Genome-Wide Identification of Litchi SPL Gene Family and Expression Analysis in Pericarp Anthocyanin Biosynthesis
Abstract
1. Introduction
2. Materials and Methods
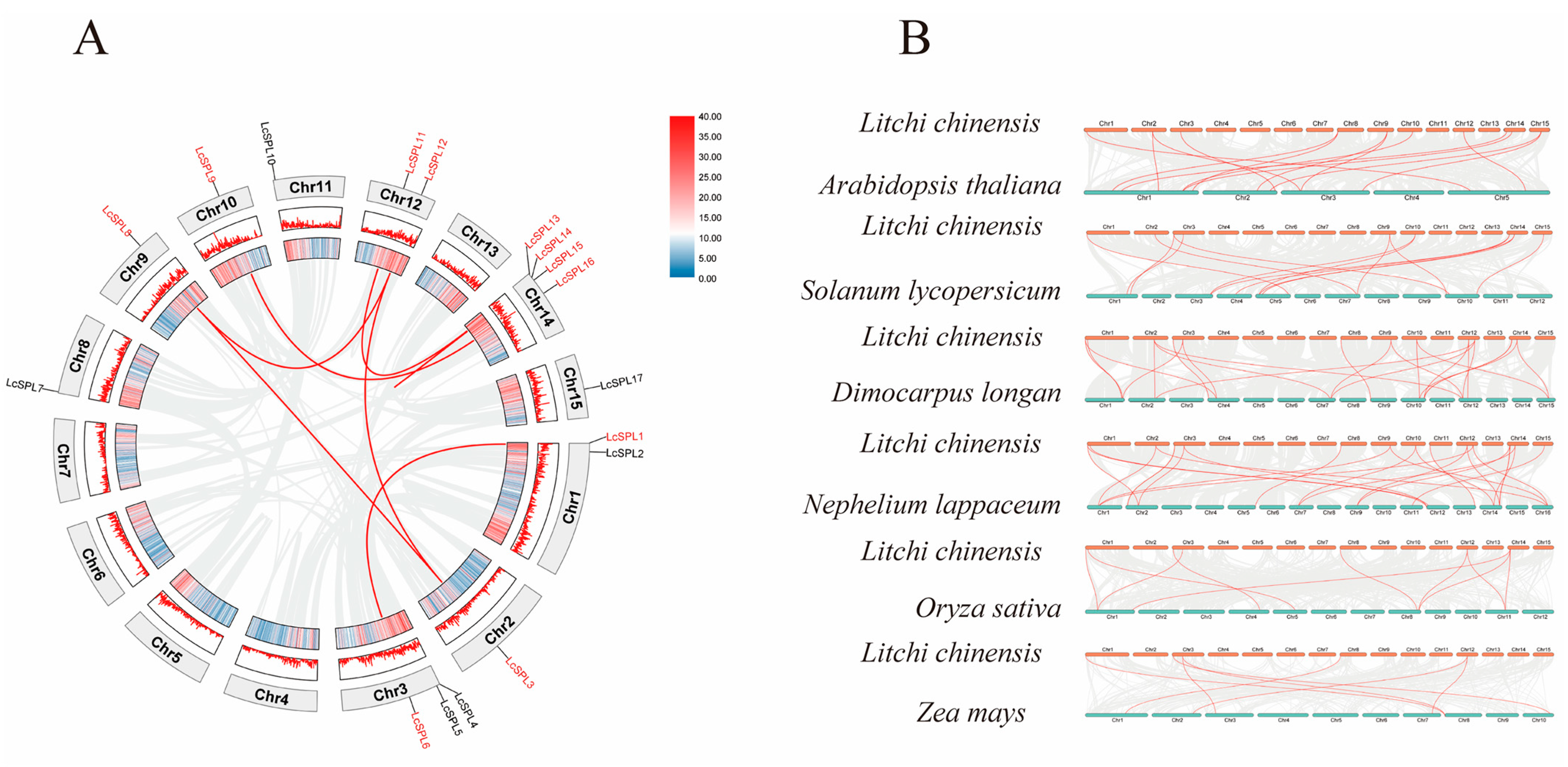
2.1. Identification of SPL Genes in Litchi and Chromosomal Location Analysis
