Construction of an Editing System for Forest Tree Genomes Based on an Efficient Visual Screening Marker in Eucalyptus urophylla × Eucalyptus grandis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Test Materials
2.2. Main Test Reagents
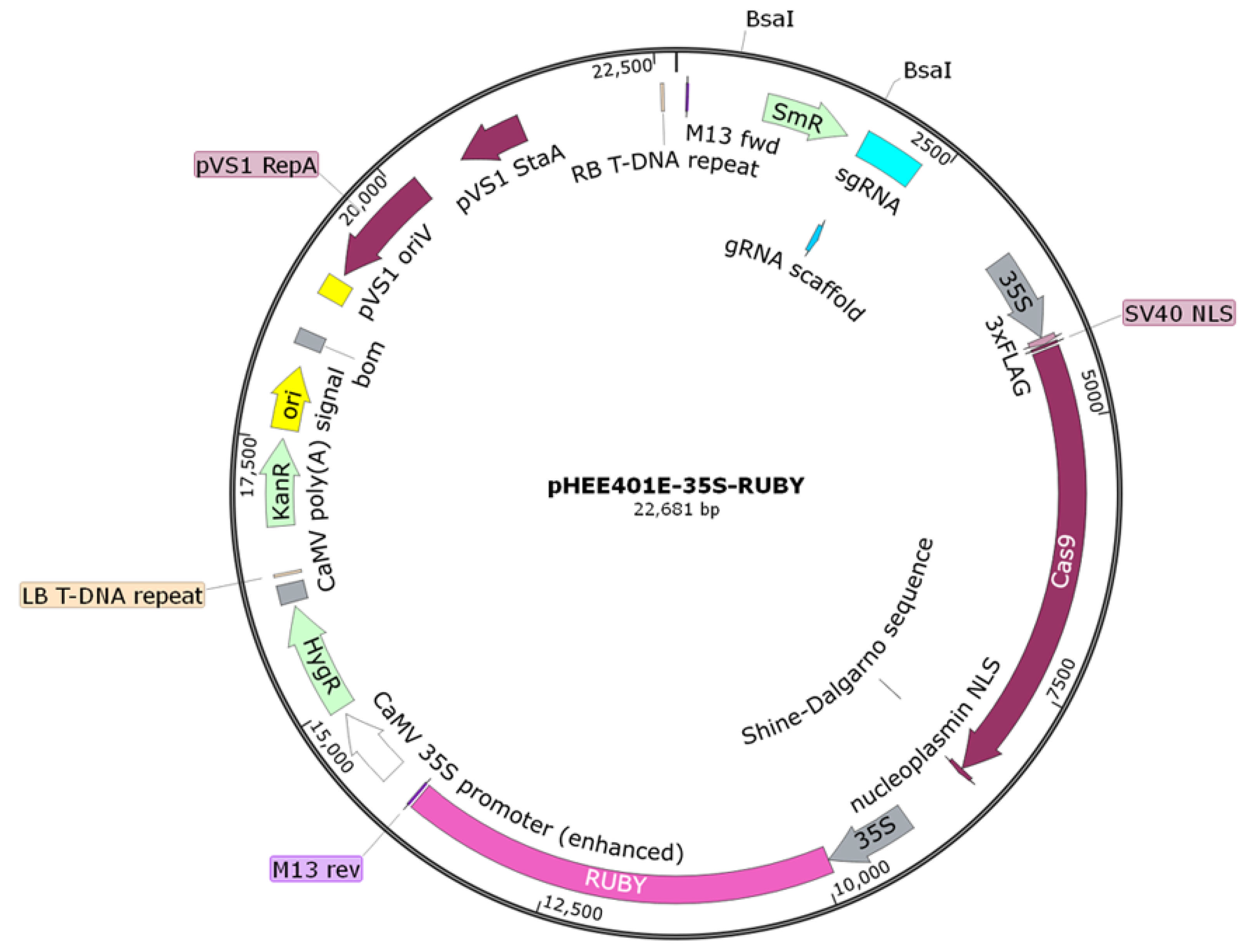
2.3. The Construction, Transformation, and Verification of the pHEE401E-35S-RUBY-EuC4H Vector
2.3.1. Primer Design
2.3.2. Construction of the pHEE401E-35S-RUBY Vector
- (1)
- Amplification of the 35S promoter and RUBY target fragment
- (2)
- Fusion PCR amplification of the 35S-RUBY fragment
- (3)
- Restriction digestion and homologous recombination of the pHEE401E-35S-Cas9 plasmid
- (4)
- Transformation of the recombinant plasmid into E. coli DH5α competent cells and the identification of transformants
2.3.3. Construction of the pHEE401E-35S-RUBY-EuC4H Vector
- (1)
- Amplification of small guide RNA (sgRNA) fragments containing two specific target sites
- (2)
- Restriction digestion and homologous recombination of the pHEE401E-35S-RUBY plasmid
- (3)
- Transformation of the recombinant plasmid into E. coli DH5α competent cells and the identification of transformants
- (4)
- Preparing the engineered bacterial culture
2.3.4. Transformation and Identification of the pHEE401E-35S-RUBY-EuC4H Plasmid
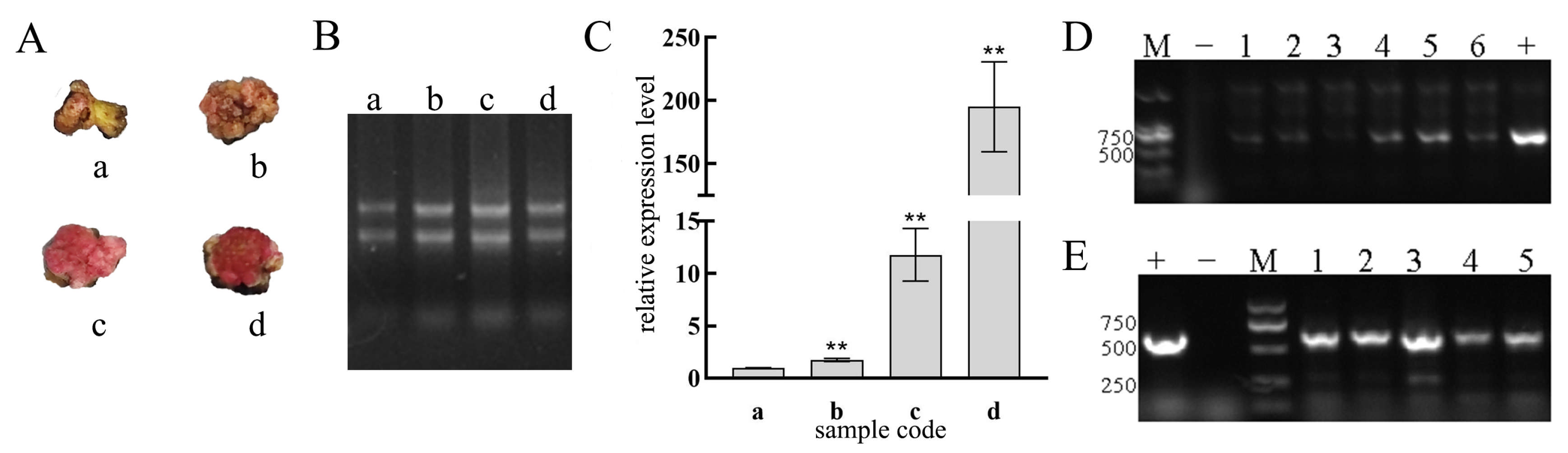
2.3.5. Correlation Between the Degree of Red Coloration and RUBY Gene Expression Levels in the Callus Tissue of E. urophylla × E. grandis
