Comprehensive Analysis of Microbiomes and Metabolomics Reveals the Mechanism of Adaptation to Cadmium Stress in Rhizosphere Soil of Rhododendron decorum subsp. Diaprepes
Abstract
1. Introduction
2. Materials and Methods
2.1. Experiments on Plant Growth and Cd Addition
2.2. Analysis of Rhizosphere Soil’s Physical Physicochemical Properties
2.3. DNA Extraction, Amplicon Sequencing, and Bioinformatics Analysis
2.4. Rhizosphere Soil Metabolomics and Data Analysis
2.5. Statistical Analysis
3. Results
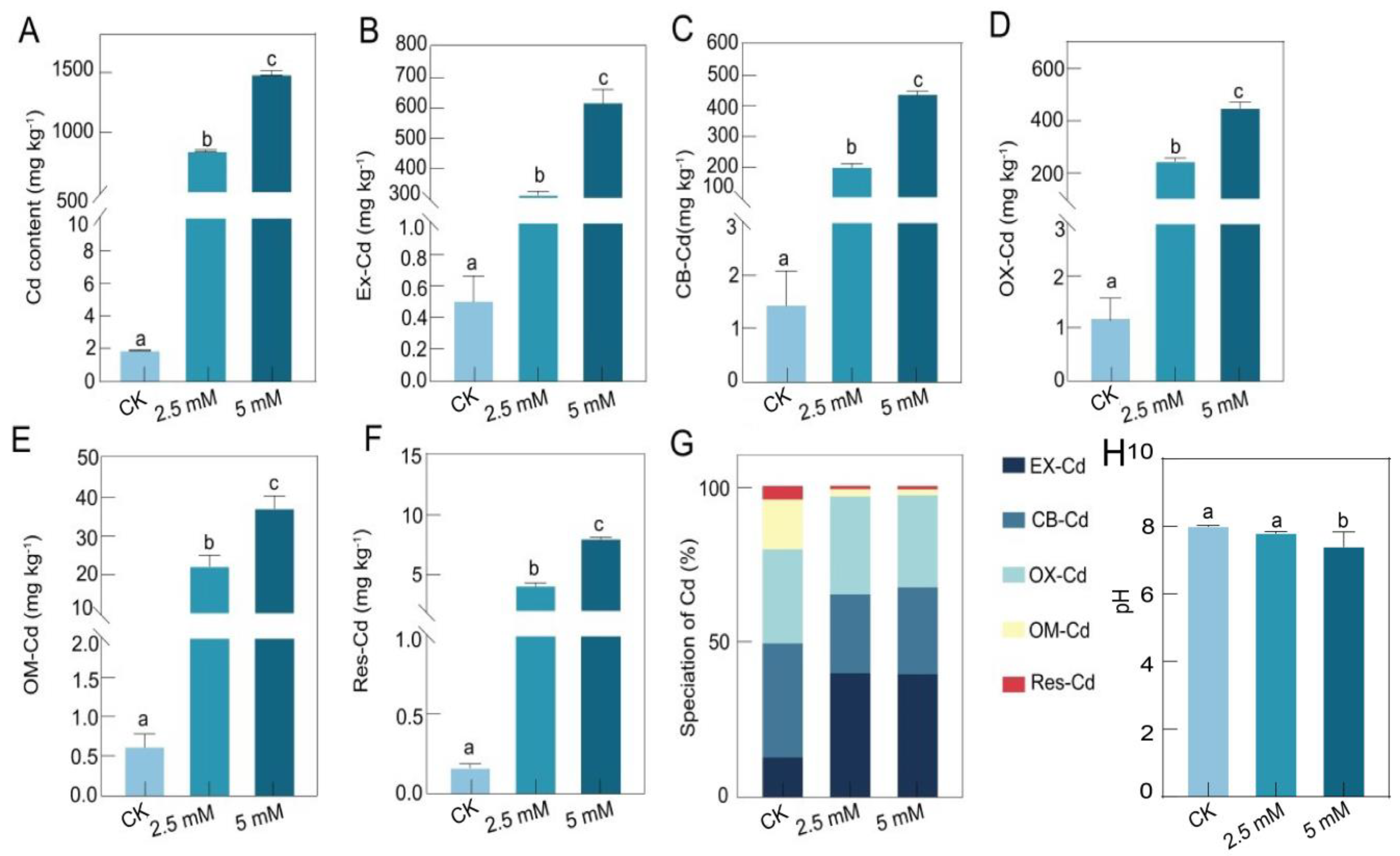
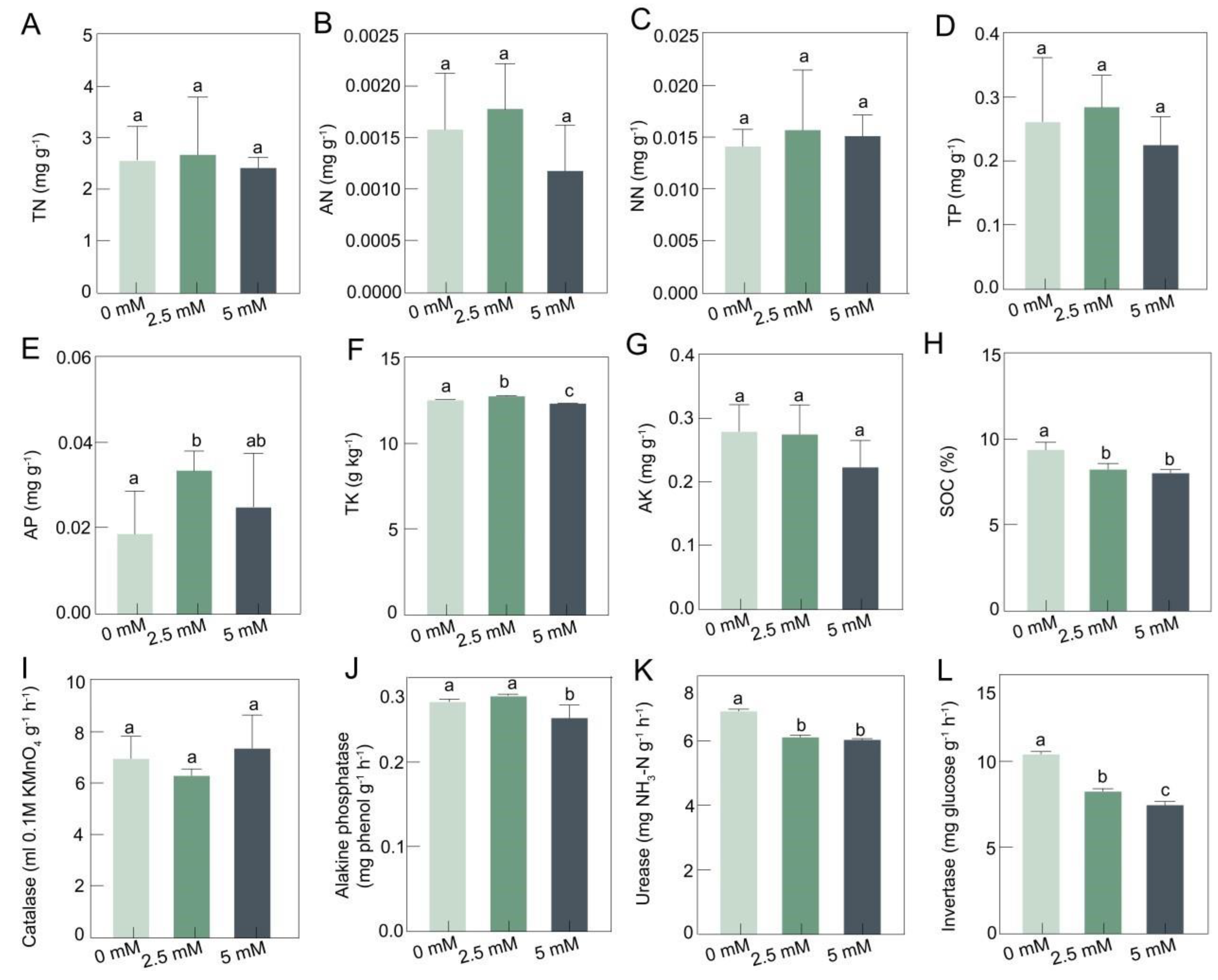
3.1. The Physicochemical Properties of Rhizosphere Soil at Different CdCl2 Concentrations
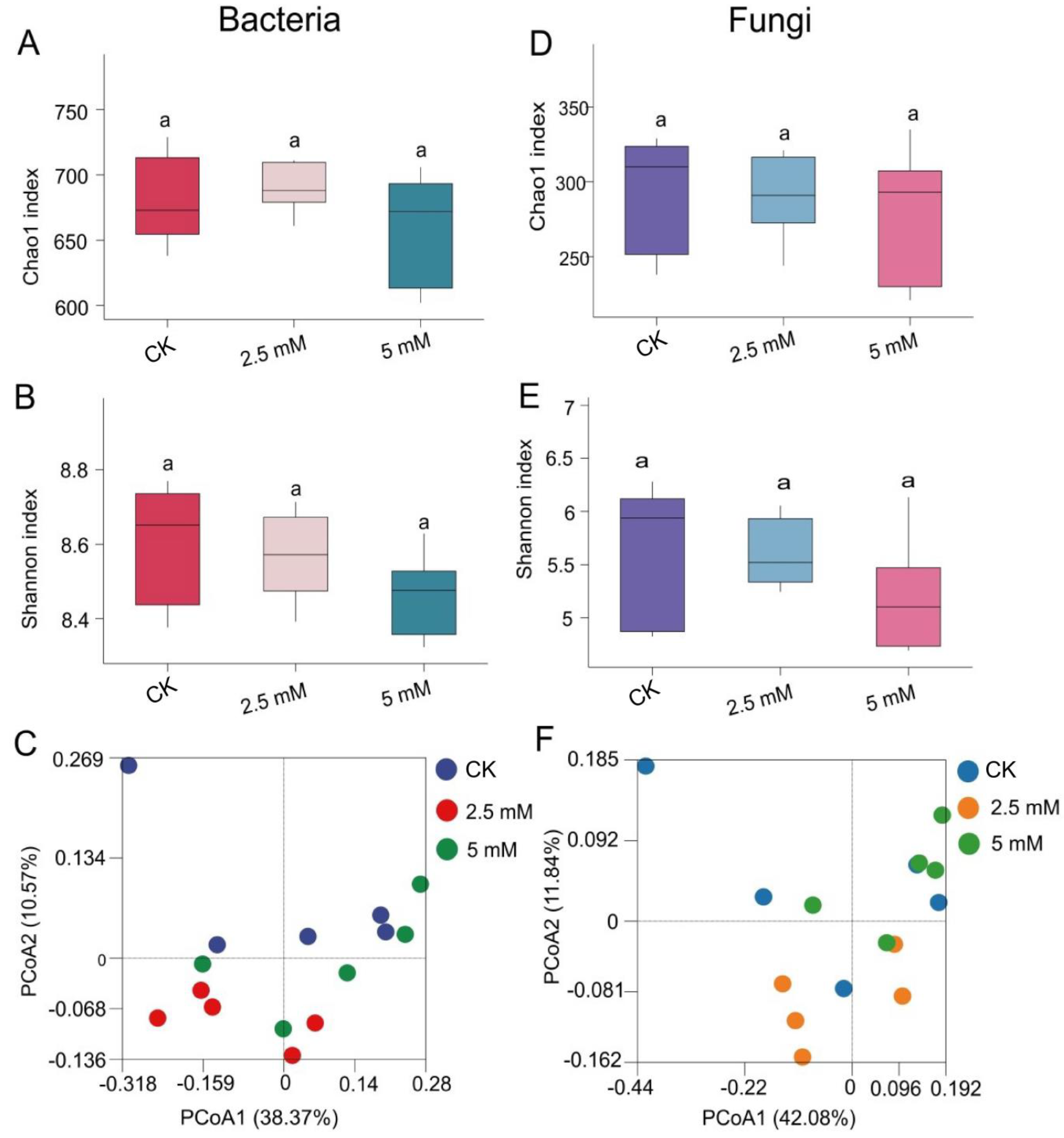
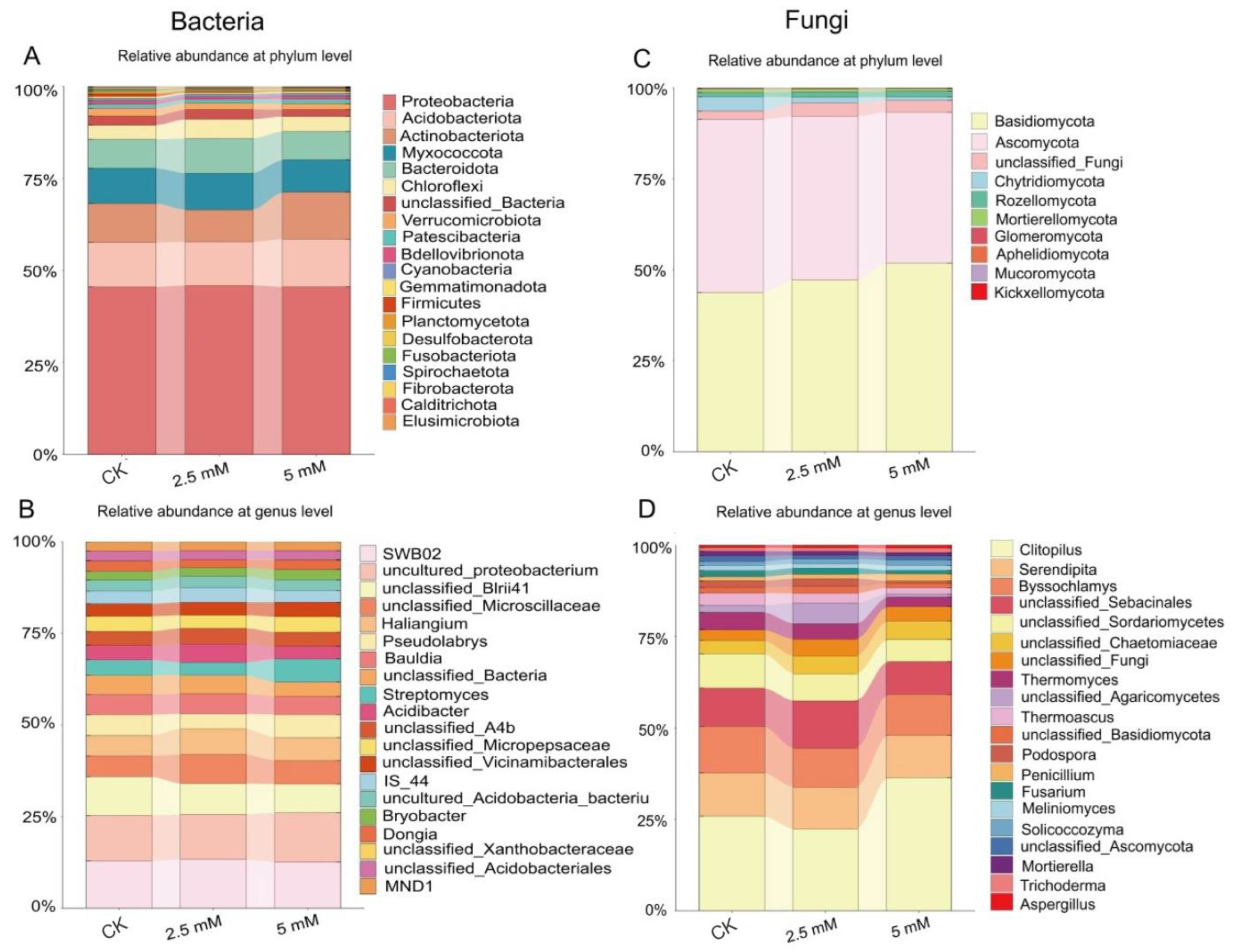
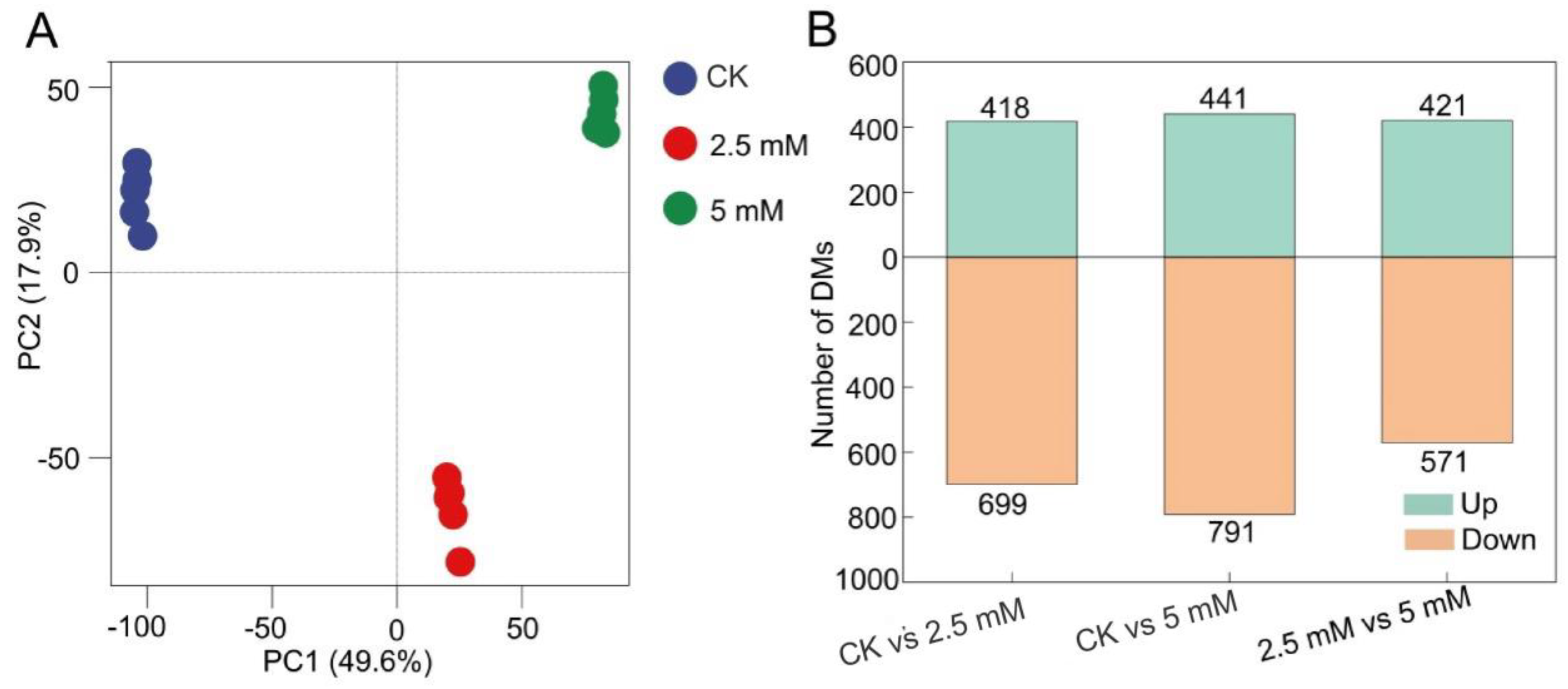
3.2. Effects of CdCl2 on Diversity and Composition of Rhizosphere Soil Bacterial and Fungal Communities
3.3. Measurement of Metabolites in Rhizosphere Soil Treated with CdCl2
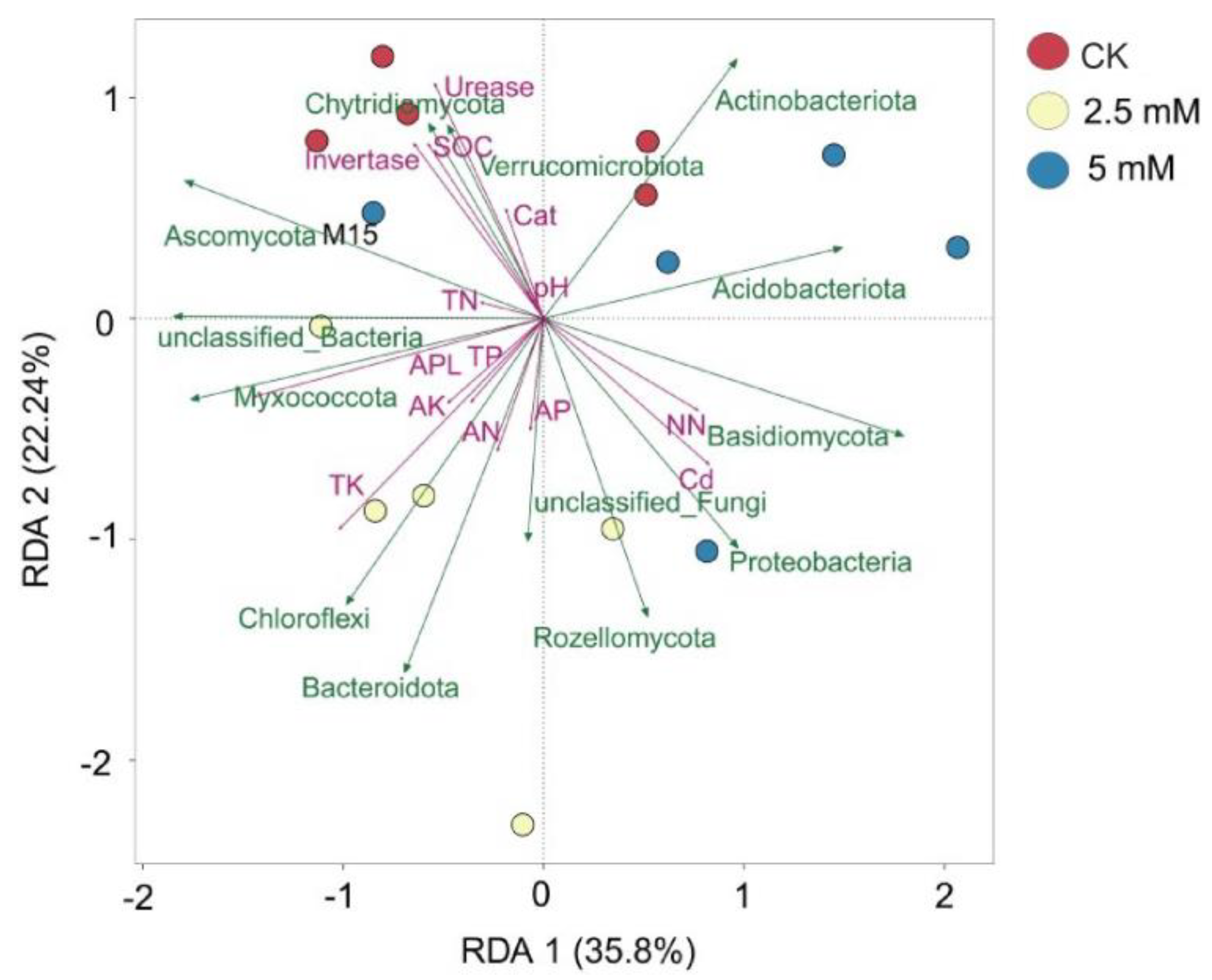
3.4. Correlations among Microbial Communities, Physicochemical Properties, and Metabolites in Rhizosphere Soil
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Mahar, A.; Ali, A.; Lahori, A.H.; Wahid, F.; Li, R.; Azeem, M.; Zhang, Z. Promising technologies for Cd-contaminated soils: Drawbacks and possibilities. In Environment, Climate, Plant and Vegetation Growth; Springer: Cham, Switzerland, 2020; pp. 63–91. [Google Scholar] [CrossRef]
- Adrees, M.; Ali, S.; Rizwan, M. The effect of excess copper on growth and physiology of important food crops: A review. Environ. Sci. Pollut. Res. Int. 2015, 22, 8148–8162. [Google Scholar] [CrossRef] [PubMed]
- Genchi, G.; Sinicropi, M.S.; Lauria, G.; Carocci, A.; Catalano, A. The Effects of Cadmium Toxicity. Int. J. Environ. Res. Public Health 2020, 17, 3782. [Google Scholar] [CrossRef]
- Ramakrishna, W.; Yadav, R.; Li, K. Plant growth promoting bacteria in agriculture: Two sides of a coin. Appl. Soil. Ecol. 2019, 138, 10–18. [Google Scholar] [CrossRef]
- Bertin, G.; Averbeck, D. Cadmium: Cellular effects, modifications of biomolecules, modulation of DNA repair and genotoxic consequences (a review). Biochimie 2006, 88, 1549–1559. [Google Scholar] [CrossRef]
- Rizwan, M.; Ali, S.; Abbas, T. Cadmium minimization in wheat: A critical review. Ecotoxicol. Environ. Saf. 2016, 130, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Rizwan, M.; Ali, S.; Adrees, M. A critical review on effects, tolerance mechanisms and management of cadmium in vegetables. Chemosphere 2017, 182, 90–105. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Wang, C.; Liu, R. Soil microbial community compositions and metabolite profiles of Achnatherum inebrians affect phytoremediation potential in Cd contaminated soil. J. Hazard. Mater. 2023, 459, 132280. [Google Scholar] [CrossRef] [PubMed]
