Functional Analysis of RMA3 in Response to Xanthomonas citri subsp. citri Infection in Citron C-05 (Citrus medica)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Inoculation of Citrus Leaves with Xcc
2.2. Construction of cDNA Library
2.3. Construction of Lob1 Promoter Bait and Detection of Autonomous Activation
2.4. Screening Xcc-Induced Citrus cDNA Library with PAbAi-Lob1-P1
2.5. Verification of Candidate Interacting Proteins by Yeast One-Hybrid System
2.6. Validation of Interaction with Dual-Luciferase Enzyme Assay
2.7. Expression Analysis of Candidate Transcription Factors
2.8. RMA3 Cloning and Sequence Comparison in Different Citrus Genotypes
2.9. Subcellular Localization of Citrus RMA3
2.10. Transient Overexpression of Candidate Genes
2.11. RMA3 Gene Silencing in Citron C-05 through VIGS
2.12. Data Analysis
3. Results
3.1. Screening Xcc-Induced Citrus cDNA Library
3.1.1. Quality Determination of Xcc-Induced cDNA Library of Yeast Hybridization
3.1.2. Screening Xcc-Induced cDNA Library with PAbAi-Lob1-P1
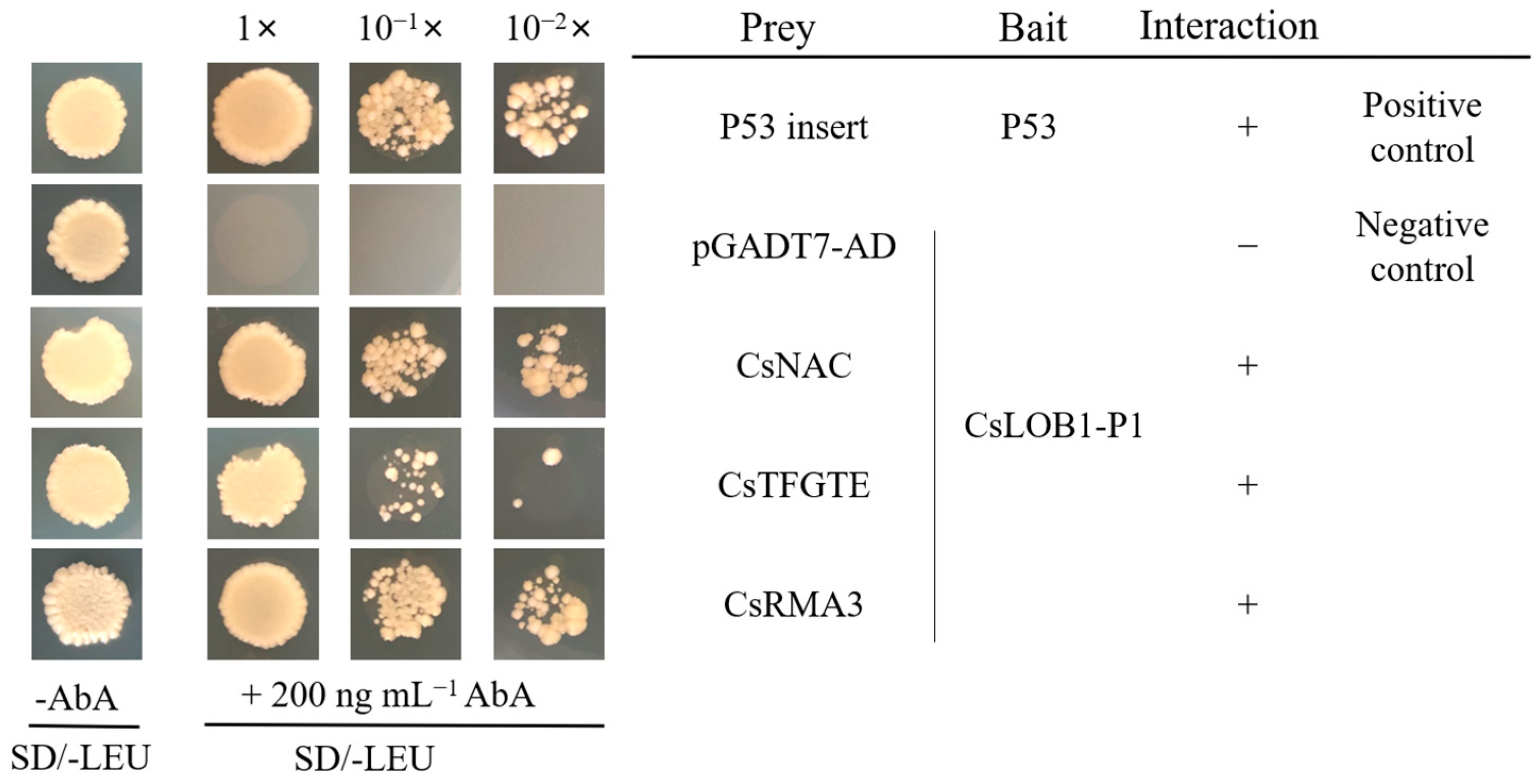
3.2. Verification of Candidate Interacting Proteins by Yeast One-Hybrid System
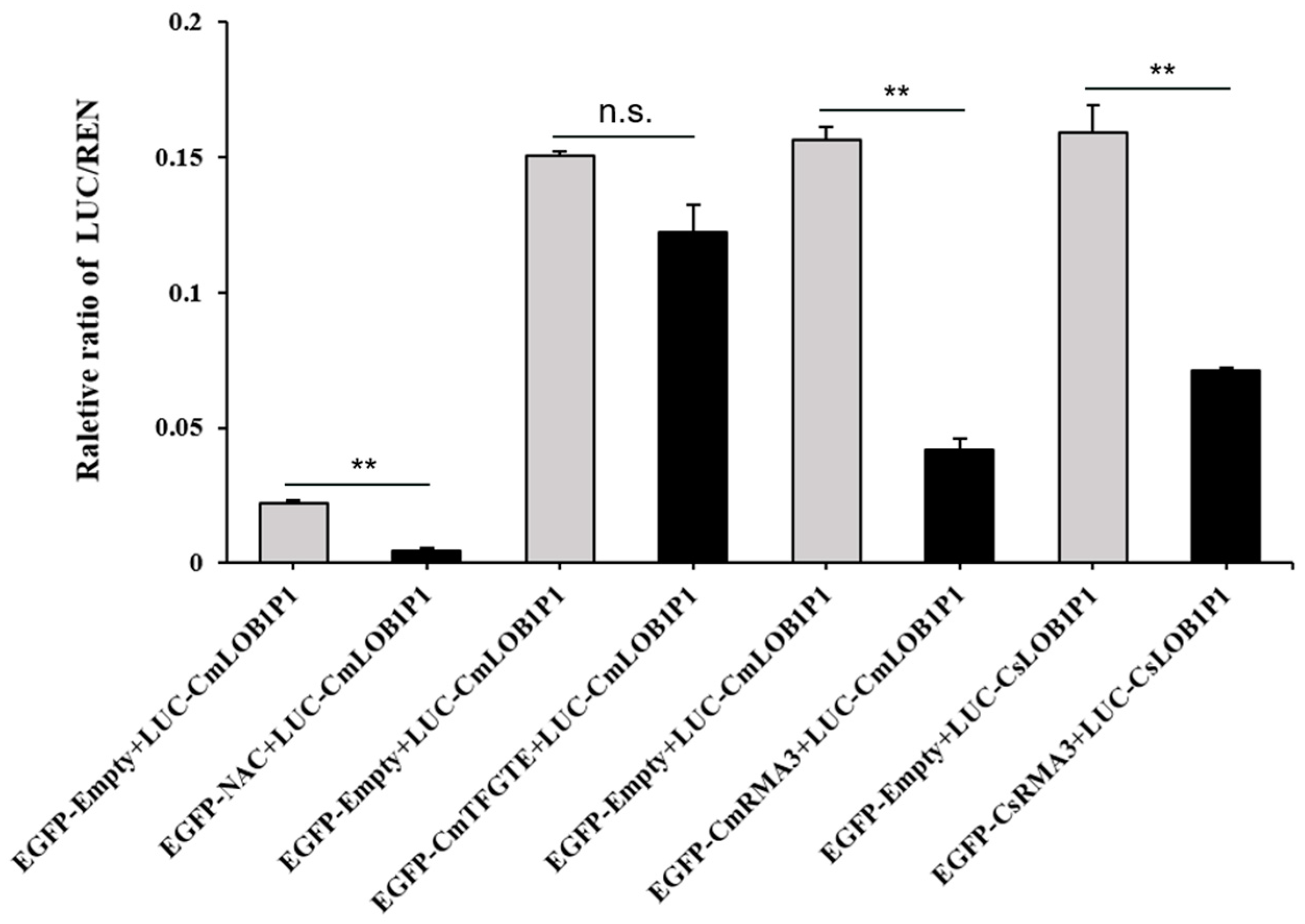
3.3. Interaction Validation with Dual-Luciferase Enzyme Assay
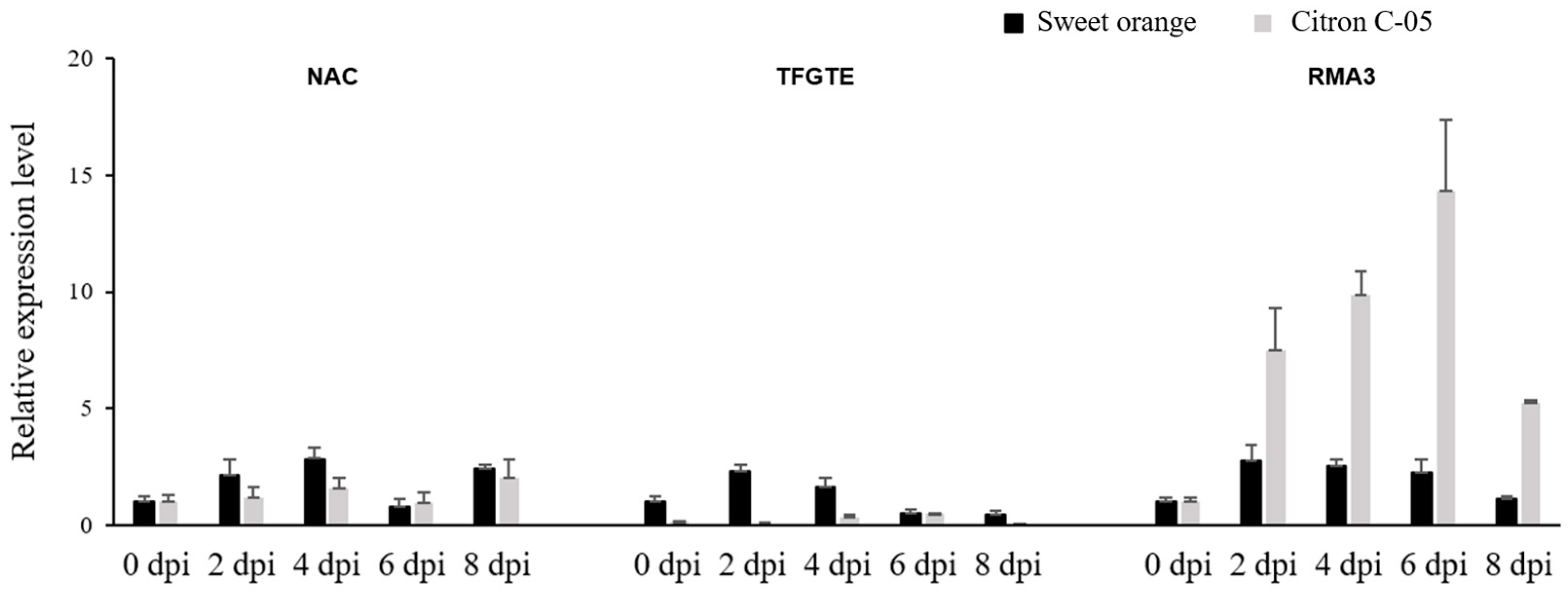
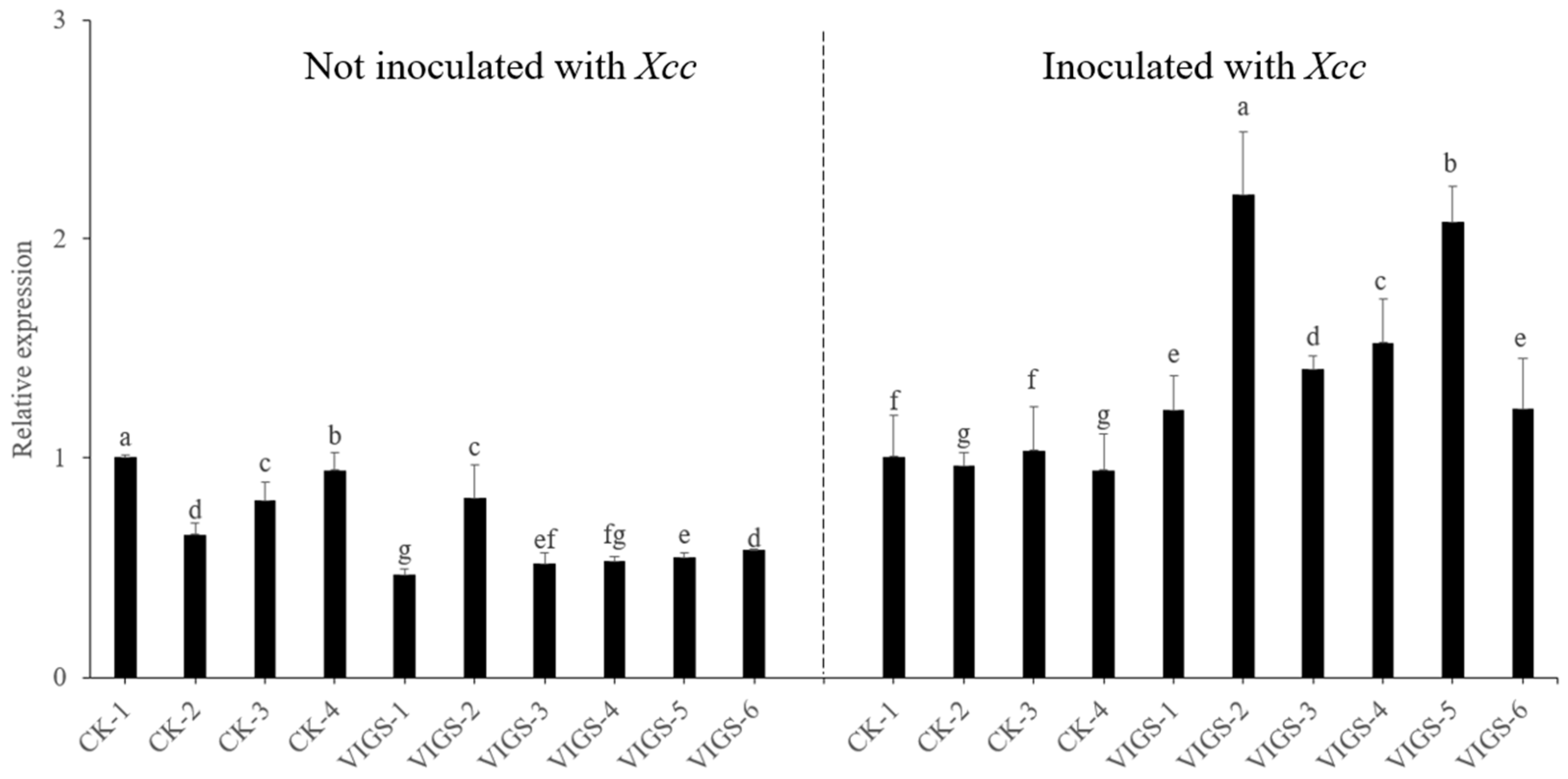
3.4. Expression Analysis of Candidate Genes following Infection with Xcc
3.5. Comparison of RMA3 Sequences among Different Citrus Genotypes
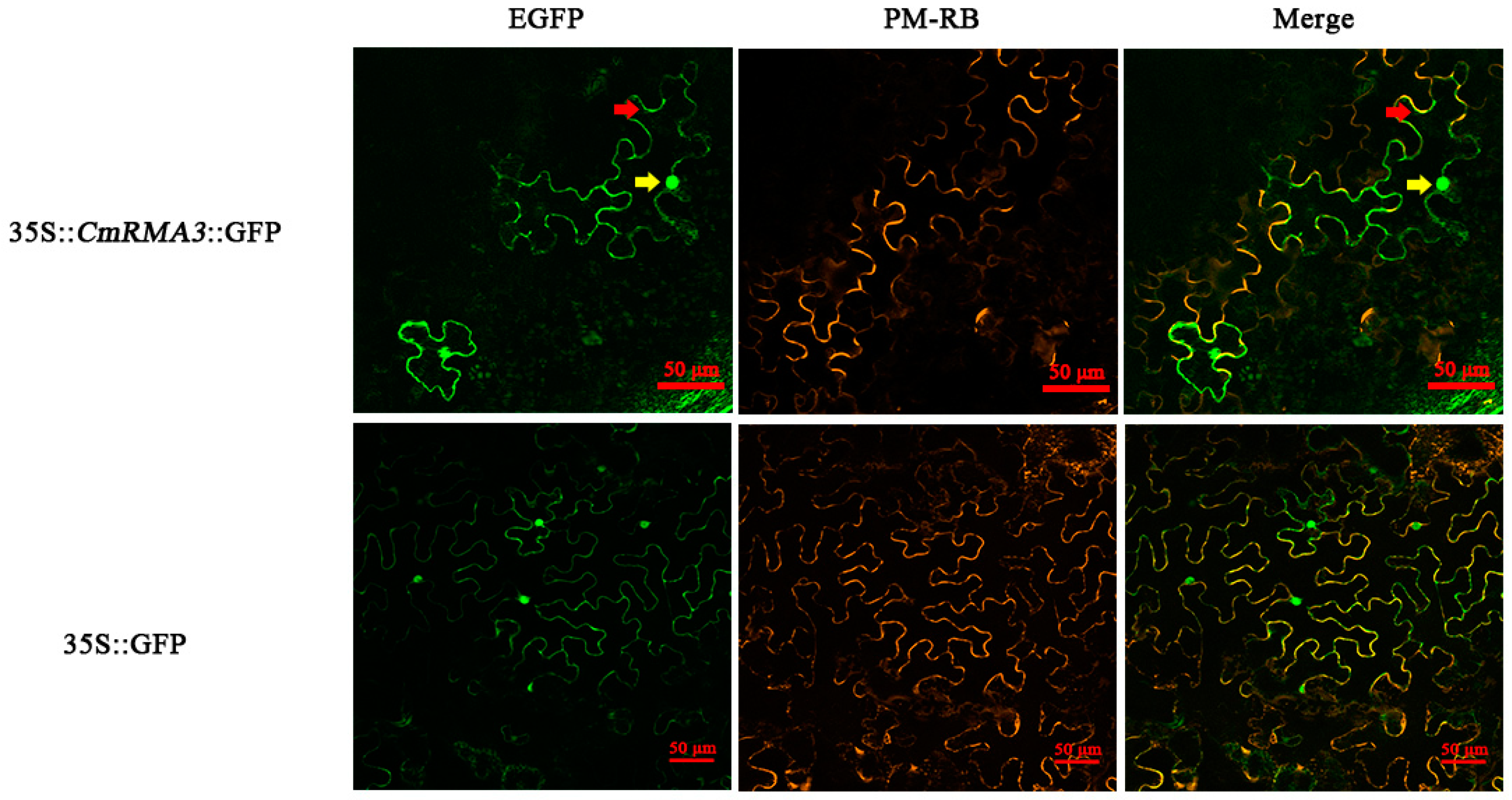
3.6. Subcellular Localization of RMA3
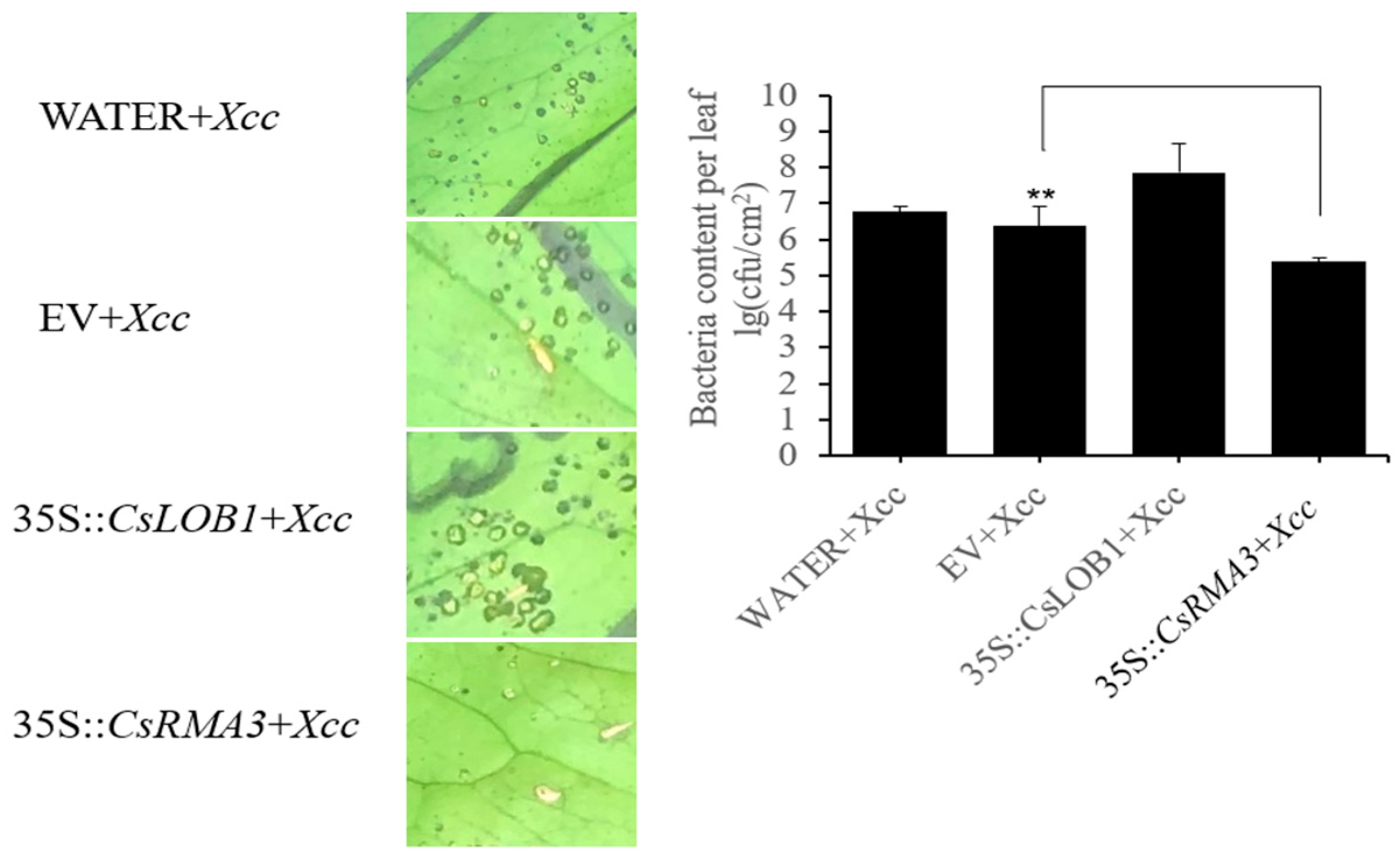
3.7. Transient Overexpression of RMA3 in ‘Bingtang’ Sweet Orange
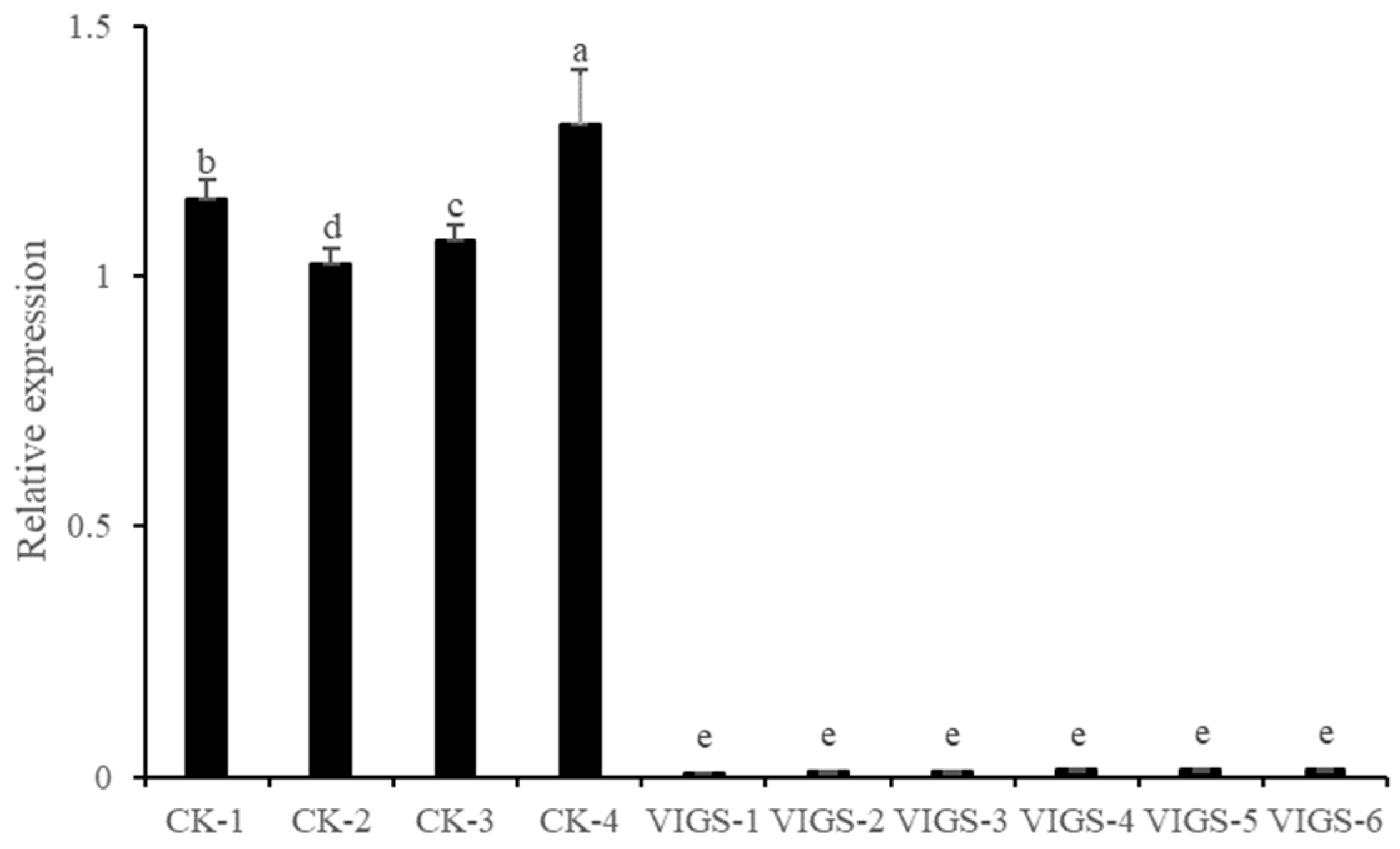
3.8. Silencing RMA3 Gene in Citron C-05 Using VIGS
3.9. Canker Symptom Observation of RMA3-Silenced Plants following Inoculation with Xcc
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Appendix A