3. Results
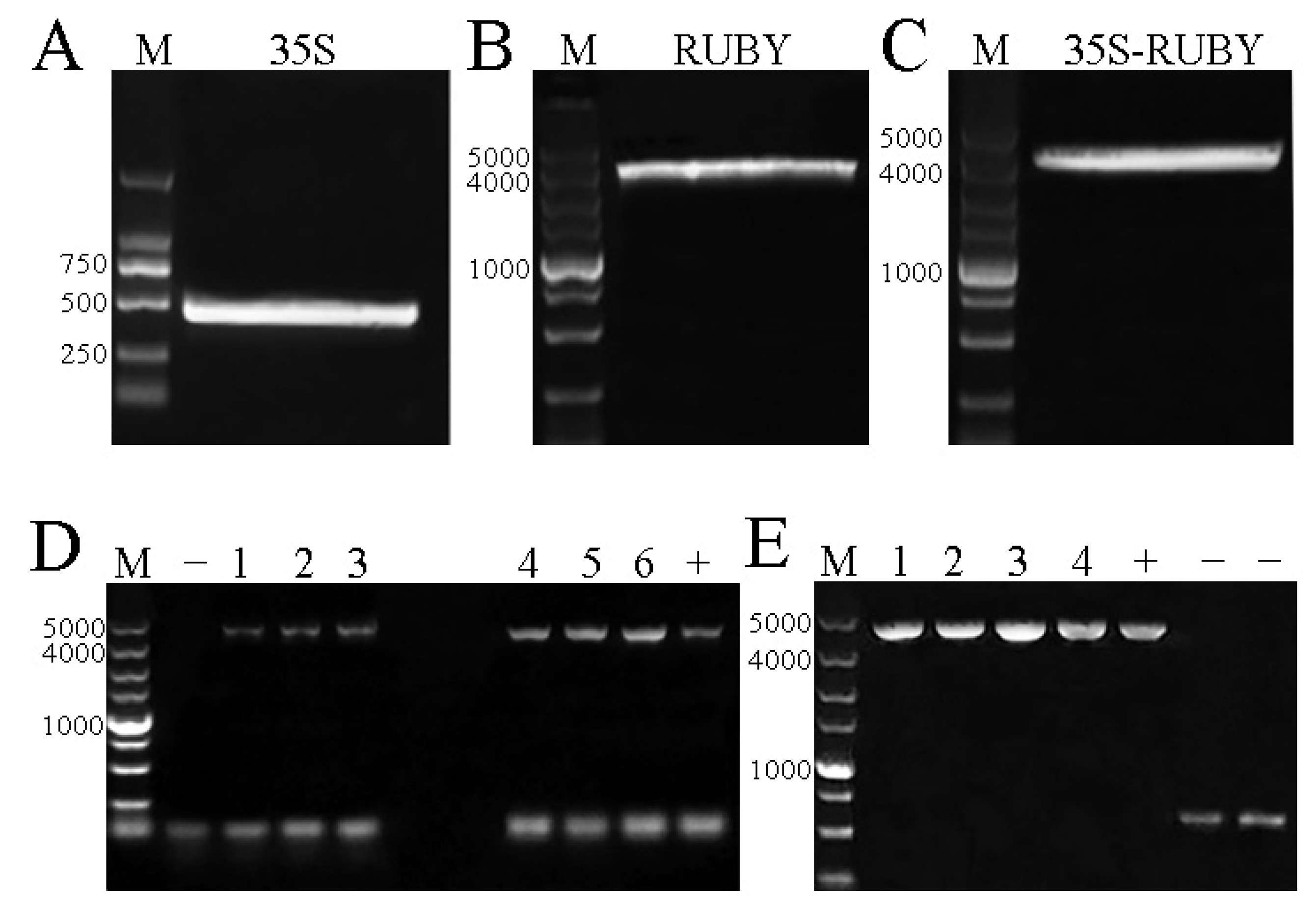
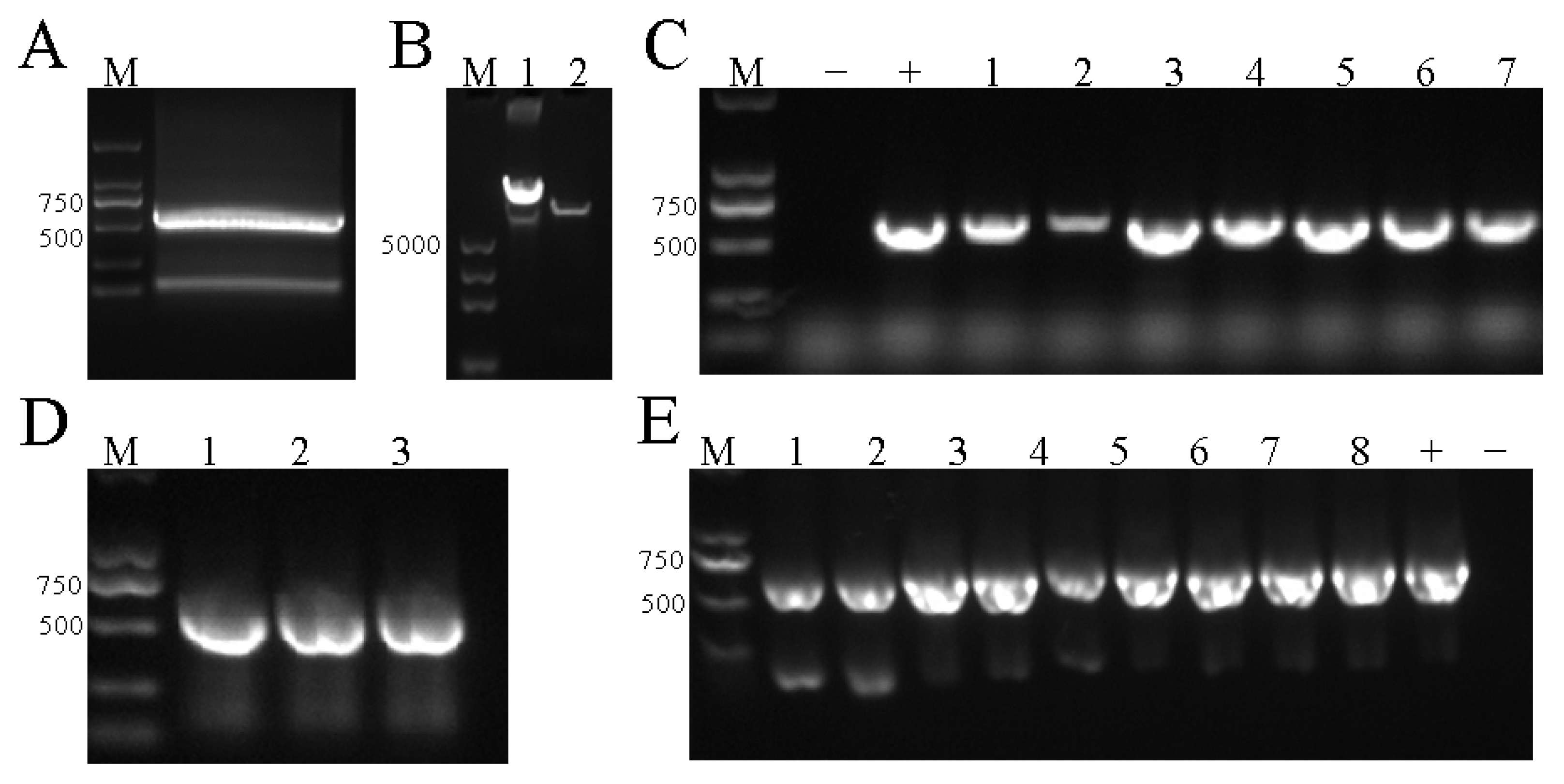
3.1. The Construction Results of the pHEE401E-35S-RUBY Vector
3.2. The Construction Results of the pHEE401E-35S-RUBY-EuC4H Vector
3.3. Transformation and Identification of the pHEE401E-35S-RUBY-EuC4H Vector
4. Discussion
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Hong, W.; Huang, J.-H.; Li, J.; Lin, H.; Chen, C.; Wu, C.-Z. Study on the δ13C Value in Different Eucalyptus Species. J. Fujian Coll. For. 2008, 28, 289–293. [Google Scholar] [CrossRef]
- Liu, T.; Xie, Y.-J. Studies on the Causes of Rapid Development of Eucalyptus Plantations in China. Eucalypt Sci. Technol. 2020, 37, 38–47. [Google Scholar] [CrossRef]
- Tang, Q.-H.; Chen, Y.-P. Status Quo and Prospect of Wood Processing and Utilization of Eucalyptus in China and Abroad. China Wood-Based Panels 2020, 27, 18–21. [Google Scholar]
- Huang, G.-Q.; Zhao, Q.-G. The History, Status Quo, Ecological Problems and Countermeasures of Eucalyptus Plantations in Guangxi. Acta Ecol. Sin. 2014, 34, 5142–5152. [Google Scholar] [CrossRef]
- Xie, Y.-J. Analysis of China’s Wood Resource Security and the Contributions of Eucalyptus Plantation. Eucalypt Sci. Technol. 2018, 35, 3–6. [Google Scholar] [CrossRef]
- Liu, Y.-T. Analysis of Enzymatic Properties of Ferulic Aid Esterase from Clostridium thermocellum and Construction of Its High-Efficiency Plant Expression Vector. Master’s Thesis, Jiangsu University, Zhenjiang, China, 2017. [Google Scholar]