- Schloter, M.; Dilly, O.; Munch, J.C. Indicators for evaluating soil quality. Agric. Ecosyst. Environ. 2003, 98, 255–262. [Google Scholar] [CrossRef]
- Sessitsch, A.; Kuffner, M.; Kidd, P. The role of plant-associated bacteria in the mobilization and phytoextraction of trace elements in contaminated soils. Soil Biol. Biochem. 2013, 60, 182–194. [Google Scholar] [CrossRef]
- Hema, T.G.; Getha, K.; Tan, G.; Sahira, H.L.; Syamil, A.M.; Fairuz, M.Y. Actinobacteria isolates from tin tailings and forest soil for bioremediation of heavy metals. J. Trop. For. Sci. 2014, 26, 153–162. [Google Scholar]
- Sathya, A.; Vijayabharathi, R.; Gopalakrishnan, S. Plant growth-promoting actinobacteria: A new strategy for enhancing sustainable production and protection of grain legumes. 3 Biotech 2017, 7, 102. [Google Scholar] [CrossRef]
- Chanclud, E.; Morel, J.B. Plant hormones: A fungal point of view. Mol. Plant Pathol. 2016, 17, 1289–1297. [Google Scholar] [CrossRef]
- Zhao, X.; Huang, J.; Lu, J.; Sun, Y. Study on the influence of soil microbial community on the long-term heavy metal pollution of different land use types and depth layers in mine. Ecotoxicol. Environ. Saf. 2019, 170, 218–226. [Google Scholar] [CrossRef] [PubMed]
- Korenblum, E.; Dong, Y.; Szymanski, J. Rhizosphere microbiome mediates systemic root metabolite exudation by root-to-root signaling. Proc. Natl. Acad. Sci. USA 2020, 117, 3874–3883. [Google Scholar] [CrossRef]
- Wang, Y.; Ren, W.; Li, Y.; Xu, Y.; Teng, Y.; Christie, P.; Luo, Y. Nontargeted metabolomic analysis to unravel the impact of di (2-ethylhexyl) phthalate stress on root exudates of alfalfa (Medicago sativa). Sci. Total Environ. 2019, 646, 212–219. [Google Scholar] [CrossRef] [PubMed]
- Zu, Y.; Li, Y.; Christian, S.; Laurent, L.; Liu, F. Accumulation of Pb, Cd, Cu and Zn in plants and hyperaccumulator choice in Lanping lead-zinc mine area, China. Environ. Int. 2004, 30, 567–576. [Google Scholar] [CrossRef]
- Tian, W.; Zhang, C.Q.; Qiao, P.; Milne, R. Diversity of culturable ericoid mycorrhizal fungi of Rhododendron decorum in Yunnan, China. Mycologia 2011, 103, 703–709. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.; Wang, C.; Wang, J.; Yang, Y.; Kong, X.; Liu, J.; Yi, Y. Integrative study of transcriptome and microbiome to reveal the response of Rhododendron decorum to cadmium stress. Ecotoxicol. Environ. Saf. 2024, 280, 116536. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.; Liu, J.; Hou, W.P.; Stubbendieck, R.M.; Xiong, H.; Jin, J.; Gong, J.Y.; Cheng, C.; Tang, X.X.; Liu, Y.L.; et al. Structural variability in the bulk soil, rhizosphere, and root endophyte fungal communities of Themeda japonica plants under different grades of karst rocky desertification. Plant Soil 2022, 475, 105–122. [Google Scholar] [CrossRef]