| Primer Name | Sequence (5′-3′) |
|---|---|
| pAbAi-CsLOB1-P-F | GCTTGAATTCGAGCTGACCTCTGCCTACCCTATTGC |
| pAbAi-CmLOB1-P-F | GCTTGAATTCGAGCTGACCTCTGCCTACCCTATTAC |
| pAbAi-LOB1-P-R | TACAGAGCACATGCCCAAAGACAGTAAGGGATGAGG |
| pAbAi-CsLOB1-P1-F | GAAAAGCTTGAATTCGGACCTCTGCCTACCCTATTGCC |
| pAbAi-CsLOB1-P1-R | ATACAGAGCACATGCCGAACTTCGAGGTTCATTGCATTG |
| pAbAi-CsLOB1-P2-F | GAAAAGCTTGAATTCGCCAGGTTATAAGTAAACAGAACGAC |
| pAbAi-CsLOB1-P2-R | ATACAGAGCACATGCCCATCAAAGACAGTAAGGGATGAGG |
| pAbAi-CmLOB1-P1-F | GAAAAGCTTGAATTCGGACCTCTGCCTACCCTATTACC |
| pAbAi-CmLOB1-P1-R | ATACAGAGCACATGCCGAACTTCGAGGTTCATTGGATTG |
| pAbAi-CmLOB1-P2-F | GAAAAGCTTGAATTCGCCAGGTTATAAGTAAACAGCACGAC |
| pAbAi-CmLOB1-P2-R | ATACAGAGCACATGCCCAAAGACAGTAAGGGATGAGG |
| pGADT7-RMA3-F | GTACCAGATTACGCTCAATGGAACAGAACCTCTTTGAGCC |
| pGADT7-RMA3-R | ATGCCCACCCGGGTGGTCAGAACAAAAGAAGACACAGGACTAGG |
| 35S RMA3-F | CACGGGGGACGAGCTCATGGAACAGAACCTCTTTGAGCC |
| 35S RMA3-R | CCTTGCTCACCATGGTACCGAACAAAAGAAGACACAGGACTAGG |
| qLOB1-F | CTGCCAGAATCTCAACGAGC |
| qLOB1-R | TTGGCTAACTGAGCCTGAAGC |
| q actin-F | CATCCCTCAGCACCTTCC |
| q actin-R | CCAACCTTAGCACTTCTCC |
| qRMA3-F | TCAATCACAGGCACCAGCAT |
| qRMA3-R | TAGAAGACGCCAAAGCTGCA |
| qNAC-F | GATGTGGCGCAAGAGAGGTA |
| qNAC-R | TGGCCCTGTTCGATCTGTTC |
| qTFGTE-F | TTGGTGCAGCACTTACCAGA |
| qTFGTE-R | AGCCTGTGTTACTCTGAGCA |
| VIGS RMA3-F | TGAGTAAGGTTACCGAATTCGAACAGAACCTCTTTGAGCCTGAGA |
| VIGS RMA3-R | GTGAGCTCGGTACCGGATCCTTGGGCTTCTTGGAGTCGGA |
References
- Wu, G.A.; Terol, J.; Ibanez, V.; López-García, A.; Pérez-Román, E.; Borredá, C.; Domingo, C.; Tadeo, F.R.; Carbonell-Caballero, J.; Alonso, R.; et al. Genomics of the origin and evolution of Citrus. Nature 2018, 554, 311–316. [Google Scholar] [CrossRef] [PubMed]
