A Novel Transcription Factor of Regulating Ag-8 Biocontrol to Grapevine Crown Gall
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant, Bacterial, and Pathogenic Culture Conditions
2.2. Construction of Deletion Strain, Complemented Strain
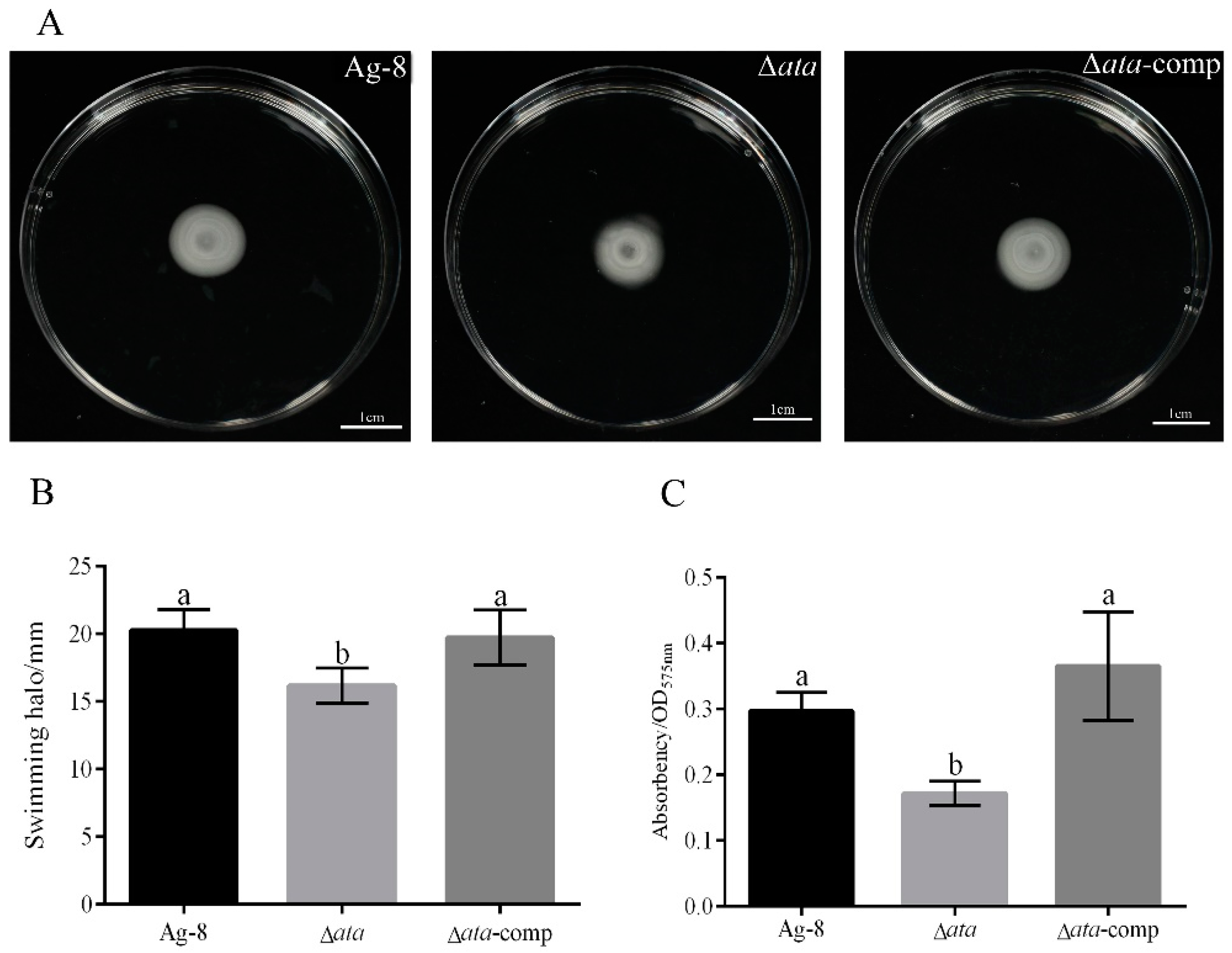
2.3. Determination of Motility and Biofilm Production
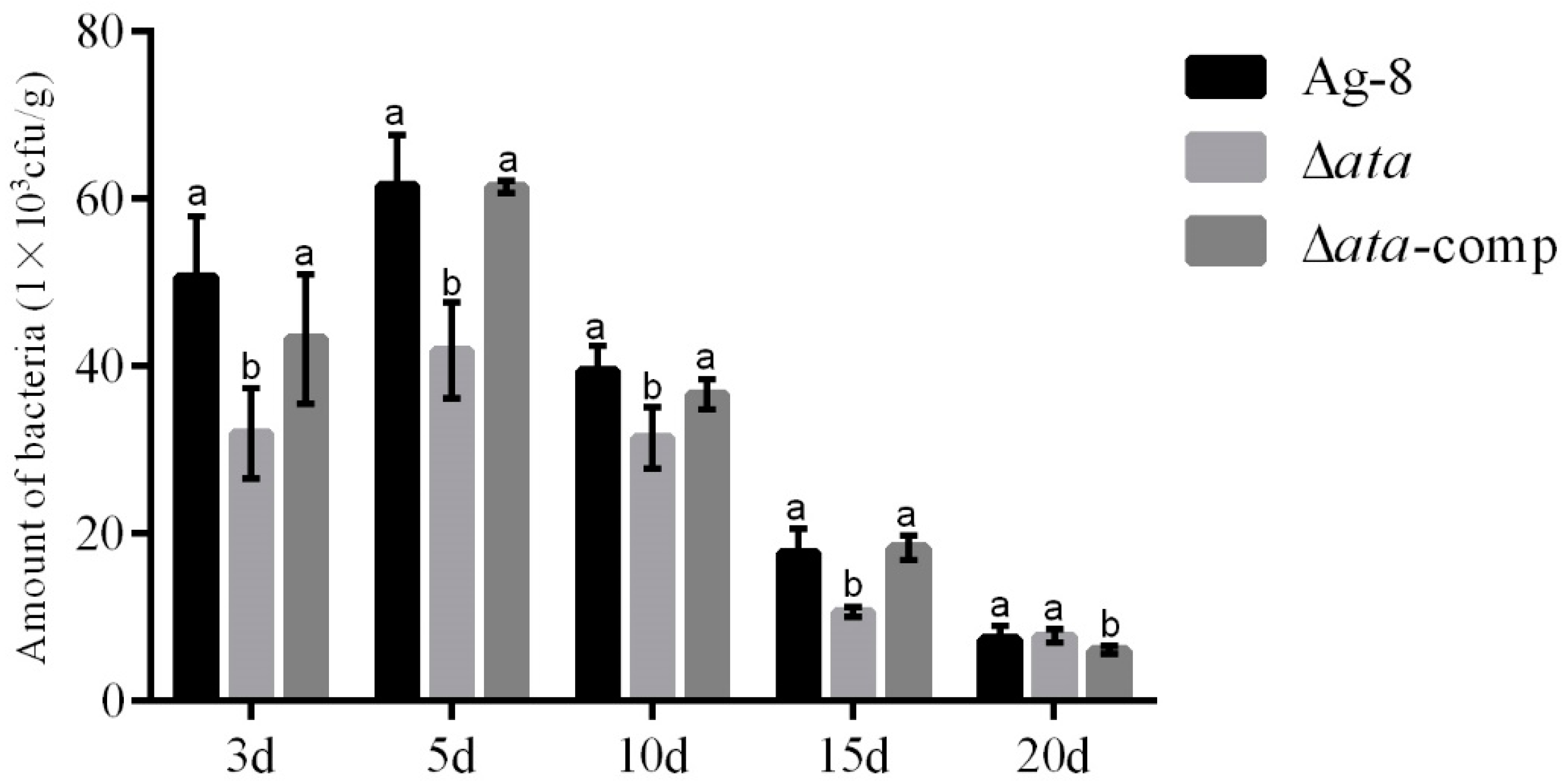
2.4. Determination of the Ability of Mutant Strains to Colonize Tomato Rhizosphere
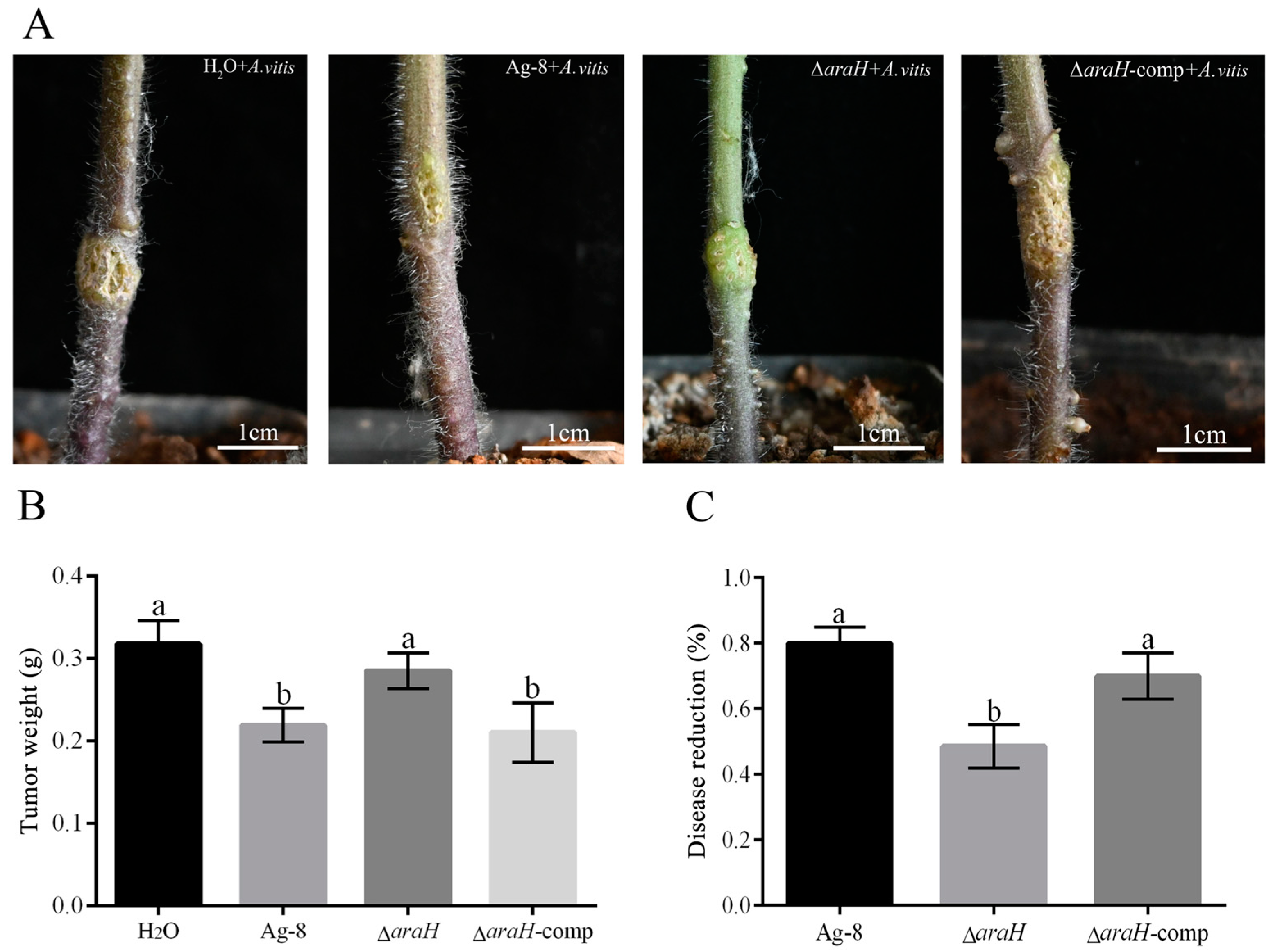
2.5. Experiments on the Efficacy of Mutant Strains Against Grapevine Crown Gall on Tomato
treatment)/average tumor weight of negative control × 100%
2.6. Construction of cDNA Libraries
2.7. Real-Time Fluorescence Quantitative PCR (RT-qPCR) Validation
2.8. Data Analysis
3. Results
3.1. ata Plays a Key Role in Ag-8 Control of Grapevine Crown Gall Disease in Tomato
3.2. ata Gene Affects the Motility and Biofilm Formation of Ag-8
3.3. ata Gene Affects Ag-8 Colonization in Tomato Rhizosphere
3.4. ata Deletion Reduces the Expression of Ag-8 and Biofilm-Related Genes
3.5. The araH Gene Has Been Demonstrated to Influence the Motility and Biofilm Formation of Ag-8
3.6. araH Gene Affects the Effectiveness of Ag-8 Against Grapevine Crown Gall Disease in Tomato
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Primer ID | Sequence |
|---|---|
| Δata-1F | GTCGACTCTAGAGGATCCCGAGGTCGGGCGCGTTGA |
| Δata-1R | AAATACATGACTGAACCATGAGGCAGAGGATAT |
| Δata-2F | TCCTCTGCCTCATGGTTCAGTCATGTATTTCCT |
| Δata-2R | GACCATGATTACGAATTCCGGGGTTCGGGCTGACAATTAG |
| ΔaraH-1F | ACCGAGCTCGAATTCACAGGAAGCGTATCATCG |
| ΔaraH-1R | ATGTCAGCGAATTGATCACCACACTGACGATCTTC |
| ΔaraH-2F | TCGTCAGTGTGGTGATCAATTCGCTGACATGCCGG |
