Chloroplast Genome Profiling and Phylogenetic Insights of the “Qixiadaxiangshui” Pear (Pyrus bretschneideri Rehd.1)
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. DNA Extraction and Sequencing
2.3. Chloroplast Genome Assembly and Annotation
2.4. Chloroplast Genome Characterization
2.5. Comparative Analysis of Chloroplast Genomes
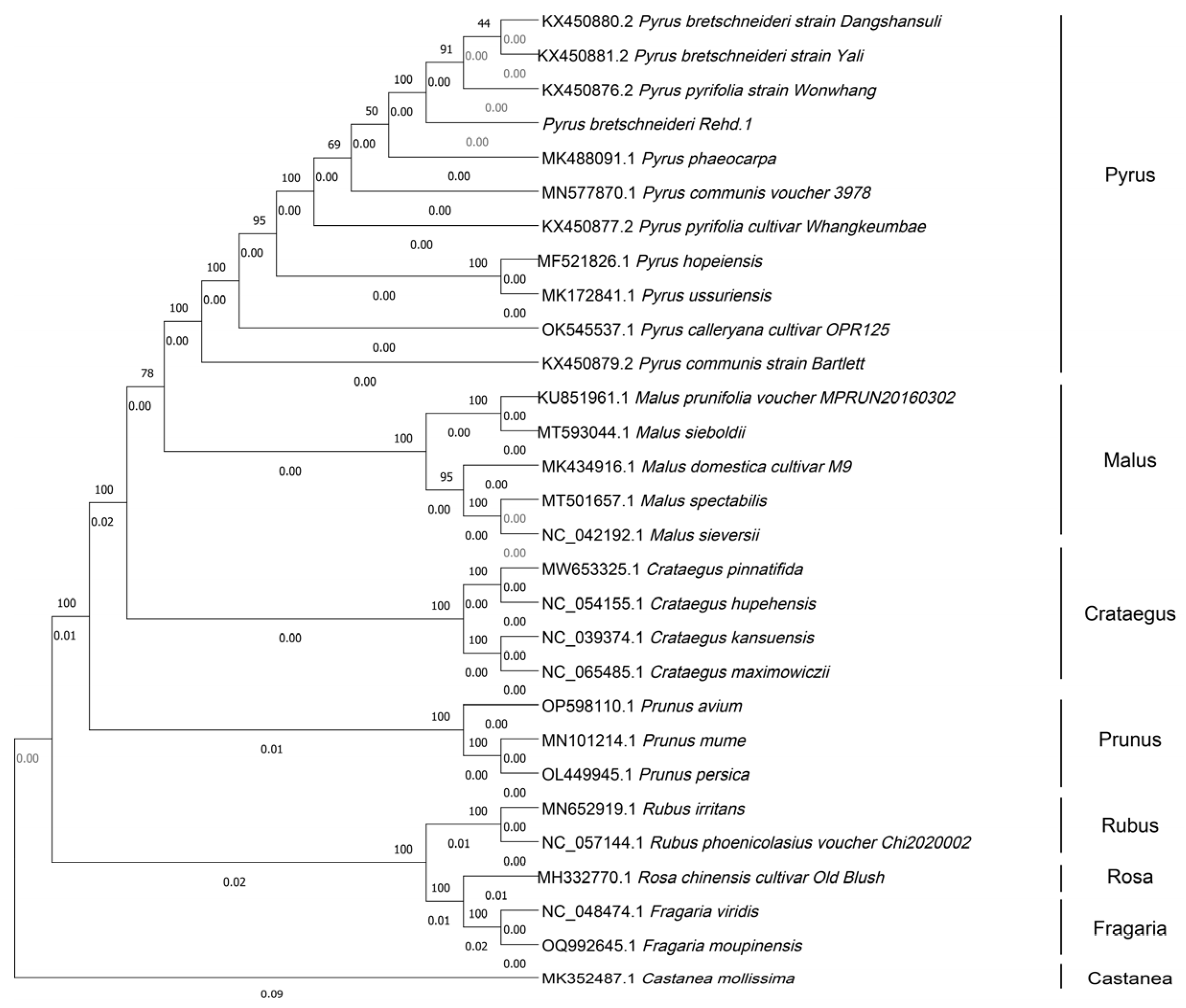
2.6. Phylogenetic Analysis
3. Results
3.1. Structure and Composition of the Chloroplast Genome of the “Qixiadaxiangshui” Pear
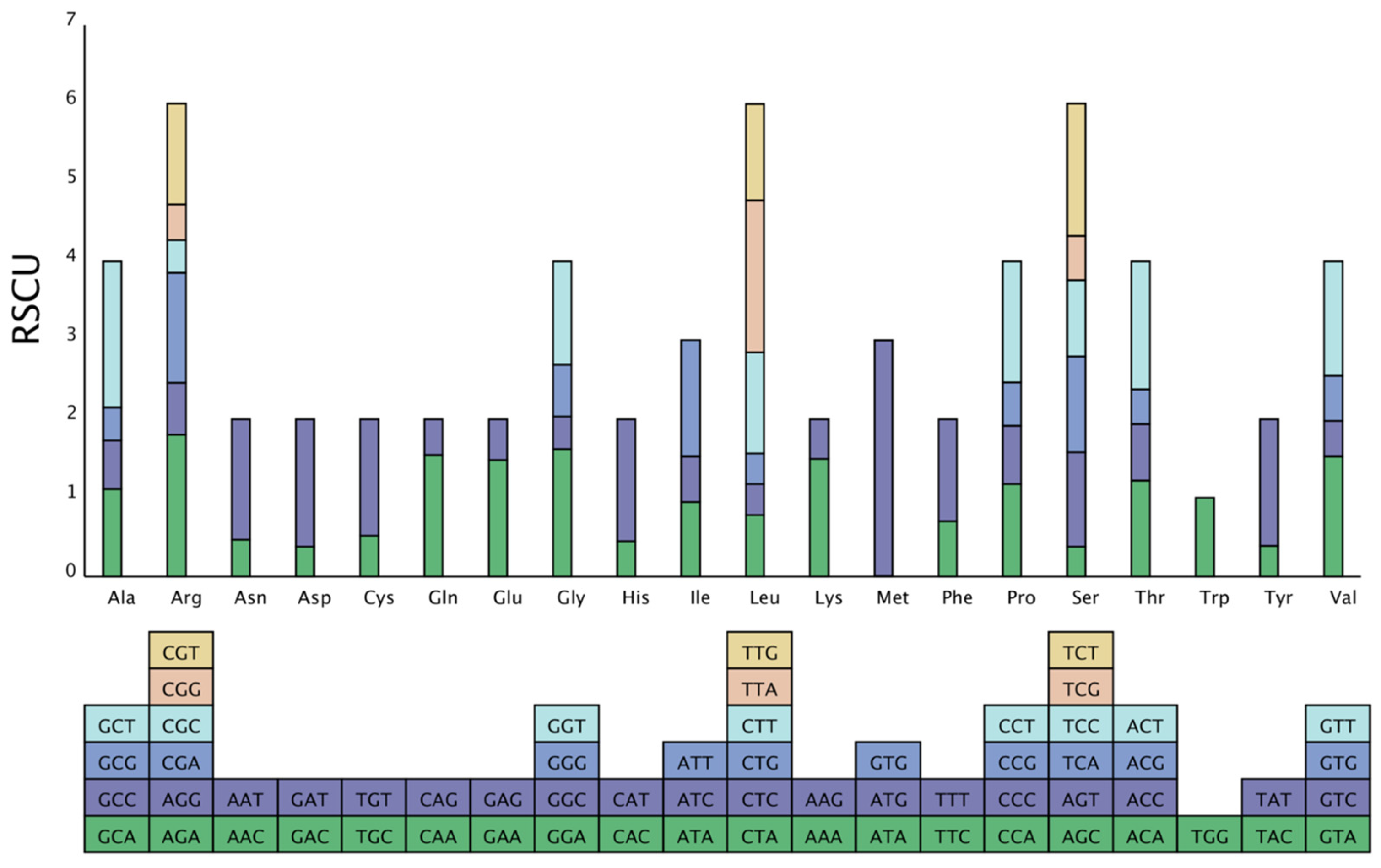
3.2. RSCU Analysis of the “Qixiadaxiangshui” Pear Genome
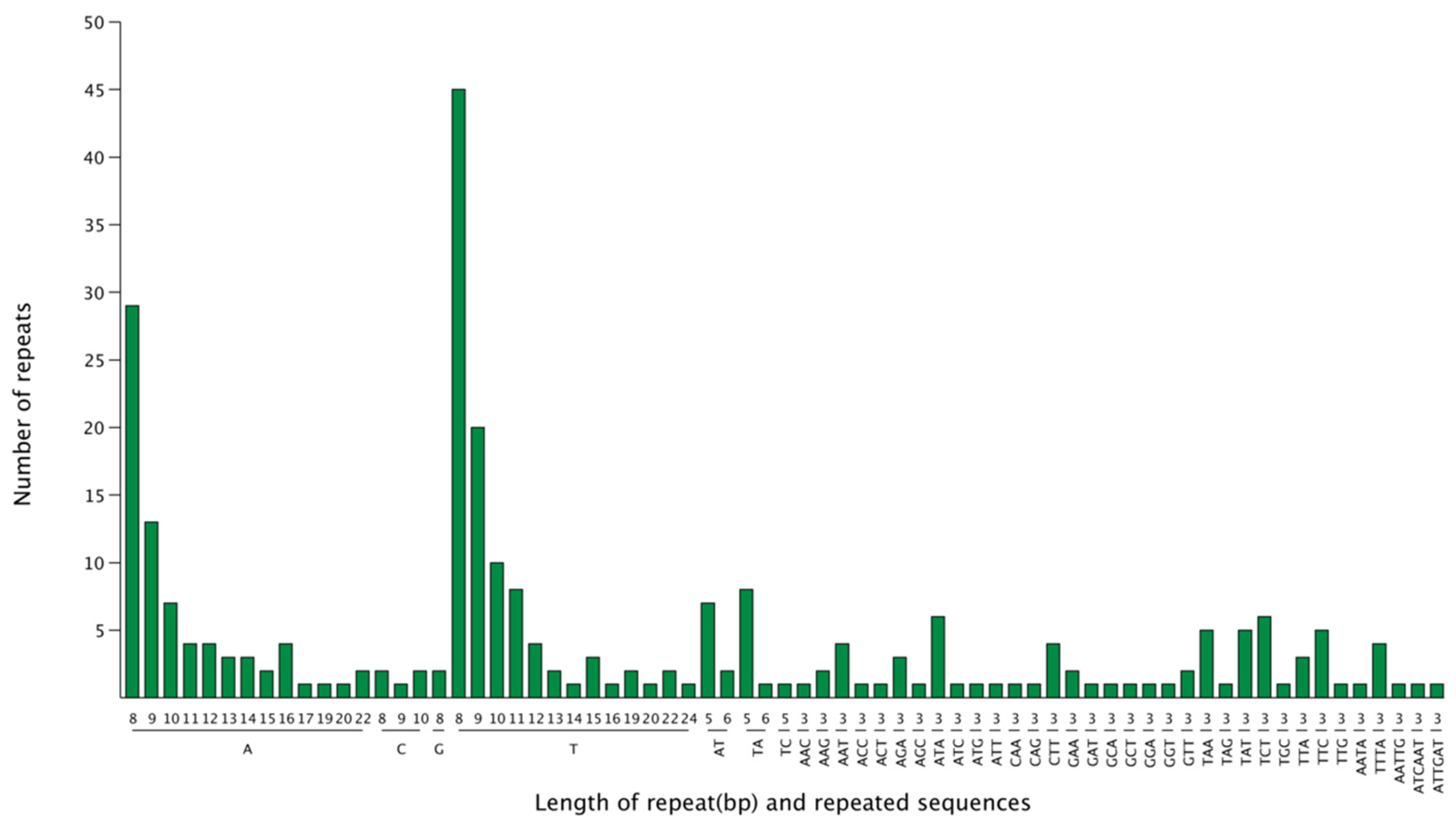
3.3. cpSSR Analysis of the “Qixiadaxiangshui” Pear Genome
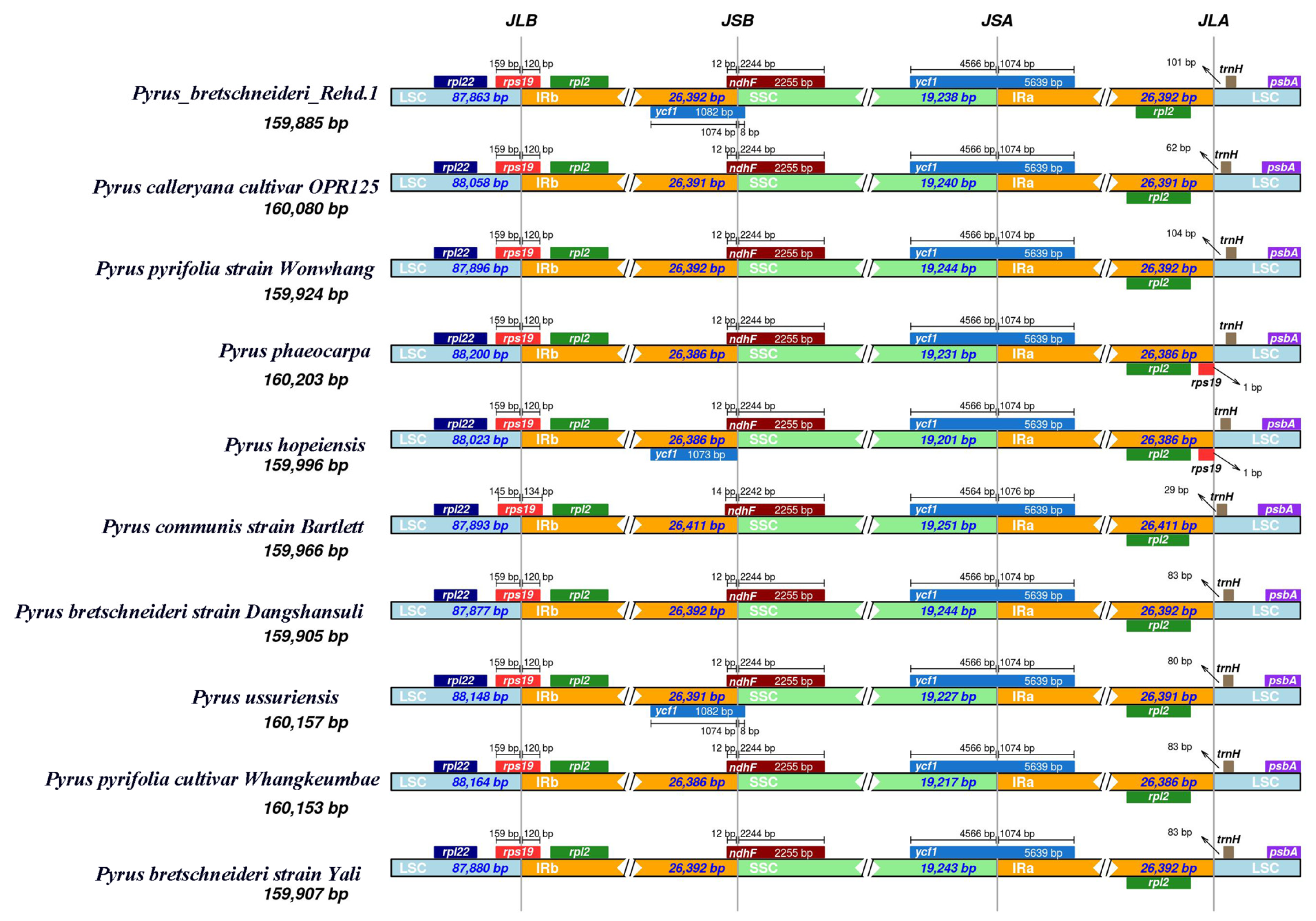
3.4. Analysis of IR Boundary Features
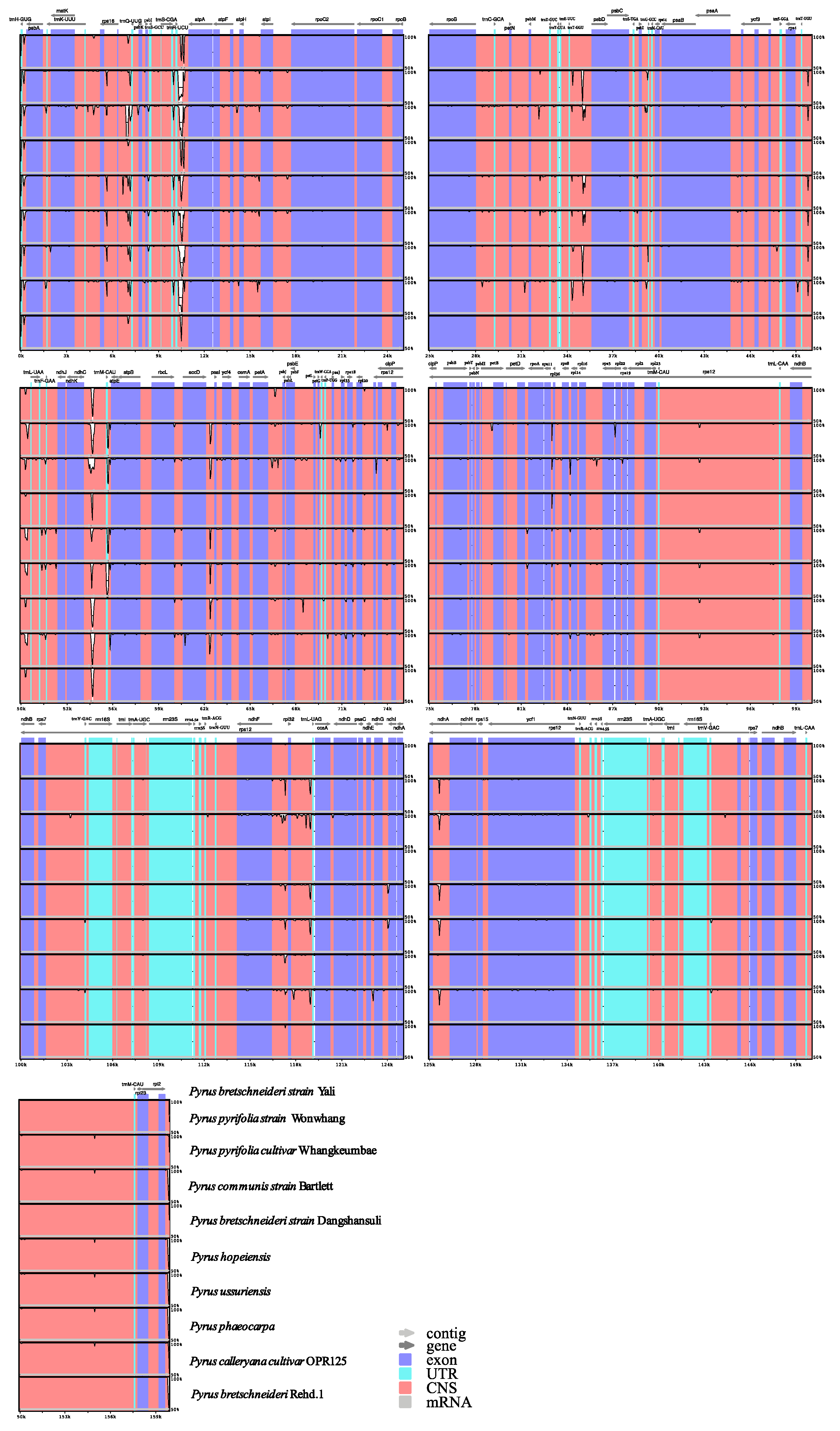
3.5. Comparative Analysis of Chloroplast Genomes among Pear Germplasm
3.6. Phylogenetic Analysis of Rosaceae Plants
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Van Dingenen, J.; Blomme, J.; Gonzalez, N.; Inzé, D. Plants grow with a little help from their organelle friends. J. Exp. Bot. 2016, 67, 6267–6281. [Google Scholar] [CrossRef] [PubMed]
- Hamsher, S.E.; Keepers, K.G.; Pogoda, C.S.; Stepanek, J.G.; Kane, N.C.; Kociolek, J.P. Extensive chloroplast genome rearrangement amongst three closely related Halamphora spp. (Bacillariophyceae), and evidence for rapid evolution as compared to land plants. PLoS ONE 2019, 14, e0217824. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Li, J.; Zhang, H.; Cai, B.; Gao, Z.; Qiao, Y.; Mi, L. The complete chloroplast genome sequence of strawberry (Fragaria × ananassa Duch.) and comparison with related species of Rosaceae. PeerJ 2017, 5, e3919. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.Y.; Lee, T.H.; Kim, Y.K.; Kim, J.S. Complete chloroplast genome sequences of Wonwhang (Pyrus pyrifolia) and its phylogenetic analysis. Mitochondrial DNA Part B 2017, 2, 325–326. [Google Scholar] [CrossRef] [PubMed]