- Rai, R.; Pasion, J.; Majumdar, T.; Green, C.E.; Hind, S.R. Genome sequencing and functional characterization of Xanthomonas cucurbitae, the causal agent of bacterial spot disease of cucurbits. Phytopathology 2021, 111, 1289–1300. [Google Scholar] [CrossRef] [PubMed]
- Ference, C.M.; Gochez, A.M.; Behlau, F.; Wang, N.; Graham, J.H.; Jones, J.B. Recent advances in the understanding of Xanthomonas citri ssp. citri pathogenesis and citrus canker disease management. Mol. Plant Pathol. 2018, 19, 1302–1318. [Google Scholar] [PubMed]
- An, S.Q.; Potnis, N.; Dow, M.; Vorhölter, F.J.; He, Y.Q.; Becker, A.; Teper, D.; Li, Y.; Wang, N.; Bleris, L.; et al. Mechanistic insights into host adaptation, virulence and epidemiology of the phytopathogen Xanthomonas. FEMS Microbiol. Rev. 2020, 44, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Graham, J.H.; Gottwald, T.R.; Cubero, J.; Achor, D.S. Xanthomonas axonopodis pv. citri: Factors affecting successful eradication of citrus canker. Mol. Plant Pathol. 2004, 5, 1–15. [Google Scholar] [PubMed]
- Favaro, M.A.; Molina, M.C.; Roeschlin, R.A.; Gadea, J.; Gariglio, N.; Marano, M.R. Different responses in mandarin cultivars uncover a role of cuticular waxes in the resistance to citrus canker. Phytopathology 2020, 110, 1791–1801. [Google Scholar] [CrossRef] [PubMed]
- Ference, C.M.; Manthey, J.A.; Narciso, J.A.; Jones, J.B.; Baldwin, E.A. Detection of phenylpropanoids in citrus leaves produced in response to Xanthomonas citri subsp. citri. Phytopathology 2020, 110, 287–296. [Google Scholar] [PubMed]
- Lanza, F.E.; Marti, W.; Silva-Junior, G.J.; Behlau, F. Characteristics of citrus canker lesions associated with premature drop of sweet orange fruit. Phytopathology 2019, 109, 44–51. [Google Scholar] [CrossRef]