| ΔaraH-2R | CAGGTCGACTCTAGAAATCGCTGGACATCCGCC |
| Primer ID | Sequence |
|---|---|
| Δata-comp-F | ACGACGGCCAGTGAATTCCCCAAGGTCTCGTGTG |
| Δata-comp-R | CTGCAGGTCGACTCTAGAGTGCTTCGCTCAAGAAGGATAC |
| ΔaraH-comp-F | CGGCCAGTGAATTCGAGCTCTCATCGCGCGGTACCTTC |
| ΔaraH-comp-R | GCCTGCAGGTCGACTCTAGAATGATGGTGGCTGTGACA |
| Primer ID | Sequence |
|---|---|
| araH QP-F | TTTGACAAAGCCCAGAACCG |
| araH QP-R | TCATCCAGACCATCAATACCG |
References
- Yuan, L.; Yin, X.; Wang, P.; Li, T.; Jiang, X.; Tang, X.; Hna, J.; Wei, Y. Research progress and control measures of grapevine crown gall. Chin. Foreign Grapes Wines 2022, 93–99. [Google Scholar] [CrossRef]
- Kawaguchi, A.; Kirino, N.; Inoue, K.; Noutoshi, Y. Biological control for grapevine crown gall through soil injection with Allorhizobium vitis strain ark-1. Eur. J. Plant Pathol. 2024, 170, 479–489. [Google Scholar] [CrossRef]
- Brown, P.J.B.; Chang, J.H.; Fuqua, C. Agrobacterium tumefaciens: A transformative agent for fundamental insights into host-microbe interactions, genome biology, chemical signaling, and cell biology. J. Bacteriol. 2023, 205, e0000523. [Google Scholar] [CrossRef]
- Stockwell, V.O.; Moore, L.W.; Loper, J.E. Fate of Agrobacterium radiobacter K84 in the environment. Appl. Environ. Microbiol. 1993, 59, 2112–2120. [Google Scholar] [CrossRef]
- Yang, Y.L.; Li, J.Y.; Wang, J.H.; Wang, H.M. Mutations affecting chemotaxis of Agrobacterium vitis strain e26 also alter attachment to grapevine roots and biocontrol of crown gall disease. Plant Pathol. 2009, 58, 594–605. [Google Scholar] [CrossRef]