- Hou, W.; Wang, J.; Nan, Z.; Christensen, M.J.; Xia, C.; Chen, T.; Niu, X. Epichloë gansuensis endophyte-infection alters soil enzymes activity and soil nutrients at different growth stages of Achnatherum inebrians. Plant Soil 2020, 455, 227–240. [Google Scholar] [CrossRef]
- Hackenberger, D.K.; Palijan, G.; Lončarić, Ž.; Jovanović Glavaš, O.; Hackenberger, B.K. Influence of soil temperature and moisture on biochemical biomarkers in earthworm and microbial activity after exposure to propiconazole and chlorantraniliprole. Ecotoxicol. Environ. Saf. 2018, 148, 480–489. [Google Scholar] [CrossRef]
- He, D.; Cui, J.; Gao, M. Effects of soil amendments applied on cadmium availability, soil enzyme activity, and plant uptake in contaminated purple soil. Sci. Total Environ. 2019, 654, 1364–1371. [Google Scholar] [CrossRef] [PubMed]
- Giller, K.E.; Witter, E.; Steve, P. McGrath. Heavy metals and soil microbes. Soil Biol. Biochem. 2009, 41, 2031–2037. [Google Scholar] [CrossRef]
- Khan, J.; Wang, L.; Zhu, L.; Wang, J. Individual and combined effects of enrofloxacin and cadmium on soil microbial biomass and the ammonia-oxidizing functional gene. Sci. Total Environ. 2018, 624, 900–907. [Google Scholar] [CrossRef]
- Boampong, E.W. Uptake of Pb and Cd by Plant Species: A Review. Int. J. Innov. Sci. Res. Technol. 2020, 5, 306–310. [Google Scholar] [CrossRef]
- Yang, L.P.; Zhu, J.; Wang, P. Effect of Cd on growth, physiological response, Cd subcellular distribution and chemical forms of Koelreuteria paniculata. Ecotoxicol. Environ. Saf. 2018, 160, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Du, P.; Zhang, J.; Ji, J.; Xu, J.; Liang, B. Dopamine alleviates cadmium stress in apple trees by recruiting beneficial microorganisms to enhance the physiological resilience revealed by high-throughput sequencing and soil metabolomics. Hortic. Res. 2023, 10, uhad112. [Google Scholar] [CrossRef]
- Arfania, H.; Samadi, A.; Asadzadeh, F.; Sepehr, E.; Jaisi, D. Distribution of phosphorous pools in western river sediments of the Urmia Lake basin, Iran. Environ. Sci. Pollut. Res. Int. 2018, 25, 1614–11625. [Google Scholar] [CrossRef]
- Rafiq, M.T.; Aziz, R.; Yang, X.; Xiao, W.; Rafiq, M.K.; Ali, B.; Li, T. Cadmium phytoavailability to rice (Oryza sativa L.) grown in representative Chinese soils. A model to improve soil environmental quality guidelines for food safety. Ecotoxicol. Environ. Saf. 2014, 103, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Hussain, B.; Ashraf, M.N.; Shafeeq-Ur-Rahman; Abbas, A.; Li, J.; Farooq, M. Cadmium stress in paddy fields: Effects of soil conditions and remediation strategies. Sci. Total Environ. 2021, 754, 142188. [Google Scholar] [CrossRef]