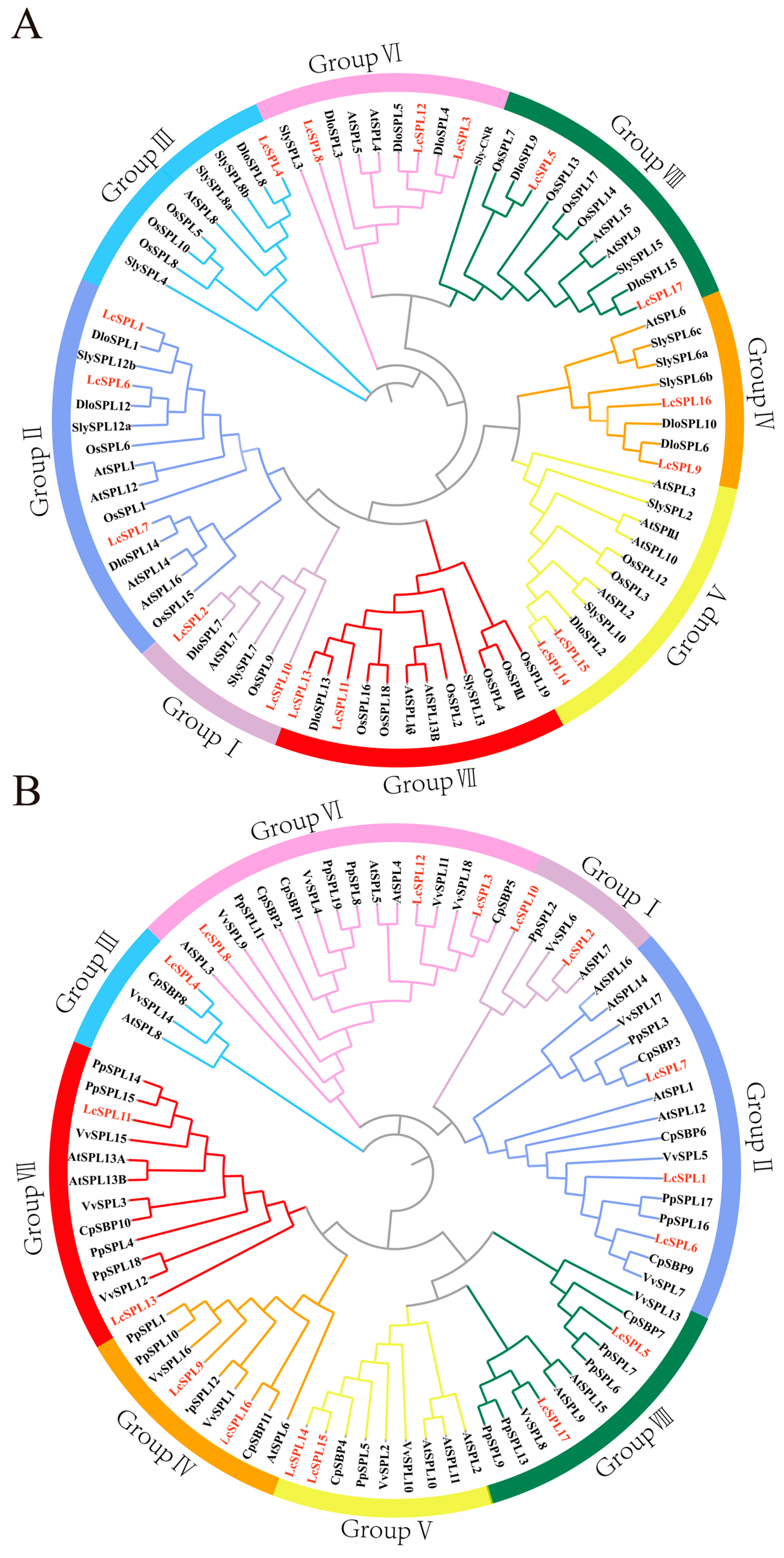
2.2. Phylogenetic Analysis of the LcSPL Family Genes
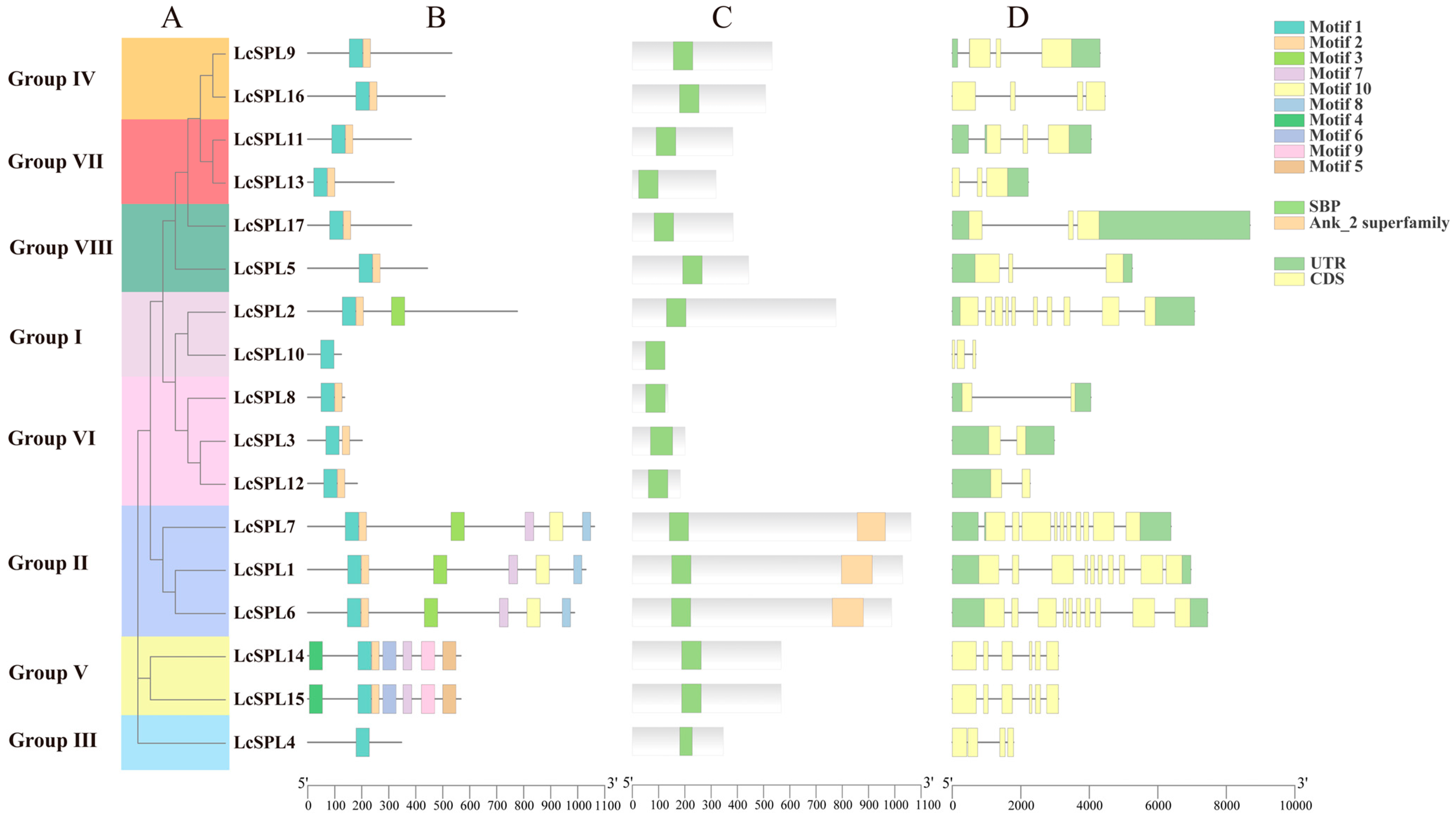
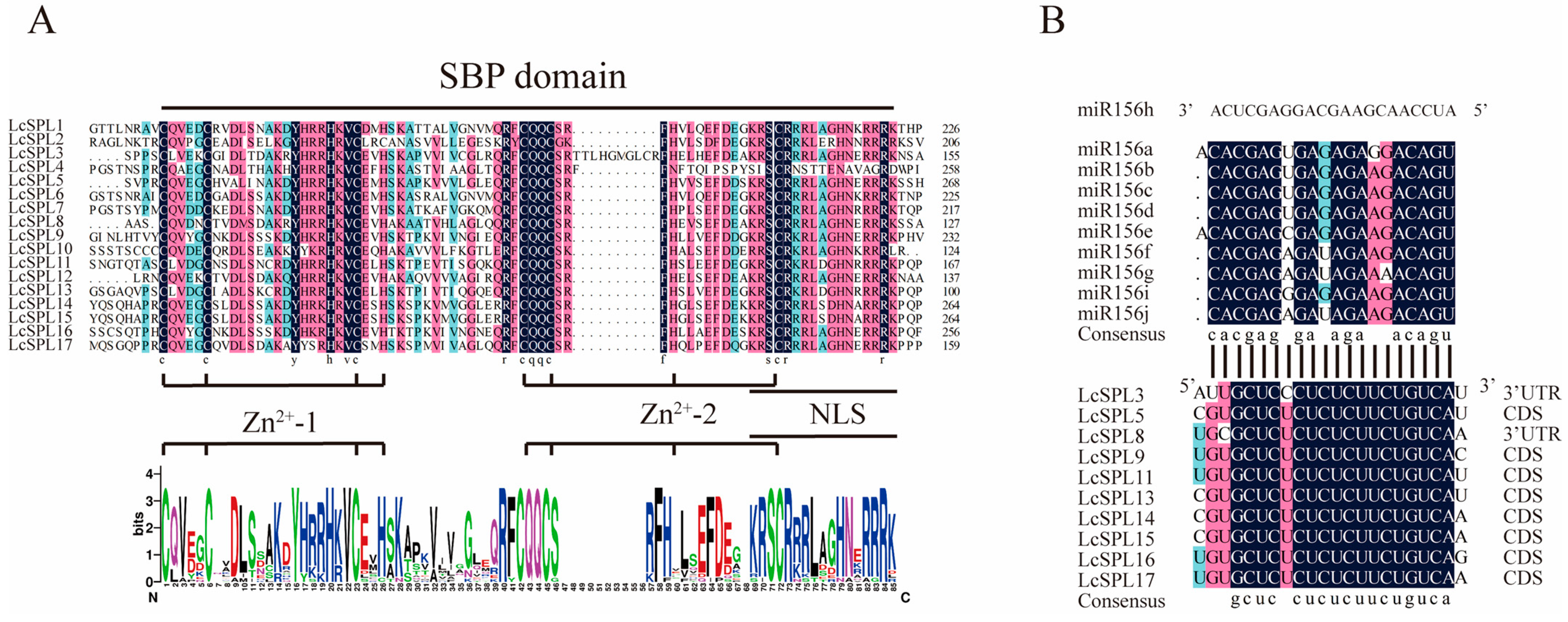
2.3. Gene Structures, Multiple Sequence Alignment Analysis and MicroRNA Target Prediction