- Feng, H.-X.; Tu, Y. Research on Lignin Biosynthesis. J. Qingdao Univ. Nat. Sci. Ed. 2018, 31, 46–54. [Google Scholar]
- Chen, B.-W. Cloning of Key Genes Involved in Lignin Monomer Biosynthesis from Eucalyptus urophylla and Transformation in Tobacco. Ph.D. Thesis, Beijing Forestry University, Beijing, China, 2017. [Google Scholar]
- Vanholme, R.; De Meester, B.; Ralph, J.; Boerjan, W. Lignin Biosynthesis and Its Integration into Metabolism. Curr. Opin. Biotechnol. 2019, 56, 230–239. [Google Scholar] [CrossRef]
- Li, W.; Yang, L.-X.; Jiang, L.-Z.; Zhang, G.-L.; Luo, Y.-G. Molecular Cloning and Functional Characterization of a Cinnamate 4-Hydroxylase-Encoding Gene from Camptotheca Acuminata. Acta Physiol. Plant. 2016, 38, 1–9. [Google Scholar] [CrossRef]
- Yan, Q.; Si, J.-R.; Cui, X.-X.; Peng, H.; Chen, X.; Xing, H.; Dou, D.-L. The Soybean Cinnamate 4-Hydroxylase Gene GmC4H1 Contributes Positively to Plant Defense via Increasing Lignin Content. Plant Growth Regul. 2019, 88, 139–141. [Google Scholar] [CrossRef]
- Blount, J.W.; Korth, K.L.; Masoud, S.A.; Rasmussen, S.; Lamb, C.; Dixon, R.A. Altering Expression of Cinnamic Acid 4-Hydroxylase in Transgenic Plants Provides Evidence for a Feedback Loop at the Entry Point into the Phenylpropanoid Pathway. Plant Physiol. Bethesda 2000, 122, 107–116. [Google Scholar] [CrossRef]