- Li, Y.; Liang, L.; Li, W.; Ashraf, U.; Ma, L.; Tang, X. ZnO nanoparticlebased seed priming modulates early growth and enhances physio-biochemical and metabolic profiles of fragrant rice against cadmium toxicity. J. Nanobiotechnol. 2021, 19, 75. [Google Scholar] [CrossRef]
- Li, W.; Xu, B.; Song, Q.; Liu, X.; Xu, J.; Brookes, P.C. The identification of ‘hotspots’ of heavy metal pollution in soil-rice systems at a regional scale in eastern China. Sci. Total Environ. 2014, 472, 407–420. [Google Scholar] [CrossRef]
- Cui, J.; Wang, W.; Peng, Y. Effects of simulated Cd deposition on soil Cd availability, microbial response, and crop Cd uptake in the passivation-remediation process of Cd-contaminated purple soil. Sci. Total Environ. 2019, 683, 782–792. [Google Scholar] [CrossRef]
- Dong, M.; Feng, R.; Wang, R.; Sun, Y.; Ding, Y.; Xu, Y.; Fan, Z.; Guo, J. Inoculation of Fe/Mn-oxidizing bacteria enhances Fe/Mn plaque formation and reduces Cd and As accumulation in Rice Plant tissues. Plant Soil 2016, 404, 75–83. [Google Scholar] [CrossRef]
- Aponte, H.; Medina, J.; Butler, B.; Meier, S.; Cornejo, P.; Kuzyakov, Y. Soil quality indices for metal(loid) contamination: An enzymatic perspective. Land Degrad. Dev. 2020, 31, 2700–2719. [Google Scholar] [CrossRef]
- Benbi, D.K.; Brar, K.; Toor, A.S.; Singh, P. Total and labile pools of soil organic carbon in cultivated and undisturbed soils in Northern India. Geoderma 2015, 237–238, 149–158. [Google Scholar] [CrossRef]
- Jia, X.; Zhao, Y.; He, Y. Glomalin-related soil protein in the rhizosphere of Robinia pseudoacacia L. seedlings under higher air temperature combined with Cd-contaminated soil. Eur. J. Soil Sci. 2018, 69, 634–645. [Google Scholar] [CrossRef]
- Kabuyah, R.N.T.M.; van Dongen, B.E.; Bewsher, A.D.; Robinson, C.H. Decomposition of lignin in wheat straw in a sand-dune grassland. Soil Biol. Biochem. 2012, 45, 128–131. [Google Scholar] [CrossRef]
- Xiao, Z.; Duan, C.; Li, S. The microbial mechanisms by which long-term heavy metal contamination affects soil organic carbon levels. Chemosphere 2023, 340, 139770. [Google Scholar] [CrossRef]
- Adetunji, A.; Lewu, F.; Mulidzi, R.; Ncube, B. The biological activities of β-glucosidase, phosphatase and urease as soil quality indicators: A review. J. Soil Sci. Plant Nutr. 2017, 17, 794–807. [Google Scholar] [CrossRef]
- Bai, Y.; Li, F.; Yang, G.; Shi, S.; Dong, F.; Liu, M.; Hai, J. Meta-analysis of experimental warming on soil invertase and urease activities. Acta Agric. Scand. Sect. B—Soil Plant Sci. 2018, 68, 104–109. [Google Scholar] [CrossRef]