- Gochez, A.M.; Huguet-Tapia, J.C.; Minsavage, G.V.; Shantaraj, D.; Jalan, N.; Strauß, A.; Lahaye, T.; Wang, N.; Canteros, B.I.; Jones, J.B.; et al. Pacbio sequencing of copper-tolerant Xanthomonas citri reveals presence of a chimeric plasmid structure and provides insights into reassortment and shuffling of transcription activator-like effectors among X. citri strains. BMC Genom. 2018, 19, 16–29. [Google Scholar] [CrossRef]
- Li, J.; Pang, Z.; Duan, S.; Lee, D.; Kolbasov, V.; Wang, N. The in planta effective concentration of oxytetracycline against Candidatus Liberibacter asiaticus for suppression of citrus Huanglongbing. Phytopathology 2019, 109, 2046–2054. [Google Scholar] [CrossRef]
- Li, J.; Kolbasov, V.; Lee, D.; Pang, Z.; Huang, Y.; Collins, N.; Wang, N. Residue dynamics of streptomycin in citrus delivered by foliar spray and trunk injection and effect on ‘Candidatus Liberibacter asiaticus’ titer. Phytopathology 2020, 111, 1095–1103. [Google Scholar] [CrossRef]
- Burlakoti, R.; Hsu, C.F.; Chen, J.R.; Wang, J.F. Population dynamics of Xanthomonads associated with bacterial spot of tomato and pepper during twenty-seven years across Taiwan. Plant Dis. 2018, 102, 1348–1356. [Google Scholar] [CrossRef]
- Strayer-Scherer, A.; Liao, Y.Y.; Young, M.; Ritchie, L.; Vallad, G.E.; Santra, S.; Freeman, J.H.; Clark, D.; Jones, J.B.; Paret, M.L. Advanced copper composites against copper-tolerant Xanthomonas perfo-rans and tomato bacterial spot. Phytopathology 2018, 108, 196–205. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Zhang, J.; Jia, H.; Sosso, D.; Li, T.; Frommer, W.B.; Yang, B. Lateral organ boundaries 1 is a disease susceptibility gene for citrus bacterial canker disease. Proc. Natl. Acad. Sci. USA 2014, 111, E521–E529. [Google Scholar] [CrossRef]
- Duan, S.; Jia, H.; Pang, Z. Functional characterization of the citrus canker susceptibility gene CsLOB1. Mol. Plant Pathol. 2018, 19, 1908–1916. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Orbovic, V.; Jones, J.B.; Wang, N. Modification of the PthA4 effector binding elements in Type I CsLOB1 promoter usingCas9/sgRNA to produce transgenic Duncan grapefruit alleviating XccDpthA4:dCsLOB1.3 infection. Plant Biotechnol. J. 2015, 14, 1291–1301. [Google Scholar] [CrossRef]