- Gil, H.Y.; Kim, Y.; Kim, S.H.; Jeon, J.H.; Kwon, Y.; Kim, S.C.; Park, J. The complete chloroplast genome of Pyrus ussuriensis Maxim. (Rosaceae). Mitochondrial DNA Part B 2019, 4, 1000–1001. [Google Scholar] [CrossRef]
- Bao, L.; Li, K.; Teng, Y.; Zhang, D. Characterization of the complete chloroplast genome of the wild Himalayan pear Pyrus pashia (Rosales: Rosaceae: Maloideae). Conserv. Genet. Resour. 2017, 9, 569–571. [Google Scholar] [CrossRef]
- Xu, Y.; Liu, Y.; Yu, Z.; Jia, X. Complete chloroplast genome sequence of the long blooming cultivar Camellia ‘Xiari Qixin’: Genome features, comparative and phylogenetic analysis. Genes 2023, 14, 460. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Wang, S.M. Effects of Bagging on Aroma of ‘Qixiadaxiangshui’ Pear Fruit. Agric. Sci. Technol. 2015, 31, 1676. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Pevzner, P.A. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef] [PubMed]
- Beier, S.; Thiel, T.; Minch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef] [PubMed]
- Amiryousefi, A.; Hyvönen, J.; Poczai, P. IRscope: An online program to visualize the junction sites of chloroplast genomes. Bioinformatics 2018, 34, 3030–3031. [Google Scholar] [CrossRef] [PubMed]
- Frazer, K.A.; Pachter, L.; Poliakov, A.; Rubin, E.M.; Dubchak, I. VISTA: Computational tools for comparative genomics. Nucleic. Acids Res. 2004, 32, W273–W279. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. Raxml-vi-hpc: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Li, X.; Hao, Z.D.; Yang, L.M.; Zhang, J.B.; Peng, Y.; Xu, H.; Lu, Y.; Zhang, J.; Shi, J.S.; et al. Phylogenetic studies and comparative chloroplast genome analyses elucidate the basal position of halophyte Nitraria sibirica (Nitrariaceae) in the Sapindales. Mitochondrial DNA Part A 2018, 29, 745–755. [Google Scholar] [CrossRef] [PubMed]