- Wang, Y.; Nie, M.; Diwu, Z.; Chang, F.; Nie, H.; Zhang, B.; Yin, Q. Toxicity evaluation of the metabolites derived from the degradation of phenanthrene by one of a soil ubiquitous PAHs-degrading strain Rhodococcus qingshengii FF. J. Hazard. Mater. 2021, 415, 125657. [Google Scholar] [CrossRef]
- Pan, J.; Yu, L. Effects of Cd or/and Pb on soil enzyme activities and microbial community structure. Ecol. Eng. 2011, 37, 1889–1894. [Google Scholar] [CrossRef]
- Ru, S.; Xing, J.; Su, D. Rhizosphere cadmium speciation and mechanisms of cadmium tolerance in different oilseed rape species. J. Plant Nutr. 2006, 29, 921–932. [Google Scholar] [CrossRef]
- Pan, X.; Zhang, S.; Zhong, Q. Effects of soil chemical properties and fractions of Pb, Cd, and Zn on bacterial and fungal communities. Sci. Total Environ. 2020, 715, 136904. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Seshadri, B.; Sarkar, B.; Wang, H.; Rumpel, C.; Sparks, D. Biochar modulates heavy metal toxicity and improves microbial carbon use efficiency in soil. Sci. Total Environ. 2018, 621, 148–159. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Nasir, M.; Lv, J.; Dai, Y.; Gao, J. Understanding the variation of microbial community in heavy metals contaminated soil using high throughput sequencing. Ecotoxicol. Environ. Saf. 2017, 144, 300–306. [Google Scholar] [CrossRef]
- Liu, H.; Wang, C.; Xie, Y.; Luo, Y.; Sheng, M.; Xu, F.; Xu, H. Ecological responses of soil microbial abundance and diversity to cadmium and soil properties in farmland around an enterprise-intensive region. J. Hazard. Mater. 2020, 392, 122478. [Google Scholar] [CrossRef]
- Meyer, A.; Lipson, D.; Martin, A.; Schadt, C.; Schmidt, S. Molecular and metabolic characterization of cold-tolerant alpine soil Pseudomonas sensu stricto. Appl. Environ. Microb. 2004, 70, 483–489. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Hou, S.; Peng, D.; Wang, Y.; Wang, C.; Xu, F.; Xu, H. Response of soil micro-ecology to different levels of cadmium in alkaline soil. Ecotoxicol. Environ. Saf. 2018, 166, 116–122. [Google Scholar] [CrossRef]
- Hou, D.; Lin, Z.; Wang, R. Cadmium Exposure-Sedum alfredii Planting Interactions Shape the Bacterial Community in the Hyperaccumulator Plant Rhizosphere. Appl. Environ. Microbiol. 2018, 84, e02797-17. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Shen, Y.; He, Y.; Gao, T.; Li, G.; Zuo, M. Effects of heavy metals on bacterial community structures in two lead–zinc tailings situated in northwestern China. Arch. Microbiol. 2022, 204, 78. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Meng, D.; Li, J.; Yin, H.; Liu, H.; Liu, X. Response of soil microbial communities and microbial interactions to long-term heavy metal contamination. Environ. Pollut. 2017, 231, 908–917. [Google Scholar] [CrossRef] [PubMed]