- Simmons, C.R.; Grant, S.; Altier, D.J.; Dowd, P.F.; Crasta, O.; Folkerts, O.; Yalpani, N. Maize rhm1 Resistance to Bipolaris Maydis Is Associated with Few Differences in Pathogenesis-Related Proteins and Global mRNA Profiles. Mol. Plant-Microbe Interact. 2001, 14, 947–954. [Google Scholar] [CrossRef] [PubMed]
- Sykes, R.W.; Gjersing, E.L.; Foutz, K.; Rottmann, W.H.; Kuhn, S.A.; Foster, C.E.; Ziebell, A.; Turner, G.B.; Decker, S.R.; Hinchee, M.A.W.; et al. Down-Regulation of p-Coumaroyl Quinate/Shikimate 3′-Hydroxylase (C3′H) and Cinnamate 4-Hydroxylase (C4H) Genes in the Lignin Biosynthetic Pathway of Eucalyptus urophylla × E. grandis Leads to Improved Sugar Release. Biotechnol. Biofuels 2015, 8, 128. [Google Scholar] [CrossRef] [PubMed]
- Franke, R.; McMichael, C.M.; Meyer, K.; Shirley, A.M.; Cusumano, J.C.; Chapple, C. Modified Lignin in Tobacco and Poplar Plants Over-expressing the Arabidopsis Gene Encoding Ferulate 5-hydroxylase. Plant J. Cell Mol. Biol. 2000, 22, 223–234. [Google Scholar] [CrossRef]
- Polturak, G.; Aharoni, A. Advances and Future Directions in Betalain Metabolic Engineering. New Phytol. 2019, 224, 1472–1478. [Google Scholar] [CrossRef]
- He, Y.-B.; Zhang, T.; Sun, H.; Zhan, H.-D.; Zhao, Y.-D. A Reporter for Noninvasively Monitoring Gene Expression and Plant Transformation. Hortic. Res. 2020, 7, 152. [Google Scholar] [CrossRef]
- Tanaka, Y.; Kojima, M.; Uritani, I. Properties, Development and Cellular-Localization of Cinnamic Acid 4-Hydroxylase in Cut-Injured Sweet Potato. Plant Cell Physiol. 1974, 15, 843–854. [Google Scholar] [CrossRef]