- Deng, Z.N.; Xu, L.; Li, D.Z.; Long, G.Y.; Liu, L.P.; Fang, F.; Shu, G.P. Screening citrus genotypes for resistance to canker disease (Xanthomonas axonopodis pv. citri). Plant Breed. 2010, 129, 341–345. [Google Scholar]
- Fu, H.Y.; Zhao, M.M.; Xu, J.; Tan, L.M.; Han, J.; Li, D.Z.; Wang, M.J.; Xiao, S.Y.; Ma, X.F.; Deng, Z.N. Citron C-05 inhibits both the penetration and colonization of Xanthomonas citri subsp. citri to achieve resistance to citrus canker disease. Hortic. Res. 2020, 7, 58–69. [Google Scholar] [PubMed]
- Wu, Q.; Zhao, M.M.; Li, Y.; Li, D.Z.; Ma, X.F.; Deng, Z.N. Identification of the transcription factors RAP2-13 activating the expression of CsBAK1 in citrus defence response to Xanthomonas citri subsp. Citri. Horticult. 2022, 8, 1012–1026. [Google Scholar]
- Sun, Y.; Li, Y.; Huang, G.; Wu, Q.; Wang, L. Application of the yeast one-hybridtechnique to plant functional genomics studies. Biotechnol. Biotechnol. Equip. 2017, 31, 1087–1092. [Google Scholar] [CrossRef]
- Jiang, W.; Liu, T.; Nan, W.; Jeewani, D.C.; Niu, Y.; Li, C.; Wang, Y.; Shi, X.; Wang, C.; Wang, J.; et al. Two transcription factors TaPpm1 and TaPpb1co-regulate anthocyanin biosynthesis in purple pericarps of wheat. J. Exp. Bot. 2018, 69, 2555–2567. [Google Scholar] [CrossRef] [PubMed]
- Lopato, S.; Bazanova, N.; Morran, S.; Milligan, A.S.; Shirley, N.; Langridge, P. Isolation of plant transcription factors using a modified yeast one-hybridsystem. Plant Methods 2006, 2, 45–66. [Google Scholar] [CrossRef] [PubMed]