2.4. Gene Duplication and Syntenic Analysis of the Litchi SPL Gene Family
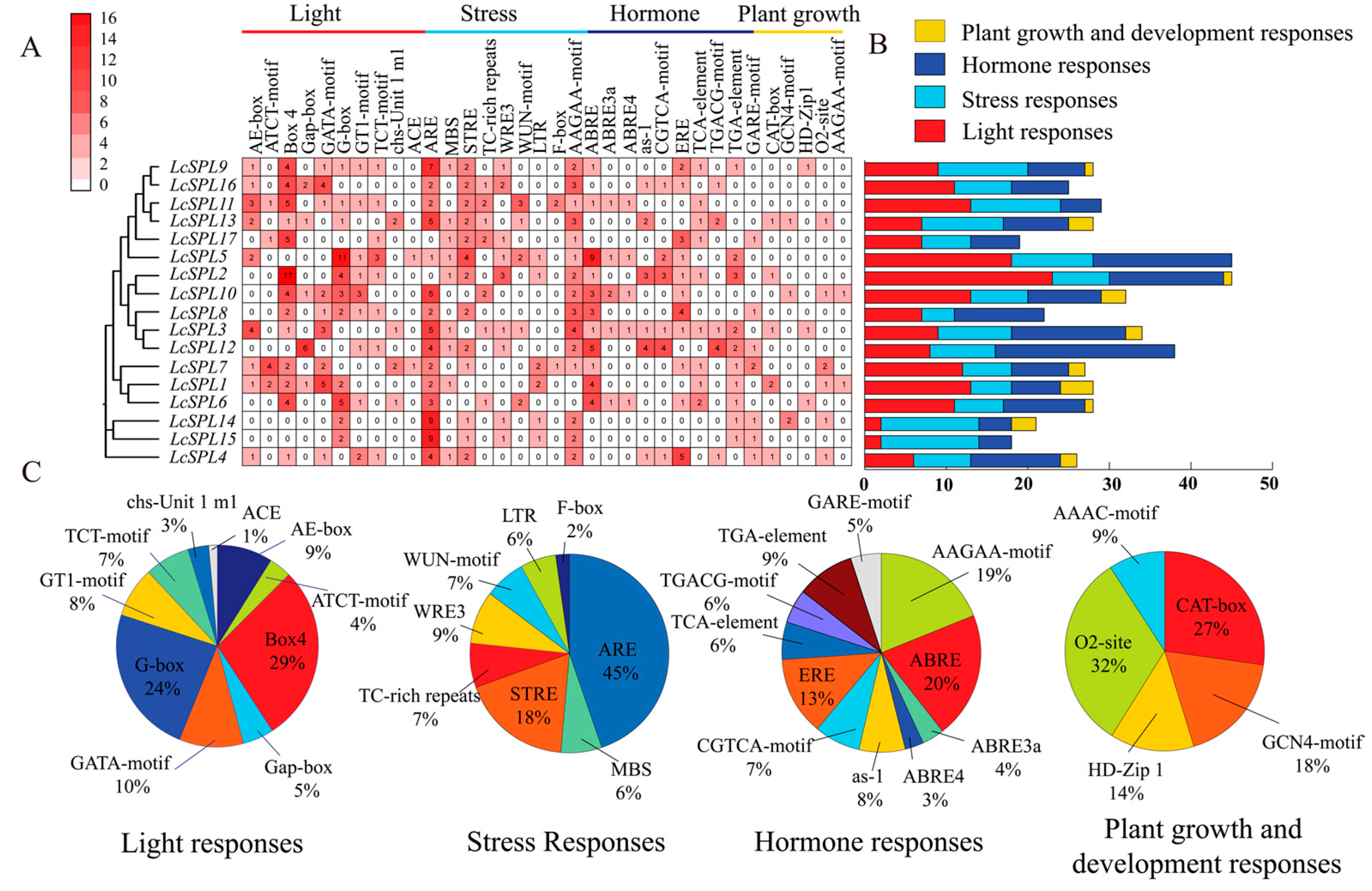
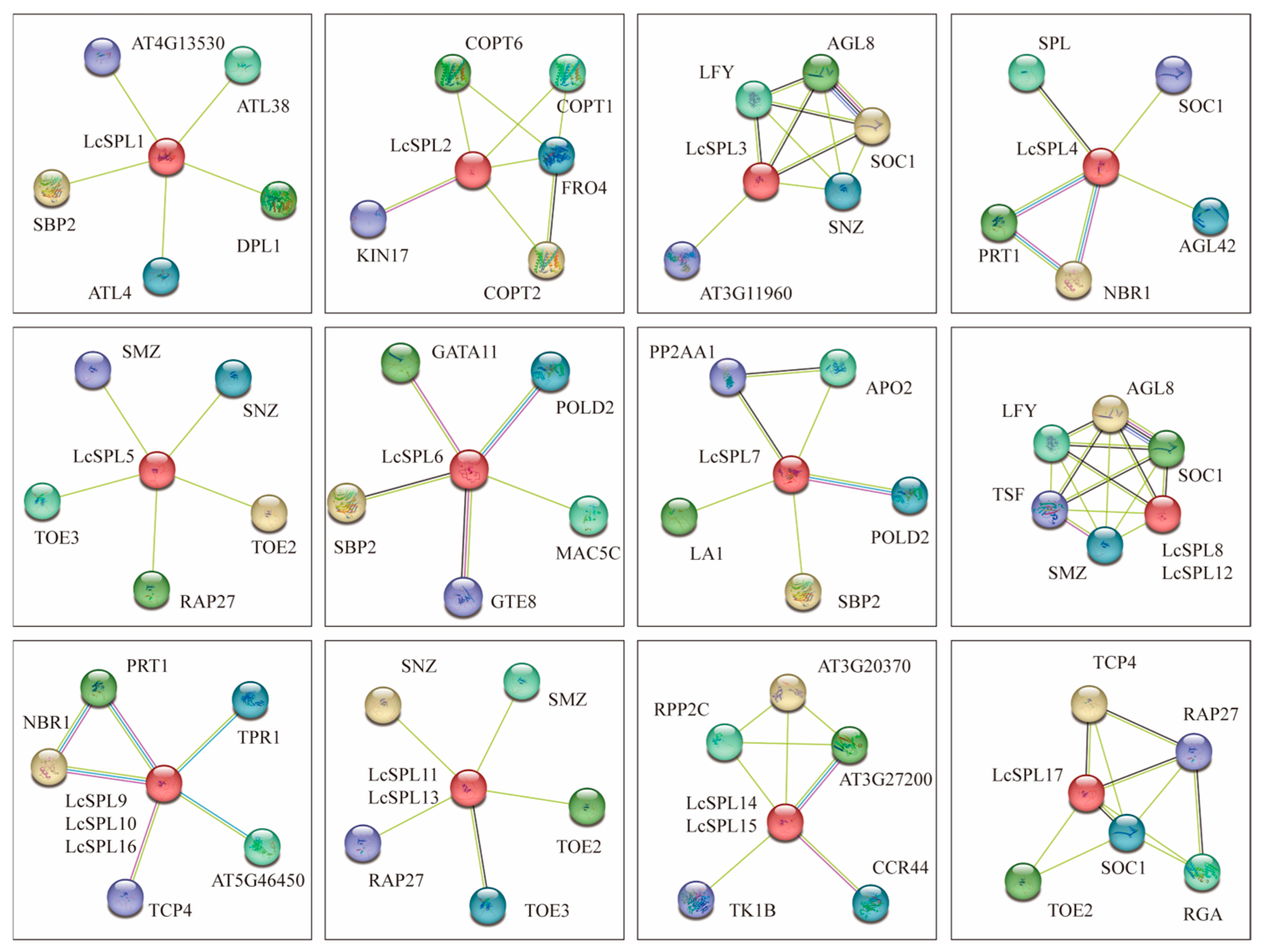
2.5. Cis-Element Identification in the Promoter and Protein Interaction Prediction Analysis
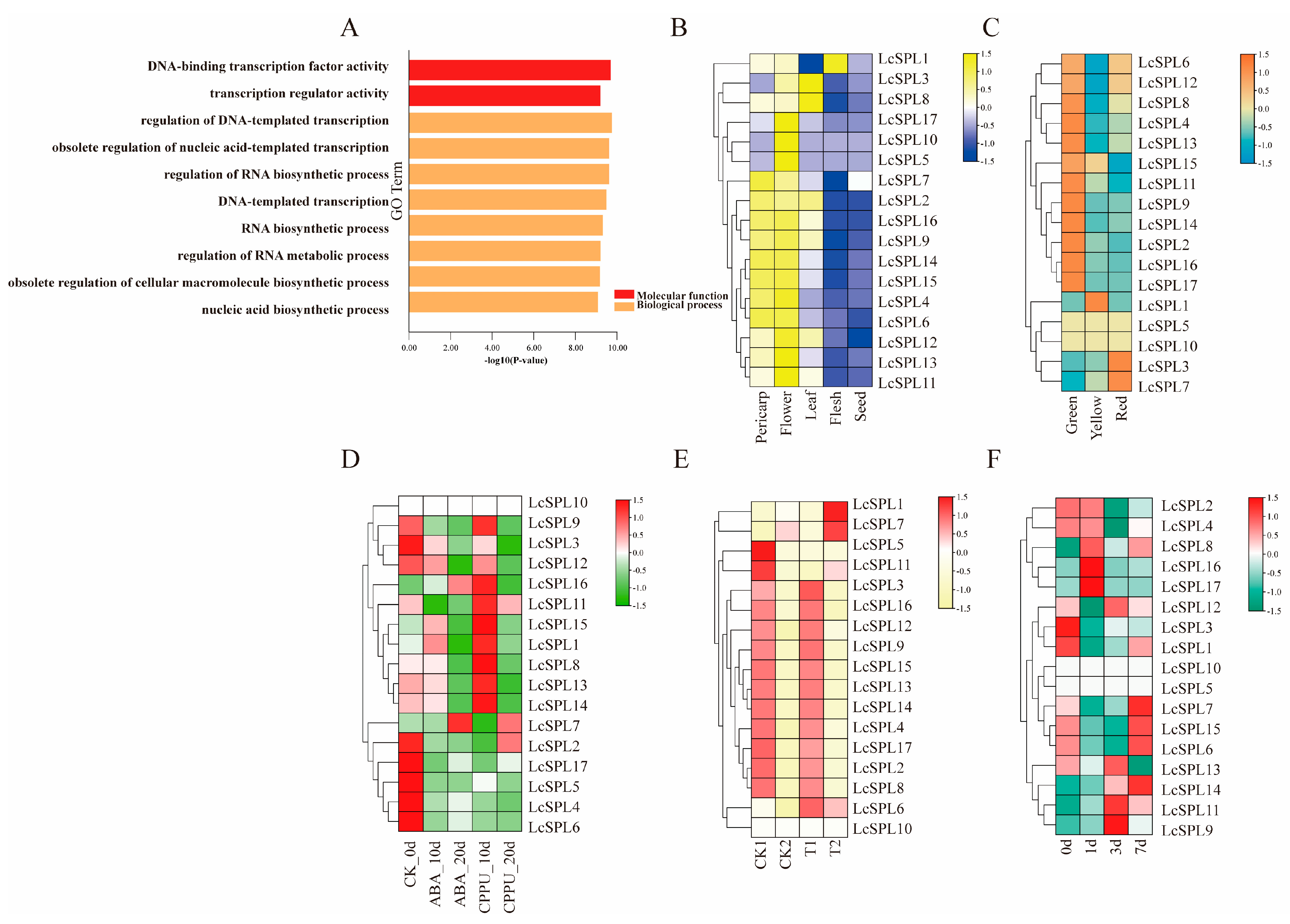