- Kawaguchi, A.; Inoue, K.; Ichinose, Y. Biological control of crown gall of grapevine, rose, and tomato by nonpathogenic Agrobacterium vitis strain var03-1. Phytopathology 2008, 98, 1218–1225. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gohlke, J.; Deeken, R. Plant responses to Agrobacterium tumefaciens and crown gall development. Front. Plant Sci. 2014, 5, 155. [Google Scholar] [CrossRef]
- Bull, C.T.; Weller, D.M.; Thomashow, L.S.J.P. Relationship between root colonization and suppression of gaeumannomyces graminis var. Tritici by pseudomonas fluorescens strain 2-79. Phytopathology 1991, 81, 954–959. [Google Scholar] [CrossRef]
- Timmusk, S.; Grantcharova, N.; Wagner, E.G.H. paenibacillus polymyxa invades plant roots and forms biofilms. Appl. Environ. Microbiol. 2005, 71, 7292–7300. [Google Scholar] [CrossRef]
- Huang, R.; Feng, H.; Xu, Z.; Zhang, N.; Liu, Y.; Shao, J.; Shen, Q.; Zhang, R. Identification of adhesins in plant beneficial rhizobacteria bacillus velezensis sqr9 and their effect on root colonization. Mol. Plant-Microbe Interact. 2022, 35, 64–72. [Google Scholar] [CrossRef]
- Gao, S.; Wu, H.; Yu, X.; Qian, L.; Gao, X. Swarming motility plays the major role in migration during tomato root colonization by bacillus subtilis swr01. Biol. Control 2016, 98, 11–17. [Google Scholar] [CrossRef]
- Zhao, A.; Sun, J.; Liu, Y. Understanding bacterial biofilms: From definition to treatment strategies. Front. Cell. Infect. Microbiol. 2023, 13, 1137947. [Google Scholar] [CrossRef]
- Zhu, M.-L.; Wu, X.-Q.; Wang, Y.-H.; Dai, Y. Role of biofilm formation bybacillus pumilushr10 in biocontrol against pine seedling damping-off disease caused byrhizoctonia solani. Forests 2020, 11, 652. [Google Scholar] [CrossRef]
- Gallegos, M.-T.; Schleif, R.; Bairoch, A.; Hofmann, K.; Ramos, J.L.J.M.; Reviews, M.B. Arac/xyls family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 1997, 61, 393–410. [Google Scholar]