- Narendrula-Kotha, R.; Nkongolo, K.K. Bacterial and fungal community structure and diversity in a mining region under long-term metal exposure revealed by metagenomics sequencing. Ecol. Genet. Genom. 2017, 2, 13–24. [Google Scholar] [CrossRef]
- Arif, S.; Nacke, H.; Schliekmann, E.; Reimer, A.; Arp, G.; Hoppert, M. Composition and niche-specific characteristics of microbial consortia colonizing Marsberg copper mine in the Rhenish massif. Biogeosciences 2022, 19, 4883–4902. [Google Scholar] [CrossRef]
- Van Bruggen, A.H.C.; Semenov, A.M. In search of biological indicators for soil health and disease suppression. Appl. Soil Ecol. 2012, 15, 13–24. [Google Scholar] [CrossRef]
- Rastogi, G.; Barua, S.; Sani, R.K.; Peyton, B.M. Investigation of microbial populations in the extremely metal-contaminated Coeur d’Alene River sediments. Microb. Ecol. 2011, 62, 1–13. [Google Scholar] [CrossRef]
- Eo, J.; Park, K. Long-term effects of imbalanced fertilization on the composition and diversity of soil bacterial community. Agric. Ecosyst. Environ. 2016, 231, 176–182. [Google Scholar] [CrossRef]
- Fierer, N.; Lauber, C.L.; Ramirez, K.S.; Zaneveld, J.; Bradford, M.A.; Knight, R. Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. ISME J. 2012, 6, 1007–1017. [Google Scholar] [CrossRef]
- Islam, Z.F.; Cordero, P.R.F.; Feng, J.; Chen, Y.J.; Bay, S.K.; Jirapanjawat, T. Two Chloroflexi classes independently evolved the ability to persist on atmospheric hydrogen and carbon monoxide. ISME J. 2019, 13, 1801–1813. [Google Scholar] [CrossRef]
- Will, C.; Thürmer, A.; Wollherr, A.; Nacke, H.; Herold, N.; Schrumpf, M. Horizon-specific bacterial community composition of German grassland soils, as revealed by pyrosequencing-based analysis of 16S rRNA genes. Appl. Environ. Microbiol. 2010, 76, 6751–6759. [Google Scholar] [CrossRef] [PubMed]
- Fernández, N.; Sierra-Alvarez, R.; Field, J.A.; Amils, R.; Sanz, J. Microbial community dynamics in a chemolithotrophic denitrification reactor inoculated with methanogenic granular sludge. Chemosphere 2008, 70, 462–474. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Saleem, M.; Xie, Z.; Wei, X.; He, T.; He, G. Sensitive or tolerant functional microorganisms under cadmium stress: Suggesting potential specific interaction network characteristics in the rhizosphere system of karst potato. Environ. Sci. Pollut. Res. Int. 2023, 30, 55932–55947. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Fan, J.; Liu, R.; Zeng, G.; Chen, A.; Zou, Z. Removal of Cd (II), Cu (II) and Zn (II) from aqueous solutions by live Phanerochaete chrysosporium. Environ. Technol. 2012, 33, 2653–2659. [Google Scholar] [CrossRef]