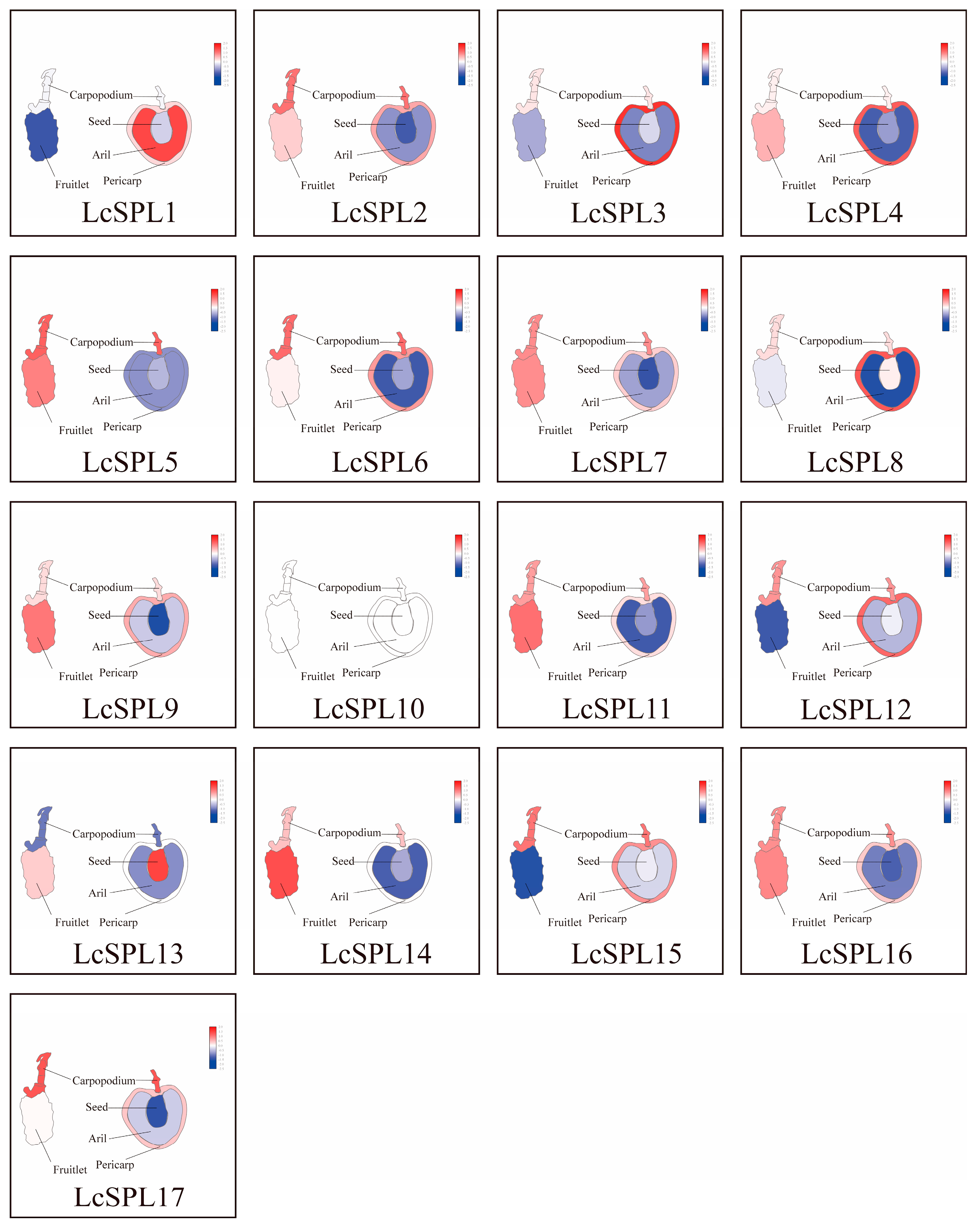
2.6. Gene Ontology (GO) Enrichment Analysis and Expression Pattern Analysis of LcSPL Genes in Litchi
- RNA-seq data relating to pericarp, flower, leaf, flesh, and seed samples of naturally grown ‘Feizixiao’ litchi (unpublished).