- Gribskov, M.; McLachlan, A.D.; Eisenberg, D. Profile analysis: Detection of distantly related proteins. Proc. Natl. Acad. Sci. USA 1987, 84, 4355–4358. [Google Scholar] [CrossRef] [PubMed]
- Greenblatt, J.; Schleif, R. Arabinose c protein: Regulation of the arabinose operon in vitro. Nat. New Biol. 1971, 233, 166–170. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Sun, L.; Srinivasan, R.; Lin, M.; Gong, L.; Lin, X. Unveiling a virulence-regulating mechanism in aeromonas hydrophila: A quantitative exoproteomic analysis of an arac-like protein. Front. Immunol. 2023, 14, 1191209. [Google Scholar] [CrossRef]
- Ueda, A.; Ogasawara, S.; Horiuchi, K. Identification of the genes controlling biofilm formation in the plant commensalpseudomonas protegenspf-5. Arch. Microbiol. 2020, 202, 2453–2459. [Google Scholar] [CrossRef]
- Chen, B.; Li, R.-F.; Zhou, L.; Qiu, J.-H.; Song, K.; Tang, J.-L.; He, Y.-W. The phytopathogenxanthomonas campestrisutilizes the divergently transcribedpoba/pobrlocus for 4-hydroxybenzoic acid recognition and degradation to promote virulence. Mol. Microbiol. 2020, 114, 870–886. [Google Scholar] [CrossRef]
- Ni, X.; Li, S.; Yuan, Y.; Chang, R.; Liu, Q.; Liu, Z.; Li, Z.; Wang, Y. Effect of siad on ag-8 to improve resistance to crown gall in grapes and related mechanisms. Plant Physiol. Biochem. 2024, 215, 108869. [Google Scholar] [CrossRef]
- Hmelo, L.R.; Borlee, B.R.; Almblad, H.; Love, M.E.; Randall, T.E.; Tseng, B.S.; Lin, C.; Irie, Y.; Storek, K.M.; Yang, J.J.; et al. Precision-engineering the Pseudomonas aeruginosa genome with two-step allelic exchange. Nat. Protoc. 2015, 10, 1820–1841. [Google Scholar] [CrossRef] [PubMed]