- Wang, Z.-C.; Liu, Y.-M.; Ouyang, L.-J.; Li, L.-M.; Liang, C.-Y.; Pan, J.-Y. Construction and Verification of an Eucalyptus Gene Editing Vector with Visual Selection Markers. Bull. Bot. Res. 2021, 41, 816–823. [Google Scholar] [CrossRef]
- Ji, X.; Yang, B.; Wang, D.-W. Achieving Plant Genome Editing While Bypassing Tissue Culture. Trends Plant Sci. 2020, 25, 427–429. [Google Scholar] [CrossRef]
- Chen, G. Common Problems and Countermeasures in Rapid Propagation Technology of Forest Tissue Culture. Prot. For. Sci. Technol. 2017, 4, 100–102. [Google Scholar] [CrossRef]
- Su, M.; Liu, Y.-M.; Li, L.-M.; Zhang, Y.-X.; Huang, H.; Guo, K.-X. Metabolomics Analysis and Differential Expression of Related Genes in Different Callus of Eucalyptus urophylla × E. grandis. Plant Physiol. J. 2022, 59, 943–953. [Google Scholar] [CrossRef]
- Stewart, D.; Yahiaoui, N.; McDougall, G.J.; Myton, K.; Marque, C.; Boudet, A.M.; Haigh, J. Fourier-Transform Infrared and Raman Spectroscopic Evidence for the Incorporation of Cinnamaldehydes into the Lignin of Transgenic Tobacco (Nicotiana tabacum L.) Plants with Reduced Expression of Cinnamyl Alcohol Dehydrogenase. Planta 1997, 201, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.-Q.; Han, G.-Y.; Liu, N.-X.; Zhou, Q. A Bioinformatics Study on Key Enzymes Involved in Tyrosine Metabolism in Beet. Chin. Agric. Sci. Bull. 2024, 40, 124–131. [Google Scholar] [CrossRef]