- RNA-seq data relating to carpopodium, aril, seed, pericarp, and fruitlet samples of naturally grown ‘Feizixiao’ litchi, accession number isPRJNA271242 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA271242/, accessed on 26 April 2024) [43].
- Normal growth of green, yellow, and red ‘Nuomici’ litchi pericarp sample RNA-seq data; the accession number is PRJNA261000 (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA261000, accessed on 26 April 2024) [44].
- RNA-seq data relating to ‘Feizixiao’ litchi pericarp samples at different periods after 5-(1-hydroxy-2,6,6-trimethy1-4-oxo-2cyclohexen-1-yl)-ethy1-(22,4E)-pentadienoic acid (ABA) and N-(2-Chloro-4-pyridyl)-N’-phenylurea (CPPU) treatment; the accession number is PRINA415698 (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA415698, accessed on 26 April 2024) [45].
- RNA-seq of CPPU-treated ‘Feizixiao’ litchi pericarp samples at the all-green and best-eat periods (unpublished).
- RNA-seq data of ‘Feizixiao’ litchi pericarp samples at different periods after lifting the shading treatment; the accession number is PRJNA303128 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA303128, accessed on 26 April 2024) [46].
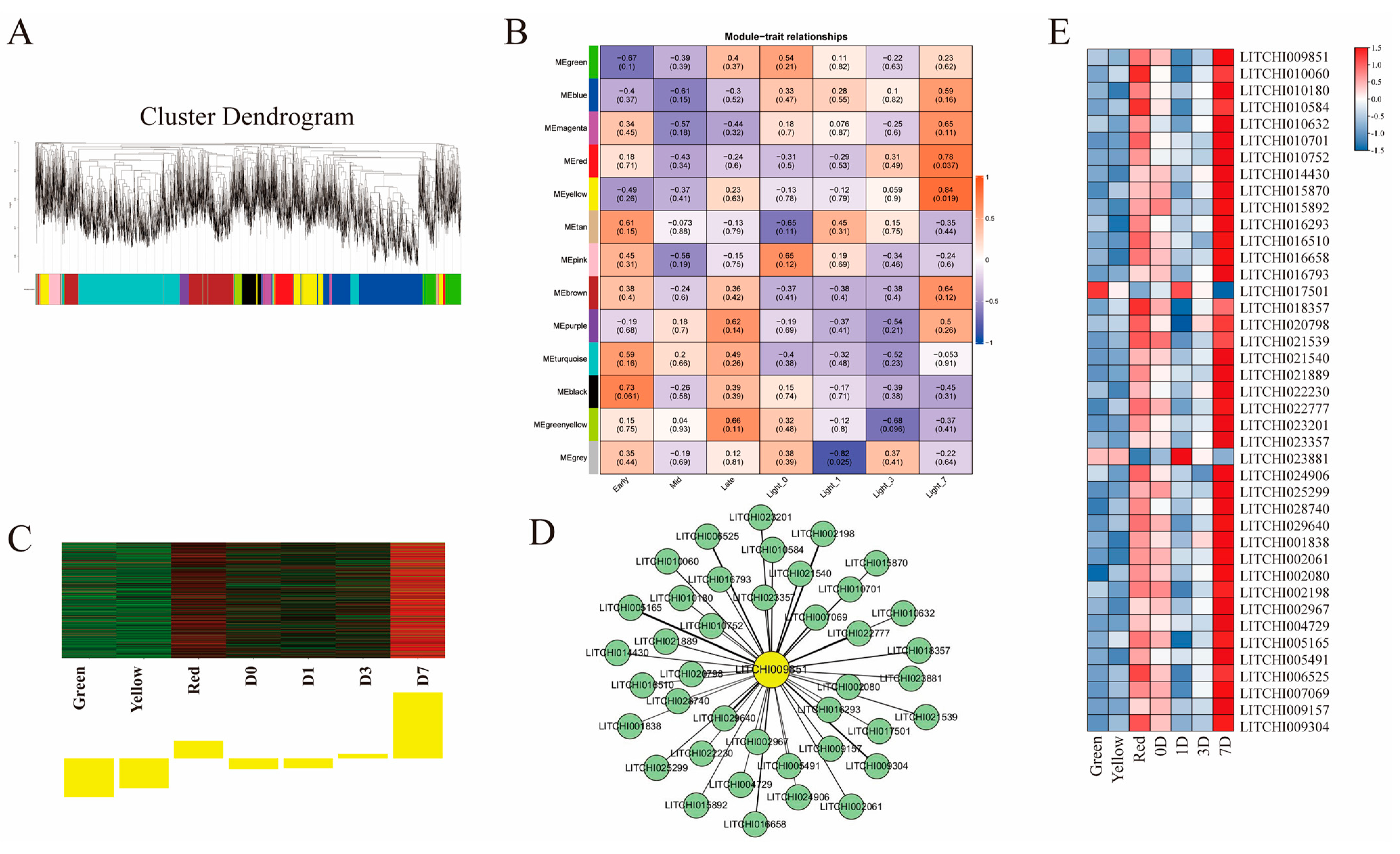
2.7. Weighted Gene Co-Expression Network Analysis (WGCNA)
3. Results
3.1. Identification of LcSPL Genes and Their Characterization in Litchi
3.2. Phylogenetic Analysis
3.3. Analysis of Gene Structures, Conserved Domains, and Motifs
3.4. Multiple Sequence Alignment Analysis and MicroRNA Target Prediction
3.5. Distribution of LcSPL Genes in the Litchi Genome and Their Evolutionary Relationships
3.6. Cis-Element Identification in the Promoter and Protein Interaction Prediction Analysis
3.7. Gene Ontology (GO) Enrichment Analysis and Analysis of Expression Profile of LcSPL Genes
3.7.1. Gene Ontology (GO) Enrichment Analysis
3.7.2. Expression Profiles of LcSPLs in Different Tissues and Fruit Organs in Litchi
3.7.3. Expression Profile of LcSPLs in Different Colored Pericarp Types
3.7.4. Expression Profile Analysis of LcSPLs under Different Hormone Treatments
3.7.5. Expression Profile Analysis of LcSPLs Released from Shade Conditions
3.8. Co-Expression Analysis of Key LcSPL Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Gong, W.; Shen, Y.-P.; Ma, L.-G.; Pan, Y.; Du, Y.-L.; Wang, D.-H.; Yang, J.-Y.; Hu, L.-D.; Liu, X.-F.; Dong, C.-X. Genome-wide ORFeome cloning and analysis of Arabidopsis transcription factor genes. Plant Physiol. 2004, 135, 773–782. [Google Scholar] [CrossRef] [PubMed]
- Shafique Khan, F.; Zeng, R.-F.; Gan, Z.-M.; Zhang, J.-Z.; Hu, C.-G. Genome-wide identification and expression profiling of the WOX gene family in Citrus sinensis and functional analysis of a CsWUS member. Int. J. Mol. Sci. 2021, 22, 4919. [Google Scholar] [CrossRef] [PubMed]
- Khan, F.S.; Goher, F.; Hu, C.-G.; Zhang, J.-Z. WUSCHEL-related homeobox (WOX) transcription factors: Key regulators in combating abiotic stresses in plants. Hortic. Adv. 2024, 2, 2. [Google Scholar] [CrossRef]
- Hu, L.; Chen, W.; Yang, W.; Li, X.; Zhang, C.; Zhang, X.; Zheng, L.; Zhu, X.; Yin, J.; Qin, P. OsSPL9 regulates grain number and grain yield in rice. Front. Plant Sci. 2021, 12, 682018. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Poethig, R.S. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 2006, 133, 3539–3547. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, A.; Wu, M.-F.; Yang, L.; Wu, G.; Poethig, R.S.; Wagner, D. The microRNA-regulated SBP-Box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1. Dev. Cell 2009, 17, 268–278. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Chen, R.; Shi, Y.; Gai, C.; Fan, K.; Li, Z. Research Advances in Biological Functions of Plant SPL Transcription Factors. Chin. Bull. Bot. 2023, 58, 982–997. [Google Scholar]
- Cui, L.; Zheng, F.; Wang, J.; Zhang, C.; Xiao, F.; Ye, J.; Li, C.; Ye, Z.; Zhang, J. miR156a-targeted SBP-Box transcription factor SlSPL13 regulates inflorescence morphogenesis by directly activating SFT in tomato. Plant Biotechnol. J. 2020, 18, 1670–1682. [Google Scholar] [CrossRef]
- Guo, S.; Zhang, M.; Feng, M.; Liu, G.; Torregrosa, L.; Tao, X.; Ren, R.; Fang, Y.; Zhang, Z.; Meng, J. miR156b-targeted VvSBP8/13 functions downstream of the abscisic acid signal to regulate anthocyanins biosynthesis in grapevine fruit under drought. Hortic. Res. 2024, 11, uhad293. [Google Scholar] [CrossRef]
- Xu, Y.; Xu, H.; Wall, M.M.; Yang, J. Roles of transcription factor SQUAMOSA promoter binding protein-like gene family in papaya (Carica papaya) development and ripening. Genomics 2020, 112, 2734–2747. [Google Scholar] [CrossRef]