- Jailani, A.; Ahmed, B.; Lee, J.-H.; Lee, J. Inhibition of Agrobacterium tumefaciens growth and biofilm formation by tannic acid. Biomedicines 2022, 10, 1619. [Google Scholar] [CrossRef] [PubMed]
- Merritt, J.H.; Kadouri, D.E.; O’Toole, G.A. Growing and analyzing static biofilms. Curr. Protoc. Microbiol. 2011, 22, 1B-1. [Google Scholar] [CrossRef]
- Molina-Henares, M.A.; Ramos-Gonzalez, M.I.; Rinaldo, S.; Espinosa-Urgel, M. Gene expression reprogramming of pseudomonas alloputida in response to arginine through the transcriptional regulator argr. Microbiol.-Sgm 2024, 170, 001449. [Google Scholar] [CrossRef]
- Sahebi, M.; Tarighi, S.; Taheri, P. The arac-like transcriptional regulator yqhc is involved in pathogenicity of Erwinia amylovora. J. Appl. Microbiol. 2022, 132, 1319–1329. [Google Scholar] [CrossRef]
- Cai, Y.-m.; Hutchin, A.; Craddock, J.; Walsh, M.A.; Webb, J.S.; Tews, I. Differential impact on motility and biofilm dispersal of closely related phosphodiesterases in Pseudomonas aeruginosa. Sci. Rep. 2020, 10, 6232. [Google Scholar] [CrossRef] [PubMed]
- Haas, D.; Défago, G. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Microbiol. 2005, 3, 307–319. [Google Scholar] [CrossRef]
- Lv, J.; Zhu, J.; Wang, T.; Xie, X.; Wang, T.; Zhu, Z.; Chen, L.; Zhong, F.; Du, H. The role of the two-component qsebc signaling system in biofilm formation and virulence of hypervirulent klebsiella pneumoniae atcc43816. Front. Microbiol. 2022, 13, 817494. [Google Scholar] [CrossRef]
- Midgett, C.R.; Talbot, K.M.; Day, J.L.; Munson, G.P.; Kull, F.J. Structure of the master regulator rns reveals an inhibitor of enterotoxigenic Escherichia coli virulence regulons. Sci. Rep. 2021, 11, 15663. [Google Scholar] [CrossRef]