- Mitsuda, N.; Ikeda, M.; Takada, S.; Takiguchi, Y.; Kondou, Y.; Yoshizumi, T.; Fujita, M.; Shinozaki, K.; Matsui, M.; Ohme-Takagi, M. Efficient yeast one−/two-hybrid screening using a library composed only of transcription factors in Arabidopsis thaliana. Plant Cell Physiol. 2010, 51, 2145–2151. [Google Scholar] [CrossRef] [PubMed]
- Zhan, Y.; Sun, X.; Rong, G.; Hou, C.; Huang, Y.; Jiang, D.; Weng, X. Identification of two transcription factors activating the expression of OsXIP in rice defense response. BMC Biotechnol. 2017, 17, 26–36. [Google Scholar] [CrossRef] [PubMed]
- Petzold, H.E.; Rigoulot, S.B.; Zhao, C.; Chanda, B.; Sheng, X.; Zhao, M.; Jia, X.; Dickerman, A.W.; Beers, E.P.; Brunner, A.M. Identification of new protein- proteinand protein- DNA interactions linked with wood formation in Populus trichocarpa. Tree Physiol. 2017, 38, 362–377. [Google Scholar] [CrossRef] [PubMed]
- Dharmaraj, K.; Cui, W.; EHA, R.; Templeton, M.D. Construction of a kiwifruit yeast two-hybrid cDNA library to identify host targets of the Pseudomonas syringae pv. actinidiae effector AvrPto5. BMC Res. Notes 2019, 12, 63–68. [Google Scholar] [PubMed]
- Hou, W.Q.; Yan, P.; Shi, T.Y.; Lu, P.Z.; Zhao, W.W.; Yang, H.M.; Zeng, L.Q.; Yang, J.; Li, Z.Y.; Fan, W.J.; et al. Modulation of anthocyanin accumulation in storage roots of sweet potato by transcription factor ibmyb1-2 through direct binding to anthocyanin biosynthetic gene promoters. Plant Physiol. Biochem. PPB 2023, 196, 868–879. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.D.; Hwang, J.G.; Han, A.R.; Park, Y.C.; Jang, C.S. Positive regulation of rice RING E3 ligase OsHIR1 in arsenic and cadmium uptakes. Plant Mol. Biol. 2014, 85, 365–379. [Google Scholar] [CrossRef]
- Stael, S.; Kmiecik, P.; Willems, P.; VD-Kelen, K.; Coll, N.S.; Teige, M.; Van Breusegem, F. Plant innate immunity-sunny side up. Trends Plant Sci. 2015, 20, 3–11. [Google Scholar] [CrossRef]
- Shu, K.; Liu, X.D.; Xie, Q.; He, Z.H. Two faces of one seed: Hormonal regulation of dormancy and germination. Mol. Plant 2016, 9, 34–45. [Google Scholar] [CrossRef]
