 | 627 | 69.04 | 4.93 | −0.137 |
| AsVINV5 | Asa6G06910.1/Asa1G04114.1 | ch6: 1914547560-1914549603 | 2044 | 1953 | 650 | 71.53 | 5.00 | −0.221 |
| AsVINV6 | Asa6G06912.1/Asa1G04116.1 | ch6: 1915034543-1915036592 | 2050 | 1956 | 651 | 72.4 | 4.98 | −0.197 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anisimova, O.K.; Shchennikova, A.V.; Kochieva, E.Z.; Filyushin, M.A. Garlic (Allium sativum L.) Invertase Genes: Genome-Wide Identification and Expression in Response to Abiotic Stresses and Phytohormones. Horticulturae 2024, 10, 581. https://doi.org/10.3390/horticulturae10060581
Anisimova OK, Shchennikova AV, Kochieva EZ, Filyushin MA. Garlic (Allium sativum L.) Invertase Genes: Genome-Wide Identification and Expression in Response to Abiotic Stresses and Phytohormones. Horticulturae. 2024; 10(6):581. https://doi.org/10.3390/horticulturae10060581
Chicago/Turabian StyleAnisimova, Olga K., Anna V. Shchennikova, Elena Z. Kochieva, and Mikhail A. Filyushin. 2024. "Garlic (Allium sativum L.) Invertase Genes: Genome-Wide Identification and Expression in Response to Abiotic Stresses and Phytohormones" Horticulturae 10, no. 6: 581. https://doi.org/10.3390/horticulturae10060581
APA StyleAnisimova, O. K., Shchennikova, A. V., Kochieva, E. Z., & Filyushin, M. A. (2024). Garlic (Allium sativum L.) Invertase Genes: Genome-Wide Identification and Expression in Response to Abiotic Stresses and Phytohormones. Horticulturae, 10(6), 581. https://doi.org/10.3390/horticulturae10060581