- Rodriguez-Valverde, D.; Giron, J.A.; Hu, Y.; Nataro, J.P.; Ruiz-Perez, F.; Santiago, A.E. Highly-conserved regulatory activity of the anr family in the virulence of diarrheagenic bacteria through interaction with master and global regulators. Sci. Rep. 2023, 13, 7024. [Google Scholar] [CrossRef]
- Rees, D.C.; Johnson, E.; Lewinson, O. Abc transporters: The power to change. Nat. Rev. Mol. Cell Biol. 2009, 10, 218–227. [Google Scholar] [CrossRef]
- Tanaka, K.J.; Song, S.; Mason, K.; Pinkett, H.W. Selective substrate uptake: The role of atp-binding cassette (abc) importers in pathogenesis. Biochim. Biophys. Acta-Biomembr. 2018, 1860, 868–877. [Google Scholar] [CrossRef]
- Horazdovsky, B.F. The High Affinity l-Arabinose Transport Operon of Escherichia coli. Ph.D. Thesis, Case Western Reserve University, Cleveland, OH, USA, 1988. [Google Scholar]
- Sheppard, D.E.; Englesberg, E. Further evidence for positive control of the l-arabinose system by gene arac. J. Mol. Biol. 1967, 25, 443–454. [Google Scholar] [CrossRef] [PubMed]
- Vasicek, E.M.; O’Neal, L.; Parsek, M.R.; Fitch, J.; White, P.; Gunn, J.S. L-arabinose transport and metabolism in salmonella influences biofilm formation. Front. Cell. Infect. Microbiol. 2021, 11, 698146. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Fan, F.; Qiu, H.; Shao, M.; Li, D.; Yu, Y.; Bi, C.; Zhang, X. New xylose transporters support the simultaneous consumption of glucose and xylose in Escherichia coli. Mlife 2022, 1, 156–170. [Google Scholar] [CrossRef] [PubMed]