- Raubeson, L.A.; Peery, R.; Chumley, T.W.; Dziubek, C.M.; Fourcade, H.M.; Boore, J.L.; Jansen, R.K. Comparative chloroplast genomics: Analyses including new sequences from the angiosperms Nuphar advena and Ranunculus macranthus. BMC Genom. 2007, 8, 174. [Google Scholar] [CrossRef] [PubMed]
- Palmer, J.D. Comparative organization of chloroplast genomes. Annu. Rev. Genet. 1985, 19, 325–354. [Google Scholar] [CrossRef]
- Dugas, D.V.; Hernandez, D.; Koenen, E.J.M.; Schwarz, E.; Straub, S.; Hughes, C.E.; Jansen, R.; Nageswara-Rao, M.; Staats, M.; Trujillo, J.T.; et al. Mimosoid legume plastome evolution: IR expansion, tandem repeat expansions, and accelerated rate of evolution in clpP. Sci. Rep. 2015, 5, 16958. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zhang, C.; Guo, X.; Liu, Q.; Wang, K. Complete chloroplast genome of Camellia japonica genome structures, comparative and phylogenetic analysis. PLoS ONE 2019, 14, e216645. [Google Scholar] [CrossRef] [PubMed]
- Zuo, L.; Shang, A.; Zhang, S.; Yu, X.; Ren, Y.; Yang, M.; Wang, J. The first complete chloroplast genome sequences of Ulmus species by de novo sequencing: Genome comparative and taxonomic position analysis. PLoS ONE 2017, 12, e171264. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Tian, J.; Xu, L.; Zhao, X.; Song, Y.; Wang, D. Comparative chloroplast genomes of six Magnoliaceae species provide new insights into intergeneric relationships and phylogeny. Biology 2022, 11, 1279. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Zheng, K.; Jiao, K.; Cai, Y.; Chen, C.; Mao, Y.; Wang, L.; Zhan, X.; Ying, Q.; Wang, H. Complete chloroplast genomes of four Physalis species (Solanaceae): Lights into genome structure, comparative analysis, and phylogenetic relationships. BMC Plant Biol. 2020, 20, 242. [Google Scholar] [CrossRef] [PubMed]