- Xu, J.-J.; Fang, X.; Li, C.-Y.; Yang, L.; Chen, X.-Y. General and Specialized Tyrosine Metabolism Pathways in Plants. aBIOTECH 2019, 1, 97–105. [Google Scholar] [CrossRef] [PubMed]



| Primer Name | Primer Sequence (5′→3′) | Purpose | Length/bp |
|---|---|---|---|
| EuC4H-gF | ctagagtcgaagtagtgattgCATATATGGGCTCGAGAAG gttttagagctagaaatagc | Constructed plasmids | 592 |
| EuC4H-gR | tgctatttctagctctaaaacACCTATGAATCGAACTCGT caatcactacttcgactcta | ||
| R35S-F | TAATGCATTTTATGACTTGCAACATGGTGGA GCACGACAC | 462 | |
| R35S-R | GCGAGGGTCGCATGATCCATCGTGTCCTCT CCAAATGAAA | ||
| RRUBY-F | TTTCATTTGGAGAGGACACGATGGATCAT GCGACCCTCG | 3957 | |
| RRUBY-F | GAATTCGTTGTCAATCAATTTCACTATCACTG GAGGCTTG | ||
| EuC4H-GT1 | TATCATCGAAAATCGCGGCGG | Identification primers | 736 |
| EuC4H-GT2 | ACCTCTCGAAAAATTGCTTGCC | ||
| RUBY-GT1 | CTCACAACTCCGCTCAACGC | 725 | |
| RUBY-GT2 | GAGTCCGGCTCTTTGAGGCT | ||
| RUBY-qF | CGCCACACTCCTCCAGTTCTTC | qPCR primers | _ |
| RUBY-qR | CGTCCTCGCCGTTCATCATCTT | ||
| EuActin-F | GCACCGCCAGAGAGGAAATA | ||
| EuActin-R | GAAGCACTTCCTGTGGACGA |
| Temperature | Step | Time |
|---|---|---|
| 98 °C | Initialization | 2 min |
| 98 °C | Denaturation | 10 s |
| 58 °C | Annealing | 15 s |
| 72 °C | Elongation | 5 s/kb |
| Temperature | Step | Time |
|---|---|---|
| 94 °C | Initialization | 5 min |
| 94 °C | Denaturation | 30 s |
| 56 °C | Annealing | 30 s |
| 72 °C | Extension | 60 s/kb |