- Jiao, Y.-Q.; Wang, Y.-H.; Xue, D.-W.; Wang, J.; Yan, M.-X.; Liu, G.-F.; Dong, G.-J.; Zeng, D.-L.; Lu, Z.-F.; Zhu, X.-D. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 2010, 42, 541–544. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.-F.; Wang, X.-F.; Gu, S.-L.; Hu, Z.-Q.; Xu, H.; Xu, C.-W. Comparative study of SBP-box gene family in Arabidopsis and rice. Gene 2008, 407, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Wu, H.; Chen, Y. Gene duplications and functional divergence analyses of the SPL gene family. J. Nanjing For. Univ. 2020, 44, 55–66. [Google Scholar]
- Birkenbihl, R.P.; Jach, G.; Saedler, H.; Huijser, P. Functional dissection of the plant-specific SBP-domain: Overlap of the DNA-binding and nuclear localization domains. J. Mol. Biol. 2005, 352, 585–596. [Google Scholar] [CrossRef] [PubMed]
- Cardon, G.; Höhmann, S.; Klein, J.; Nettesheim, K.; Saedler, H.; Huijser, P. Molecular characterisation of the Arabidopsis SBP-box genes. Gene 1999, 237, 91–104. [Google Scholar] [CrossRef] [PubMed]
- Xie, K.; Wu, C.; Xiong, L. Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice. Plant Physiol. 2006, 142, 280–293. [Google Scholar] [CrossRef] [PubMed]
- Salinas, M.; Xing, S.; Höhmann, S.; Berndtgen, R.; Huijser, P. Genomic organization, phylogenetic comparison and differential expression of the SBP-box family of transcription factors in tomato. Planta 2012, 235, 1171–1184. [Google Scholar] [CrossRef] [PubMed]
- Hou, H.; Li, J.; Gao, M.; Singer, S.D.; Wang, H.; Mao, L.; Fei, Z.; Wang, X. Genomic organization, phylogenetic comparison and differential expression of the SBP-box family genes in grape. PLoS ONE 2013, 8, e59358. [Google Scholar] [CrossRef] [PubMed]
- Qian, M.; Ni, J.; Niu, Q.; Bai, S.; Bao, L.; Li, J.; Sun, Y.; Zhang, D.; Teng, Y. Response of miR156-SPL module during the red peel coloration of bagging-treated Chinese sand pear (Pyrus pyrifolia Nakai). Front. Physiol. 2017, 8, 550. [Google Scholar] [CrossRef]
- Shao, F.-j.; Lu, Q.; Wilson, I.W.; Qiu, D.-y. Genome-wide identification and characterization of the SPL gene family in Ziziphus jujuba. Gene 2017, 627, 315–321. [Google Scholar] [CrossRef]
- Zhou, Q.; Zhang, S.; Chen, F.; Liu, B.-J.; Wu, L.; Li, F.; Zhang, J.-Q.; Bao, M.-Z.; Liu, G.-F. Genome-wide identification and characterization of the SBP-box gene family in Petunia. BMC Genom. 2018, 19, 193. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, R.K.; Goel, R.; Kumari, S.; Dahuja, A. Genomic organization, phylogenetic comparison, and expression profiles of the SPL family genes and their regulation in soybean. Dev. Genes Evol. 2017, 227, 101–119. [Google Scholar] [CrossRef] [PubMed]
- Li, C.-L.; Lu, S.-F. Molecular characterization of the SPL gene family in Populus trichocarpa. BMC Plant Biol. 2014, 14, 131. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.-S.; Li, J.-T.; Liu, Y.; Zhang, Q.; Gao, Y.-R.; Fang, K.-F.; Cao, Q.-Q.; Qin, L.; Xing, Y. Roles of the GA-mediated SPL gene family and miR156 in the floral development of Chinese chestnut (Castanea mollissima). Int. J. Mol. Sci. 2019, 20, 1577. [Google Scholar] [CrossRef] [PubMed]
- Schwab, R.; Palatnik, J.F.; Riester, M.; Schommer, C.; Schmid, M.; Weigel, D. Specific effects of microRNAs on the plant transcriptome. Dev. Cell 2005, 8, 517–527. [Google Scholar] [CrossRef] [PubMed]
- Samad, A.F.; Sajad, M.; Nazaruddin, N.; Fauzi, I.A.; Murad, A.M.; Zainal, Z.; Ismail, I. MicroRNA and transcription factor: Key players in plant regulatory network. Front. Plant Sci. 2017, 8, 565. [Google Scholar] [CrossRef] [PubMed]
- Cho, E.; Seddon, J.M.; Rosner, B.; Willett, W.C.; Hankinson, S.E. Prospective study of intake of fruits, vegetables, vitamins, and carotenoidsand risk of age-related maculopathy. Arch. Ophthalmol. 2004, 122, 883–892. [Google Scholar] [CrossRef] [PubMed]
- Gou, J.; Felippes, F.; Liu, C.; Weigel, D.; Wang, J. Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 2011, 23, 1512–1522. [Google Scholar] [CrossRef] [PubMed]
- Luo, Q.-J.; Mittal, A.; Jia, F.; Rock, C.D. An autoregulatory feedback loop involving PAP1 and TAS4 in response to sugars in Arabidopsis. Plant Mol. Biol. 2012, 80, 117–129. [Google Scholar] [CrossRef]
- Munusamy, P.; Zolotarov, Y.; Meteignier, L.-V.; Moffett, P.; Strömvik, M.V. De novo computational identification of stress-related sequence motifs and microRNA target sites in untranslated regions of a plant translatome. Sci. Rep. 2017, 7, 43861. [Google Scholar] [CrossRef]
- Rhoades, M.W.; Reinhart, B.J.; Lim, L.P.; Burge, C.B.; Bartel, B.; Bartel, D.P. Prediction of plant microRNA targets. Cell 2002, 110, 513–520. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, W.; Wang, X.; Yang, R.; Wu, Z.; Wang, H.; Wang, L.; Hu, Z.; Guo, S.; Zhang, H. MiR156 regulates anthocyanin biosynthesis through SPL targets and other microRNAs in poplar. Hortic. Res. 2020, 7, 118. [Google Scholar] [CrossRef]