- Amiteye, S. Basic concepts and methodologies of DNA marker systems in plant molecular breeding. Heliyon 2021, 7, e08093. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Fang, Z.; Zhou, J.; Chen, H.; Hu, Z.; Gao, L.; Chen, L.; Ren, S.; Ma, H.; Zhang, L.; et al. An accurate and efficient method for large-scale SSR genotyping and applications. Nucleic. Acids Res. 2017, 45, e88. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Qu, D.; Ma, Y. Characterization of the chloroplast genome of Argyranthemum frutescens and a comparison with other species in Anthemideae. Genes 2022, 13, 1720. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zhou, T.; Su, X.; Wang, G.; Zhang, X.; Guo, Q.; Cao, F. Structural characterization and comparative analysis of the chloroplast genome of Ginkgo biloba and other gymnosperms. J. For. Res. 2021, 32, 765–778. [Google Scholar] [CrossRef]
- Liu, S.X.; Xue, D.Y.; Cheng, R.; Han, H.X. The complete mitogenome of Apocheima cinerarius (Lepidoptera: Geometridae: Ennominae) and comparison with that of other lepidopteran insects. Gene 2014, 547, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Yu, H.; Wang, J.; Lei, W.; Gao, J.; Qiu, X.; Wang, J. The complete chloroplast genome sequences of the medicinal plant Forsythia suspensa (Oleaceae). Int. J. Mol. Sci. 2017, 18, 2288. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhan, D.F.; Jia, X.; Mei, W.L.; Dai, H.F.; Chen, X.T.; Peng, S.Q. Complete chloroplast genome sequence of Aquilaria sinensis (Lour.) Gilg and evolution analysis within the Malvales order. Front. Plant Sci. 2016, 7, 280. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Lin, F.; Huang, P.; Guo, W.; Zheng, Y. Complete chloroplast genome sequence of Decaisnea insignis: Genome organization, genomic resources and comparative analysis. Sci. Rep. 2017, 7, 10073. [Google Scholar] [CrossRef]