- Li, C.; Huang, H.; Gu, X.; Zhong, K.; Yin, J.; Mao, J.; Zhang, C. Accumulation of heavy metals in rice and the microbial response in a contaminated paddy field. J. Soils Sediments 2024, 24, 644–656. [Google Scholar] [CrossRef]
- James, T.Y.; Pelin, A.; Bonen, L.; Ahrendt, S.; Sain, D.; Corradi, N.; Stajich, J. Shared signatures of parasitism and phylogenomics unite Cryptomycota and microsporidia. Curr. Biol. 2013, 23, 548–1553. [Google Scholar] [CrossRef]
- Gundacker, C.; Gencik, M.; Hengstschläger, M. The relevance of the individual genetic background for the toxicokinetics of two significant neurodevelopmental toxicants: Mercury and lead. Mutat. Res. 2010, 705, 130–140. [Google Scholar] [CrossRef]
- Mohapatra, B.R.; Gould, W.D.; Dinardo, O.; Koren, D.W. Tracking the prokaryotic diversity in acid mine drainage-contaminated environments: A review of molecular methods. Miner. Eng. 2011, 24, 709–718. [Google Scholar] [CrossRef]
- Anjitha, K.S.; Sameena, P.P.; Puthur, J.T. Functional aspects of plant secondary metabolites in metal stress tolerance and their importance in pharmacology. Plant Stress 2021, 2, 100038. [Google Scholar] [CrossRef]
- Li, X.; Wang, S.; Guo, L.; Huang, L. Effect of cadmium in the soil on growth, secondary metabolites and metal uptake in Salvia miltiorrhiza. Toxicol. Environ. Chem. 2013, 95, 1525–1538. [Google Scholar] [CrossRef]
- Xiong, H.; Duan, C.; Xinxiang, A.; Chen, M. Response of Scutellarin Content to Heavy Metals in Erigeron breviscapus. Int. J. Environ. Sci. Dev. 2013, 4, 277–281. [Google Scholar] [CrossRef]
- Haichar, F.Z.; Marol, C.; Berge, O.; Rangel-Castro, J.I.; Prosser, J.I.; Balesdent, J.; Heulin, T.; Achouak, W. Plant host habitat and root exudates shape soil bacterial community structure. ISME J. 2008, 2, 1221–1230. [Google Scholar] [CrossRef]
- Chaparro, J.M.; Badri, D.V.; Vivanco, J.M. Rhizosphere microbiome assemblage is affected by plant development. ISME J. 2014, 8, 790–803. [Google Scholar] [CrossRef]
- Lareen, A.; Burton, F.; Schäfer, P. Plant root-microbe communication in shaping root microbiomes. Plant Mol. Biol. 2016, 90, 575–587. [Google Scholar] [CrossRef]
- Santolini, M.; Barabási, A.L. Predicting perturbation patterns from the topology of biological networks. Proc. Natl. Acad. Sci. USA 2018, 115, E6375–E6383. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Zhong, S.; Luo, K.; Yang, S.; Xia, J.; Chen, Q. Effect of metal pollution on the distribution and co-occurrence pattern of bacterial, archaeal and fungal communities throughout the soil profiles. Chemosphere 2023, 315, 137692. [Google Scholar] [CrossRef]
- Mougi, A.; Kondoh, M. Diversity of interaction types and ecological community stability. Science 2012, 337, 349–351. [Google Scholar] [CrossRef]