- Li, H.; Wang, S.; Zhai, L.; Cui, Y.; Tang, G.; Huo, J.; Li, X.; Bian, S. The miR156/SPL12 module orchestrates fruit colour change through directly regulating ethylene production pathway in blueberry. Plant Biotechnol. J. 2024, 22, 386–400. [Google Scholar] [CrossRef]
- Singh, S.K.; Marboh, E.; Nath, V. Litchi. In Fruit and Nut Crops; Springer: Berlin/Heidelberg, Germany, 2023; pp. 1–28. [Google Scholar]
- Ghosh, S. World trade in litchi: Past, present and future. In Proceedings of the I International Symposium on Litchi and Longan, Guangzhou, China, 16 June 2000; Volume 558, pp. 23–30. [Google Scholar]
- Underhill, S.; Critchley, C. Anthocyanin decolorisation and its role in lychee pericarp browning. Aust. J. Exp. Agric. 1994, 34, 115–122. [Google Scholar] [CrossRef]
- Wei, Y.-Z.; Hu, F.-C.; Hu, G.-B.; Li, X.-J.; Huang, X.-M.; Wang, H.-C. Differential expression of anthocyanin biosynthetic genes in relation to anthocyanin accumulation in the pericarp of Litchi chinensis Sonn. PLoS ONE 2011, 6, e19455. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, H.; Zhou, B.; Zhao, M.; Li, C.; Xia, R.; Huang, X. Research advances in physiology and molecular biology of flower and fruit development in litchi. J. South China Agric. Univ. 2019, 40, 119–127. [Google Scholar]
- Li, J.; Chen, C.; Zeng, Z.; Wu, F.; Feng, J.; Liu, B.; Mai, Y.; Chu, X.; Wei, W.; Li, X. SapBase (Sapinaceae Genomic DataBase): A central portal for functional and comparative genomics of Sapindaceae species. bioRxiv 2022. bioRvix:2022.2011. 2025.517904. [Google Scholar]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J.; Tamura, K. eggNOG-mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y. TBtools-II: A one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Li, C.; Wang, Y.; Huang, X.; Li, J.; Wang, H.; Li, J. De novo assembly and characterization of fruit transcriptome in Litchi chinensis Sonn and analysis of differentially regulated genes in fruit in response to shading. BMC Genom. 2013, 14, 552. [Google Scholar] [CrossRef] [PubMed]
- Lai, B.; Hu, B.; Qin, Y.; Zhao, J.; Wang, H.; Hu, G. Transcriptomic analysis of Litchi chinensis pericarp during maturation with a focus on chlorophyll degradation and flavonoid biosynthesis. BMC Genom. 2015, 16, 225. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Li, J.; Wang, D.; Wang, H.; Qin, Y.; Hu, G.; Zhao, J. Transcriptome profiling of Litchi chinensis pericarp in response to exogenous cytokinins and abscisic acid. Plant Growth Regul. 2018, 84, 437–450. [Google Scholar] [CrossRef]
- Zhang, H.-N.; Li, W.-C.; Wang, H.-C.; Shi, S.-Y.; Shu, B.; Liu, L.-Q.; Wei, Y.-Z.; Xie, J.-H. Transcriptome profiling of light-regulated anthocyanin biosynthesis in the pericarp of litchi. Front. Plant Sci. 2016, 7, 963. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, K.; Du, J.; Weng, Y.; Xu, Y.; Xiong, J.; Deng, Z.; Liu, H.; Liu, Y.; Song, H.; Li, H.; et al. Genome-Wide Study of the Rice SBP Gene Family. Acta Agric. Univ. Jiangxiensis 2023, 45, 1315–1323. [Google Scholar]
- Lu, B.; Zhu, Y.; Zhang, S.; Lü, Y.; Li, X.; Song, Y.; Lai, Z.; Lin, Y. Whole-genome identification and expression analysis of SPL gene family in Dimocarpus Longan. Sci. Agric. Sin. 2020, 53, 4259–4270. [Google Scholar]
- Cheng, H.; Hao, M.; Wang, W.; Mei, D.; Tong, C.; Wang, H.; Liu, J.; Fu, L.; Hu, Q. Genomic identification, characterization and differential expression analysis of SBP-box gene family in Brassica napus. BMC Plant Biol. 2016, 16, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Liu, Y.; Ma, L.; Wang, X.; Zhang, D.; Han, Y.; Ding, Q.; Ma, L. Genome-wide identification, phylogeny and expression analysis of the SPL gene family in wheat. BMC Plant Biol. 2020, 20, 420. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Gao, H.; Chen, H.; Fu, C. The involvement of papaya CpSBP1 in modulating fruit softening and carotenoid accumulation by repressing CpPME1/2 and CpPDS4. Sci. Hortic. 2019, 256, 108582. [Google Scholar] [CrossRef]
- Yamasaki, K.; Kigawa, T.; Inoue, M.; Tateno, M.; Yamasaki, T.; Yabuki, T.; Aoki, M.; Seki, E.; Matsuda, T.; Nunokawa, E. A novel zinc-binding motif revealed by solution structures of DNA-binding domains of Arabidopsis SBP-family transcription factors. J. Mol. Biol. 2004, 337, 49–63. [Google Scholar] [CrossRef]
- Hu, G.; Feng, J.; Xiang, X.; Wang, J.; Salojärvi, J.; Liu, C.; Wu, Z.; Zhang, J.; Liang, X.; Jiang, Z. Two divergent haplotypes from a highly heterozygous lychee genome suggest independent domestication events for early and late-maturing cultivars. Nat. Genet. 2022, 54, 73–83. [Google Scholar] [CrossRef]