- Knight, R.D.; Freeland, S.J.; Landweber, L.F. A simple model based on mutation and selection explains trends in codon and amino-acid usage and GC composition within and across genomes. Genome Biol. 2001, 2, H10. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.L.; Lee, W.; Hottes, A.K.; Shapiro, L.; Mcadams, H.H. Codon usage between genomes is constrained by genome-wide mutational processes. Proc. Natl. Acad. Sci. USA 2004, 101, 3480–3485. [Google Scholar] [CrossRef] [PubMed]
- Quax, T.E.F.; Claassens, N.J.; Söll, D.; van der Oost, J. Codon Bias as a Means to Fine-Tune Gene Expression. Mol. Cell 2015, 59, 149–161. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, S.; Liu, Y.; Yuan, Q.; Sun, J.; Guo, L. Chloroplast genome variation and phylogenetic relationships of Atractylodes species. BMC Genom. 2021, 22, 103. [Google Scholar] [CrossRef] [PubMed]
- Jeon, J.; Kim, S. Comparative Analysis of the complete chloroplast genome sequences of three closely related East-Asian wild roses (Rosa sect. Synstylae; Rosaceae). Genes 2019, 10, 23. [Google Scholar] [CrossRef] [PubMed]
- Katayama, H.; Uematsu, C. Comparative analysis of chloroplast DNA in Pyrus species: Physical map and gene localization. Theor. Appl. Genet. 2003, 106, 303–310. [Google Scholar] [CrossRef] [PubMed]

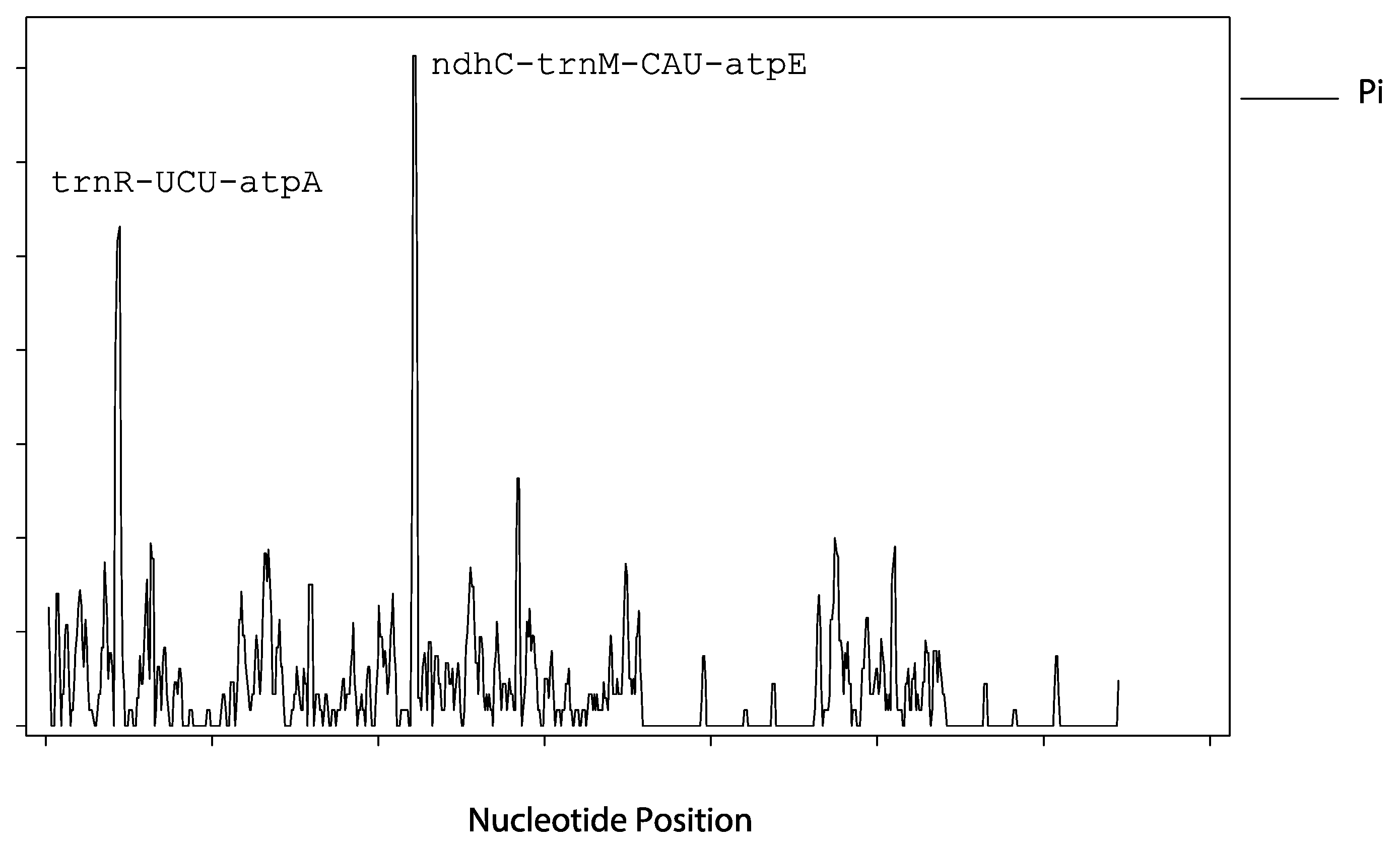
| Region | A (%) | C (%) | G (%) | T (%) | GC (%) | Base Size (bp) |
|---|---|---|---|---|---|---|
| IRa | 28.6 | 22.1 | 20.54 | 28.76 | 42.64 | 26,392 |
| IRb | 28.76 | 20.54 | 22.1 | 28.6 | 42.64 | 26,392 |
| LSC | 32.12 | 17.64 | 16.65 | 33.6 | 34.28 | 87,863 |
| SSC | 34.77 | 15.9 | 14.5 | 34.83 | 30.4 | 19,238 |