- Zhang, X.; Garreton, V.; Chua, N.H. The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genes Dev. 2005, 19, 1532–1543. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.H.; Choi, H.W.; Hwang, B.K. The pepper E3 ubiquitin ligase ring1 gene, CaRING1, is required for cell death and the salicylic acid-dependent defense response. Plant Physiol. 2011, 156, 2011–2025. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.S.; Martin, R.; Mongrand, S.; Vandenabeele, S.; Chen, K.C.; Jang, I.C.; Chua, N.H. RING1 E3 ligase localizes to plasma membrane lipid rafts to trigger FB1-induced programmed cell death in Arabidopsis. Plant J. 2008, 56, 550–561. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Huang, Y.; Deng, X.X. The definition, classification and evolution of citrus based on whole genome sequence information. Sci. Sin. Vitae 2024, 54, 525–536. (In Chinese) [Google Scholar] [CrossRef]




| Bait Name | Library Genotype | Gene Code | Function |
|---|---|---|---|
| PAbAi-CsLob1-P1 | ‘Bingtang’ sweet orange | CsLP1-1 | Bispecific inhibitors; lipid transfer proteins; seed storage 2S albumins. |
| CsLP1-2 | Oxygen-evolving enhancer protein 2; chloroplast. | ||
| CsLP1-3 | Composition of ribosomal protein S9; cytoplasmic 80S ribosome. | ||
| CsLP1-4 | E3 ubiquitin protein ligase RING1; RING finger protein 126; E3 ubiquitin ligase. | ||
| CsLP1-5 | Zinc finger protein and putative TFIIIA-like zinc finger protein. | ||
| CsLP1-6 | Oxygen-evolving enhancer protein 3; chloroplast. | ||
| CsLP1-7 | Wound-induced protein 1; putative unidentified protein. | ||
| CsLP1-8 | Uncharacterized protein At4g01150 in chloroplast. | ||
| PAbAi-CmLob1-P1 | Citron C-05 | CmLP1-1 | Protein binding chlorophyll a and b. |
| CmLP1-2 | Alkaline chitinase A and lectin. | ||
| CmLP1-3 | Unidentified protein. | ||