- Wagg, C.; Schlaeppi, K.; Banerjee, S.; Kuramae, E.E.; van der Heijden, M.G.A. Fungal-bacterial diversity and microbiome complexity predict ecosystem functioning. Nat. Commun. 2019, 10, 4841. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zhou, C.; Wu, Y.; An, Q.; Zhang, J.; Fang, Y.; Li, J.Q.; Pan, C. Nanoselenium integrates soil-pepper plant homeostasis by recruiting rhizosphere beneficial microbiomes and allocating signaling molecule levels under Cd stress. J. Hazard. Mater. 2022, 432, 128763. [Google Scholar] [CrossRef]
- Hanrahan-Tan, D.G.; Henderson, L.; Kertesz, M.A.; Lilje, O. The Effects of Nitrogen and Phosphorus on Colony Growth and Zoospore Characteristics of Soil Chytridiomycota. J. Fungi 2022, 8, 341. [Google Scholar] [CrossRef]

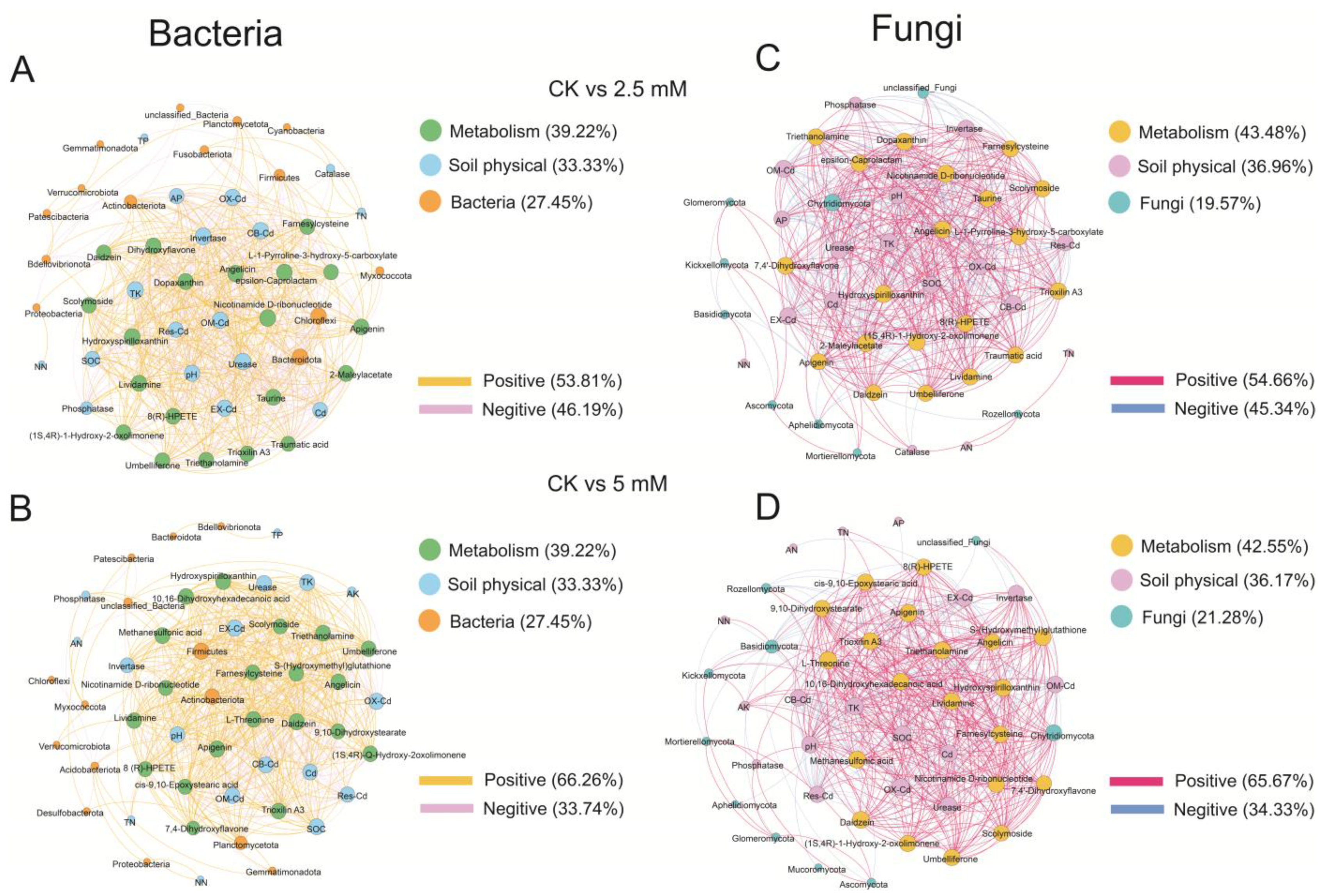
| Total Edges | Positive Edges | Negative Edges | Connectance | Average Degree | Average Pathway Length | Diameter | Clustering Coefficient | ||
|---|---|---|---|---|---|---|---|---|---|
| Bac | CK vs. 2.5 mM | 630 | 339 | 291 | 0.49 | 24.7 | 1.31 | 4.79 | 0.87 |
| CK vs. 5 mM | 569 | 377 | 192 | 0.45 | 22.3 | 1.33 | 4.20 | 0.90 | |
| Fun | CK vs. 2.5 mM | 558 | 305 | 253 | 0.54 | 24.3 | 0.96 | 2.88 | 0.90 |
| CK vs. 5 mM | 533 | 350 | 183 | 0.49 | 22.7 | 1.15 | 2.87 | 0.89 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, M.; Chen, L.; Wang, L.; Yi, Y.; Wang, J.; Wang, C.; Chen, X.; Liu, J.; Yang, Y.; Malik, K.; et al. Comprehensive Analysis of Microbiomes and Metabolomics Reveals the Mechanism of Adaptation to Cadmium Stress in Rhizosphere Soil of Rhododendron decorum subsp. Diaprepes. Horticulturae 2024, 10, 884. https://doi.org/10.3390/horticulturae10080884
Tang M, Chen L, Wang L, Yi Y, Wang J, Wang C, Chen X, Liu J, Yang Y, Malik K, et al. Comprehensive Analysis of Microbiomes and Metabolomics Reveals the Mechanism of Adaptation to Cadmium Stress in Rhizosphere Soil of Rhododendron decorum subsp. Diaprepes. Horticulturae. 2024; 10(8):884. https://doi.org/10.3390/horticulturae10080884
Chicago/Turabian StyleTang, Ming, Lanlan Chen, Li Wang, Yin Yi, Jianfeng Wang, Chao Wang, Xianlei Chen, Jie Liu, Yongsong Yang, Kamran Malik, and et al. 2024. "Comprehensive Analysis of Microbiomes and Metabolomics Reveals the Mechanism of Adaptation to Cadmium Stress in Rhizosphere Soil of Rhododendron decorum subsp. Diaprepes" Horticulturae 10, no. 8: 884. https://doi.org/10.3390/horticulturae10080884
APA StyleTang, M., Chen, L., Wang, L., Yi, Y., Wang, J., Wang, C., Chen, X., Liu, J., Yang, Y., Malik, K., & Gong, J. (2024). Comprehensive Analysis of Microbiomes and Metabolomics Reveals the Mechanism of Adaptation to Cadmium Stress in Rhizosphere Soil of Rhododendron decorum subsp. Diaprepes. Horticulturae, 10(8), 884. https://doi.org/10.3390/horticulturae10080884