| Total | 31.3 | 18.64 | 17.93 | 32.12 | 36.58 | 159,885 |
| Gene Function | Gene Category | Gene Name | Number |
|---|---|---|---|
| Photosynthesis | Subunits of photosystem I | psaA, psaB, psaC, psaI, psaJ | 5 |
| Subunits of photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | 15 | |
| Subunits of NADH dehydrogenase | ndhA*, ndhB*(2), ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | 12 | |
| Subunits of cytochrome b/f complex | petA, petB*, petD*, petG, petL, petN | 6 | |
| Subunits of ATP synthase | atpA, atpB, atpE, atpF*, atpH, atpI | 6 | |
| Large subunit of rubis CO | rbcL | 1 | |
| Proteins of large ribosomal subunit | #rpl2*, rpl14, rpl16*, rpl2*, rpl20, rpl22, rpl23(2), rpl32, rpl33 | 10 | |
| Self-replication | Proteins of small ribosomal subunit | rps11, rps12**(2), rps14, rps15, rps16*, rps18, rps19, rps2, rps3, rps4, rps7(2), rps8 | 14 |
| Subunits of RNA polymerase | rpoA, rpoB, rpoC1*, rpoC2 | 4 | |
| Ribosomal RNAs | rrn16(2), rrn23(2), rrn4.5(2), rrn5(2) | 8 | |
| Transfer RNAs | trnA-UGC*(2), trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnG-GCC, trnG-UCC*, trnH-GUG, trnI-CAU(2), trnI-GAU*(2), trnK-UUU*, trnL-CAA(2), trnL-UAA*, trnL-UAG, trnM-CAU, trnN-GUU(2), trnP-UGG, trnQ-UUG, trnR-ACG(2), trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC(2), trnV-UAC*, trnW-CCA, trnY-GUA, trnfM-CAU | 37 | |
| Maturase | matK | 1 | |
| Other genes | Protease | clpP** | 1 |
| Envelope membrane protein | cemA | 1 | |
| Acetyl-CoA carboxylase | accD | 1 | |
| c-type cytochrome synthesis gene | ccsA | 1 | |
| Genes of unknown function | Conserved hypothetical chloroplast ORF | #ycf1, ycf1, ycf15(2), ycf2(2), ycf3**, ycf4 | 8 |
| Amino Acid | Codon | No. | Total | RSCU | Amino Acid | Codon | No. | Total | RSCU |
|---|---|---|---|---|---|---|---|---|---|