| CmLP1-4 | Bifunctional dihydroflavonol 4-reductase plays crucial role in reduction of flavonoids. | ||
| CmLP1-5 | 60S ribosomal protein L10, a fragment of the same. | ||
| CmLP1-6 | Nuclear transcription factor Y subunit C-1. | ||
| CmLP1-7 | Transcription factor GTE1; chloroplast transcription factor GTE3. | ||
| CmLP1-8 | The first variation could be ‘E3 ubiquitin protein ligase RMA3, peroxisome generating factor 10, and helicase-like transcription factor are the proteins under study’. | ||
| CmLP1-9 | Receptor kinase 1 associated with brassinosteroid insensitivity 1. | ||
| CmLP1-10 | 2-Acetamido-2-deoxy-d-galactose-bound seed agglutinin 2, also known as lectin. | ||
| CmLP1-11 | NAC domain referred to as Protein 29. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, M.; Ye, R.; Li, Y.; Liu, L.; Su, H.; Ma, X.; Deng, Z. Functional Analysis of RMA3 in Response to Xanthomonas citri subsp. citri Infection in Citron C-05 (Citrus medica). Horticulturae 2024, 10, 693. https://doi.org/10.3390/horticulturae10070693
Zhao M, Ye R, Li Y, Liu L, Su H, Ma X, Deng Z. Functional Analysis of RMA3 in Response to Xanthomonas citri subsp. citri Infection in Citron C-05 (Citrus medica). Horticulturae. 2024; 10(7):693. https://doi.org/10.3390/horticulturae10070693
Chicago/Turabian StyleZhao, Mingming, Rongchun Ye, Yi Li, Lian Liu, Hanying Su, Xianfeng Ma, and Ziniu Deng. 2024. "Functional Analysis of RMA3 in Response to Xanthomonas citri subsp. citri Infection in Citron C-05 (Citrus medica)" Horticulturae 10, no. 7: 693. https://doi.org/10.3390/horticulturae10070693
APA StyleZhao, M., Ye, R., Li, Y., Liu, L., Su, H., Ma, X., & Deng, Z. (2024). Functional Analysis of RMA3 in Response to Xanthomonas citri subsp. citri Infection in Citron C-05 (Citrus medica). Horticulturae, 10(7), 693. https://doi.org/10.3390/horticulturae10070693