| 72 °C | Complete extension | 7 min |
| Experiment | Explants | Transgenic Buds | Edited Buds | Transformation Efficiency | Gene-Editing Efficiency |
|---|---|---|---|---|---|
| I | 600 | 413 | 1 | 68.8% | 0.0876% |
| II | 600 | 357 | 0 | 59.5% | |
| III | 600 | 371 | 0 | 61.8% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, M.; Wu, X.; Wang, Z.; Li, L.; Ouyang, L. Construction of an Editing System for Forest Tree Genomes Based on an Efficient Visual Screening Marker in Eucalyptus urophylla × Eucalyptus grandis. Horticulturae 2025, 11, 406. https://doi.org/10.3390/horticulturae11040406
Su M, Wu X, Wang Z, Li L, Ouyang L. Construction of an Editing System for Forest Tree Genomes Based on an Efficient Visual Screening Marker in Eucalyptus urophylla × Eucalyptus grandis. Horticulturae. 2025; 11(4):406. https://doi.org/10.3390/horticulturae11040406
Chicago/Turabian StyleSu, Min, Xinlin Wu, Zechen Wang, Limei Li, and Lejun Ouyang. 2025. "Construction of an Editing System for Forest Tree Genomes Based on an Efficient Visual Screening Marker in Eucalyptus urophylla × Eucalyptus grandis" Horticulturae 11, no. 4: 406. https://doi.org/10.3390/horticulturae11040406
APA StyleSu, M., Wu, X., Wang, Z., Li, L., & Ouyang, L. (2025). Construction of an Editing System for Forest Tree Genomes Based on an Efficient Visual Screening Marker in Eucalyptus urophylla × Eucalyptus grandis. Horticulturae, 11(4), 406. https://doi.org/10.3390/horticulturae11040406