| * (Ter) | UAA | 47 | 84 | 1.6785 | M (Met) | AUG | 616 | 2.9904 | |
| * (Ter) | UAG | 22 | 0.7857 | M (Met) | GUG | 1 | 0.0048 | ||
| * (Ter) | UGA | 15 | 0.5358 | N (Asn) | AAC | 298 | 1269 | 0.4696 | |
| A (Ala) | GCA | 384 | 1384 | 1.11 | N (Asn) | AAU | 971 | 1.5304 | |
| A (Ala) | GCC | 213 | 0.6156 | P (Pro) | CCA | 314 | 1068 | 1.176 | |
| A (Ala) | GCG | 146 | 0.422 | P (Pro) | CCC | 197 | 0.738 | ||
| A (Ala) | GCU | 641 | 1.8528 | P (Pro) | CCG | 148 | 0.5544 | ||
| C (Cys) | UGC | 77 | 299 | 0.515 | P (Pro) | CCU | 409 | 1.532 | |
| C (Cys) | UGU | 222 | 1.485 | Q (Gln) | CAA | 714 | 927 | 1.5404 | |
| D (Asp) | GAC | 207 | 1091 | 0.3794 | Q (Gln) | CAG | 213 | 0.4596 | |
| D (Asp) | GAU | 884 | 1.6206 | R (Arg) | AGA | 470 | 1569 | 1.7976 | |
| E (Glu) | GAA | 1010 | 1367 | 1.4776 | R (Arg) | AGG | 174 | 0.6654 | |
| E (Glu) | GAG | 357 | 0.5224 | R (Arg) | CGA | 363 | 1.3884 | ||
| F (Phe) | UUC | 518 | 1483 | 0.6986 | R (Arg) | CGC | 109 | 0.417 | |
| F (Phe) | UUU | 965 | 1.3014 | R (Arg) | CGG | 119 | 0.4548 | ||
| G (Gly) | GGA | 701 | 1736 | 1.6152 | R (Arg) | CGU | 334 | 1.2774 | |
| G (Gly) | GGC | 181 | 0.4172 | S (Ser) | AGC | 127 | 2011 | 0.3792 | |
| G (Gly) | GGG | 284 | 0.6544 | S (Ser) | AGU | 401 | 1.1964 | ||
| G (Gly) | GGU | 570 | 1.3132 | S (Ser) | UCA | 407 | 1.2144 | ||
| H (His) | CAC | 140 | 626 | 0.4472 | S (Ser) | UCC | 324 | 0.9666 | |
| H (His) | CAU | 486 | 1.5528 | S (Ser) | UCG | 190 | 0.567 | ||
| I (Ile) | AUA | 709 | 2244 | 0.948 | S (Ser) | UCU | 562 | 1.677 | |
| I (Ile) | AUC | 433 | 0.579 | T (Thr) | ACA | 408 | 1343 | 1.2152 | |
| I (Ile) | AUU | 1102 | 1.4733 | T (Thr) | ACC | 242 | 0.7208 | ||
| K (Lys) | AAA | 1029 | 1379 | 1.4924 | T (Thr) | ACG | 148 | 0.4408 | |
| K (Lys) | AAG | 350 | 0.5076 | T (Thr) | ACU | 545 | 1.6232 | ||