- Suchan, D.M.; Bergsveinson, J.; Manzon, L.; Pierce, A.; Kryachko, Y.; Korber, D.; Tan, Y.; Tambalo, D.D.; Khan, N.H.; Whiting, M.; et al. Transcriptomics reveal core activities of the plant growth-promoting bacterium delftia acidovorans ray209 during interaction with canola and soybean roots. Microb. Genom. 2020, 6, e000462. [Google Scholar] [CrossRef]
- Benda, M.; Schulz, L.M.; Stuelke, J.; Rismondo, J. Influence of the abc transporter ytrbcdef of Bacillus subtilis on competence, biofilm formation and cell wall thickness. Front. Microbiol. 2021, 12, 587035. [Google Scholar] [CrossRef]
- Zeng, Y.; Charkowski, A.O. The role of atp-binding cassette transporters in bacterial phytopathogenesis. Phytopathology 2021, 111, 600–610. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, S.; Zhang, Y.; Liu, Z.; Gu, Y.; Bi, Y.; Yang, J.; Yu, W.; Li, Z.; Wang, Y. A Novel Transcription Factor of Regulating Ag-8 Biocontrol to Grapevine Crown Gall. Horticulturae 2025, 11, 465. https://doi.org/10.3390/horticulturae11050465
Li S, Zhang Y, Liu Z, Gu Y, Bi Y, Yang J, Yu W, Li Z, Wang Y. A Novel Transcription Factor of Regulating Ag-8 Biocontrol to Grapevine Crown Gall. Horticulturae. 2025; 11(5):465. https://doi.org/10.3390/horticulturae11050465
Chicago/Turabian StyleLi, Shiyu, Yaping Zhang, Zhenxing Liu, Yilin Gu, Yue Bi, Jianyu Yang, Weiwei Yu, Zhuoran Li, and Yuanhong Wang. 2025. "A Novel Transcription Factor of Regulating Ag-8 Biocontrol to Grapevine Crown Gall" Horticulturae 11, no. 5: 465. https://doi.org/10.3390/horticulturae11050465
APA StyleLi, S., Zhang, Y., Liu, Z., Gu, Y., Bi, Y., Yang, J., Yu, W., Li, Z., & Wang, Y. (2025). A Novel Transcription Factor of Regulating Ag-8 Biocontrol to Grapevine Crown Gall. Horticulturae, 11(5), 465. https://doi.org/10.3390/horticulturae11050465