- Zhong, H.; Kong, W.; Gong, Z.; Fang, X.; Deng, X.; Liu, C.; Li, Y. Evolutionary analyses reveal diverged patterns of SQUAMOSA promoter binding protein-like (SPL) gene family in Oryza genus. Front. Plant Sci. 2019, 10, 565. [Google Scholar] [CrossRef] [PubMed]
- Ling, L.-Z.; Zhang, S.-D. Unraveling the Distribution and Evolution of miR156 targeted SPLs in Plants by Phylogenetic Analysis. Plant Divers. 2012, 34, 33. [Google Scholar] [CrossRef]
- Zhuang, W.; Liu, T.; Shu, X.; Qu, S.; Zhai, H.; Wang, T.; Zhang, F.; Wang, Z. The molecular regulation mechanism of anthocyanin biosynthesis and coloration in plants. Plant Physiol. J. 2018, 54, 1630–1644. [Google Scholar]
- Preston, J.C.; Hileman, L.C. Functional evolution in the plant SQUAMOSA-PROMOTER BINDING PROTEIN-LIKE (SPL) gene family. Front. Plant Sci. 2013, 4, 80. [Google Scholar] [CrossRef] [PubMed]
- Panchy, N.; Lehti-Shiu, M.; Shiu, S.-H. Evolution of gene duplication in plants. Plant Physiol. 2016, 171, 2294–2316. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Wang, Q.; Zhao, Y.; Li, X.; Ma, Q. Comparative genome analysis of the SPL gene family reveals novel evolutionary features in maize. Genet. Mol. Biol. 2019, 42, 380–394. [Google Scholar] [CrossRef]
- He, F.; Long, R.; Wei, C.; Zhang, Y.; Li, M.; Kang, J.; Yang, Q.; Wang, Z.; Chen, L. Genome-wide identification, phylogeny and expression analysis of the SPL gene family and its important role in salt stress in Medicago sativa L. BMC Plant Biol. 2022, 22, 295. [Google Scholar] [CrossRef]
- Lee, H.; Wicker, L. Anthocyanin pigments in the skin of lychee fruit. J. Food Sci. 1991, 56, 466–468. [Google Scholar] [CrossRef]
- Liu, R.; Lai, B.; Hu, B.; Qin, Y.; Hu, G.; Zhao, J. Identification of microRNAs and their target genes related to the accumulation of anthocyanins in Litchi chinensis by high-throughput sequencing and degradome analysis. Front. Plant Sci. 2017, 7, 241260. [Google Scholar] [CrossRef]


| Gene Name | Gene ID | Protein/AA | Molecular Weight (kDa) | pI | Instability Index | Aliphatic Index | GRAVY | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|
| LcSPL1 | LITCHI014255.m1 | 1030 | 114.61 | 6.64 | 49.72 | 82.68 | −0.367 | Nucleus |
| LcSPL2 | LITCHI014612.m1 | 776 | 86.75 | 6.29 | 50.85 | 86.51 | −0.267 | Nucleus |
| LcSPL3 | LITCHI013083.m1 | 201 | 22.87 | 8.95 | 70.47 | 54.23 | −1.002 | Nucleus |
| LcSPL4 | LITCHI025415.m1 | 347 | 38.41 | 8.37 | 51.02 | 51.24 | −0.678 | Nucleus |
| LcSPL5 | LITCHI025416.m1 | 443 | 48.98 | 6.91 | 50.46 | 59.03 | −0.679 | Nucleus |
| LcSPL6 | LITCHI026667.m1 | 988 | 109.72 | 5.91 | 51.8 | 78.83 | −0.434 | Nucleus |
| LcSPL7 | LITCHI009851.m3 | 1062 | 118.46 | 7.87 | 61.09 | 72.82 | −0.528 | Nucleus |
| LcSPL8 | LITCHI029170.m1 | 136 | 15.40 | 7.04 | 75.31 | 43.09 | −1.281 | Nucleus |
| LcSPL9 | LITCHI023329.m1 | 533 | 58.22 | 7 | 52.76 | 65.25 | −0.54 | Nucleus |
| LcSPL10 | LITCHI006358.m1 | 124 | 14.57 | 8.97 | 97.99 | 34.6 | −1.305 | Nucleus |
| LcSPL11 | LITCHI020317.m1 | 383 | 42.23 | 7.64 | 57.52 | 58.56 | −0.746 | Nucleus |
| LcSPL12 | LITCHI021016.m1 | 183 | 20.62 | 9.21 | 51.5 | 59.07 | −0.945 | Nucleus |
| LcSPL13 | LITCHI004632.m1 | 319 | 34.55 | 8.74 | 57.11 | 60.85 | −0.571 | Nucleus |
| LcSPL14 | LITCHI004647.m1 | 567 | 62.27 | 9.31 | 42.54 | 65.54 | −0.554 | Nucleus |
| LcSPL15 | LITCHI004652.m1 | 567 | 62.30 | 9.31 | 42.32 | 65.54 | −0.554 | Nucleus |
| LcSPL16 | LITCHI005348.m1 | 508 | 56.45 | 7.51 | 54.35 | 70.02 | −0.57 | Nucleus |
| LcSPL17 | LITCHI019149.m1 | 384 | 40.65 | 9.23 | 59.98 | 50.55 | −0.666 | Nucleus |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Z.; Wu, J.; Jing, X.; Khan, F.S.; Chen, Y.; Chen, Z.; Zhang, H.; Wei, Y. Genome-Wide Identification of Litchi SPL Gene Family and Expression Analysis in Pericarp Anthocyanin Biosynthesis. Horticulturae 2024, 10, 762. https://doi.org/10.3390/horticulturae10070762
Xu Z, Wu J, Jing X, Khan FS, Chen Y, Chen Z, Zhang H, Wei Y. Genome-Wide Identification of Litchi SPL Gene Family and Expression Analysis in Pericarp Anthocyanin Biosynthesis. Horticulturae. 2024; 10(7):762. https://doi.org/10.3390/horticulturae10070762
Chicago/Turabian StyleXu, Ziqiang, Jiayun Wu, Xiangyang Jing, Faiza Shafique Khan, Yanzhao Chen, Zhe Chen, Hongna Zhang, and Yongzan Wei. 2024. "Genome-Wide Identification of Litchi SPL Gene Family and Expression Analysis in Pericarp Anthocyanin Biosynthesis" Horticulturae 10, no. 7: 762. https://doi.org/10.3390/horticulturae10070762
APA StyleXu, Z., Wu, J., Jing, X., Khan, F. S., Chen, Y., Chen, Z., Zhang, H., & Wei, Y. (2024). Genome-Wide Identification of Litchi SPL Gene Family and Expression Analysis in Pericarp Anthocyanin Biosynthesis. Horticulturae, 10(7), 762. https://doi.org/10.3390/horticulturae10070762