| L (Leu) | CUA | 357 | 2752 | 0.7782 | V (Val) | GUA | 539 | 1415 | 1.5236 |
| L (Leu) | CUC | 181 | 0.3948 | V (Val) | GUC | 161 | 0.4552 | ||
| L (Leu) | CUG | 179 | 0.39 | V (Val) | GUG | 202 | 0.5712 | ||
| L (Leu) | CUU | 588 | 1.2822 | V (Val) | GUU | 513 | 1.45 | ||
| L (Leu) | UUA | 884 | 1.9272 | W (Trp) | UGG | 452 | 452 | 1 | |
| L (Leu) | UUG | 563 | 1.2276 | Y (Tyr) | UAC | 189 | 971 | 0.3892 | |
| M (Met) | AUA | 1 | 618 | 0.0048 | Y (Tyr) | UAU | 782 | 1.6108 |
| Region | Exon | Intron | Intergenic | Total |
|---|---|---|---|---|
| LSC | 31 | 16 | 82 | 129 |
| SSC | 22 | 3 | 13 | 38 |
| IR | 21 | 3 | 18 | 42 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiao, H.; Chen, Q.; Xiong, C.; Wang, H.; Ran, K.; Dong, R.; Dong, X.; Guan, Q.; Wei, S. Chloroplast Genome Profiling and Phylogenetic Insights of the “Qixiadaxiangshui” Pear (Pyrus bretschneideri Rehd.1). Horticulturae 2024, 10, 744. https://doi.org/10.3390/horticulturae10070744
Jiao H, Chen Q, Xiong C, Wang H, Ran K, Dong R, Dong X, Guan Q, Wei S. Chloroplast Genome Profiling and Phylogenetic Insights of the “Qixiadaxiangshui” Pear (Pyrus bretschneideri Rehd.1). Horticulturae. 2024; 10(7):744. https://doi.org/10.3390/horticulturae10070744
Chicago/Turabian StyleJiao, Huijun, Qiming Chen, Chi Xiong, Hongwei Wang, Kun Ran, Ran Dong, Xiaochang Dong, Qiuzhu Guan, and Shuwei Wei. 2024. "Chloroplast Genome Profiling and Phylogenetic Insights of the “Qixiadaxiangshui” Pear (Pyrus bretschneideri Rehd.1)" Horticulturae 10, no. 7: 744. https://doi.org/10.3390/horticulturae10070744
APA StyleJiao, H., Chen, Q., Xiong, C., Wang, H., Ran, K., Dong, R., Dong, X., Guan, Q., & Wei, S. (2024). Chloroplast Genome Profiling and Phylogenetic Insights of the “Qixiadaxiangshui” Pear (Pyrus bretschneideri Rehd.1). Horticulturae, 10(7), 744. https://doi.org/10.3390/horticulturae10070744






