Genome-Wide Identification of CaGA20ox Gene Family Members Related to Floral Organ Development in Pepper (Capsicum annuum) at Different Temperatures
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Identification and Characterization of CaGA20ox Genes
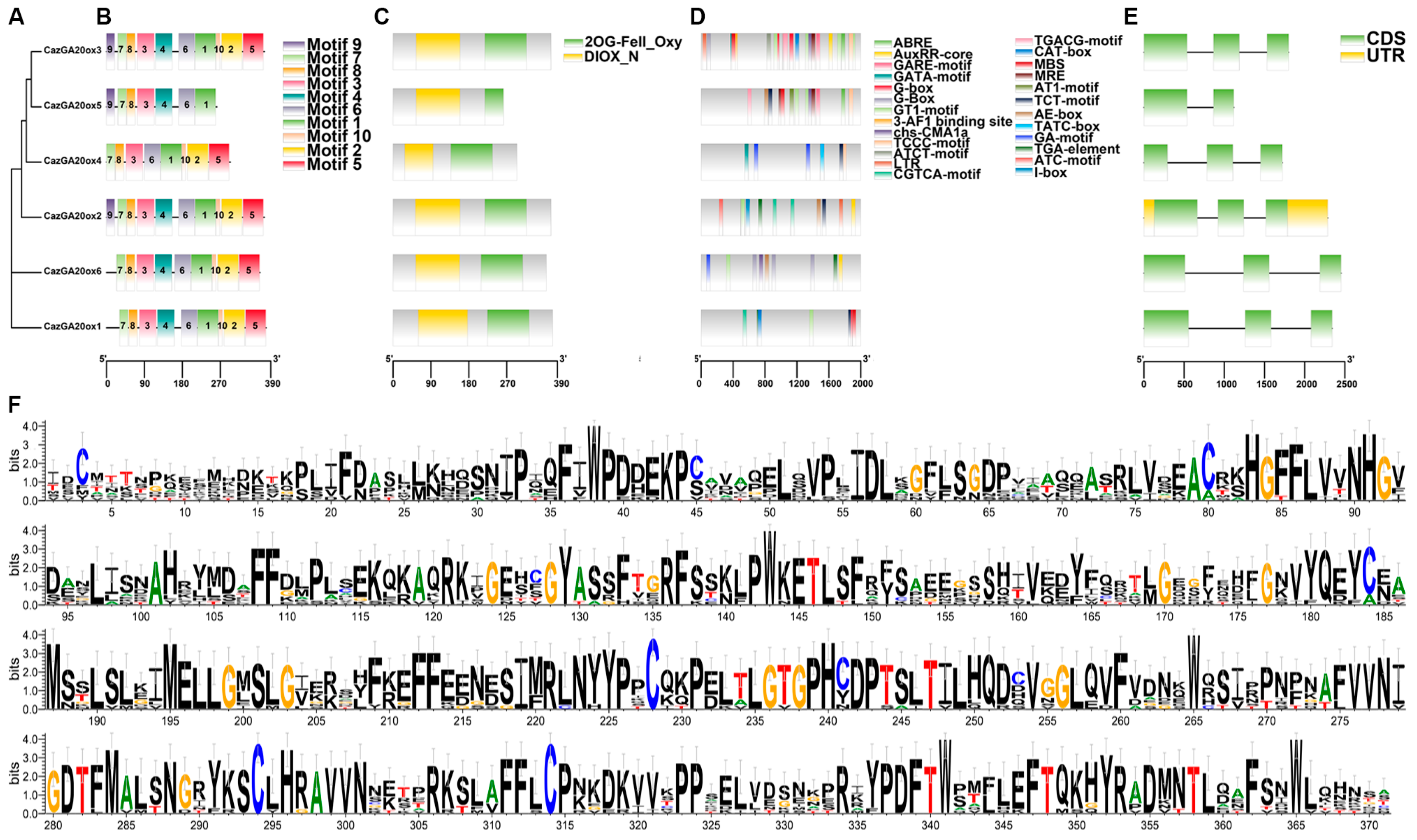
2.3. Phylogenetic, Gene Structure, Conserved Motif, and Cis-Acting Element Analyses
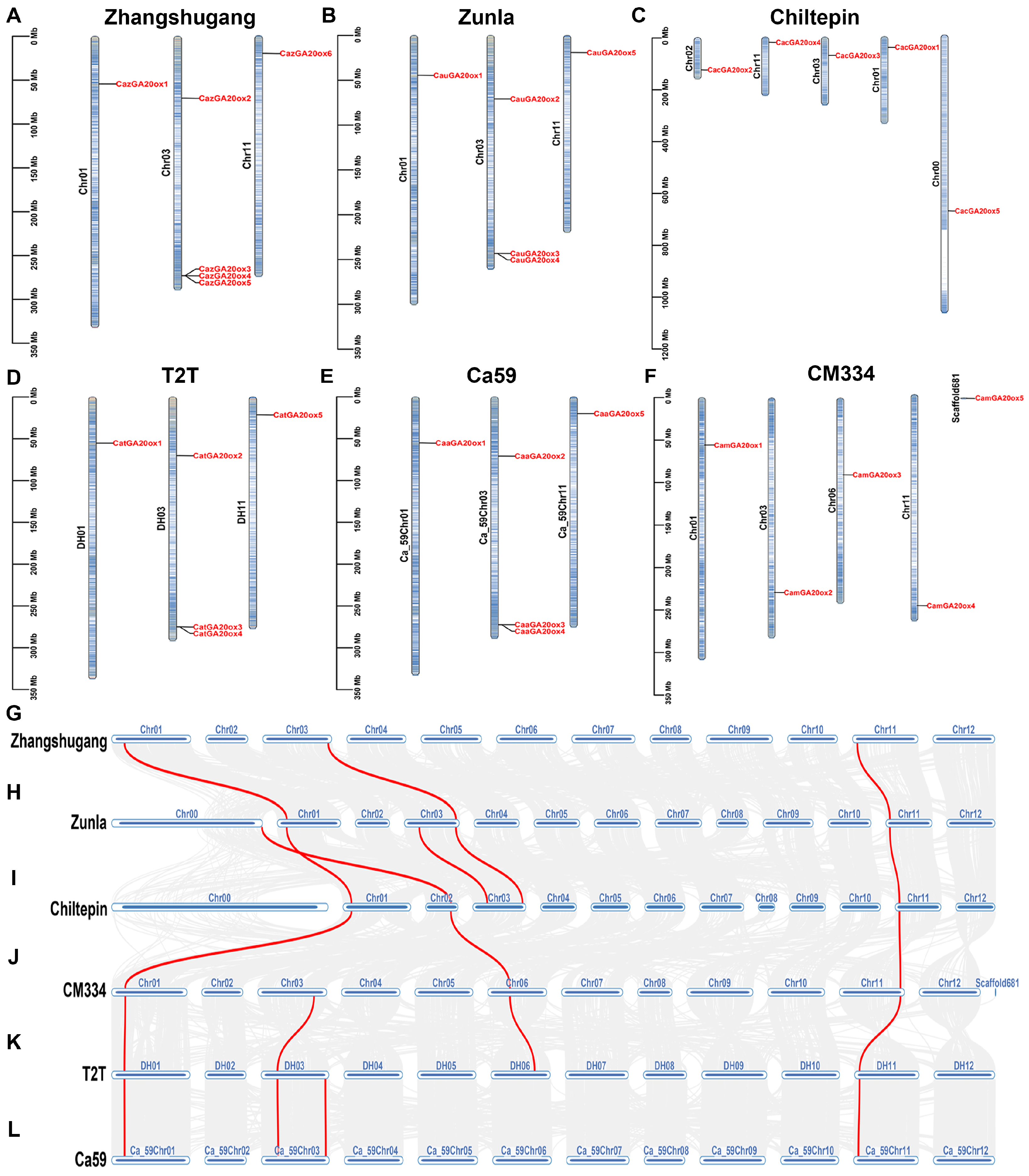
2.4. Chromosome Mapping and Synteny Analysis
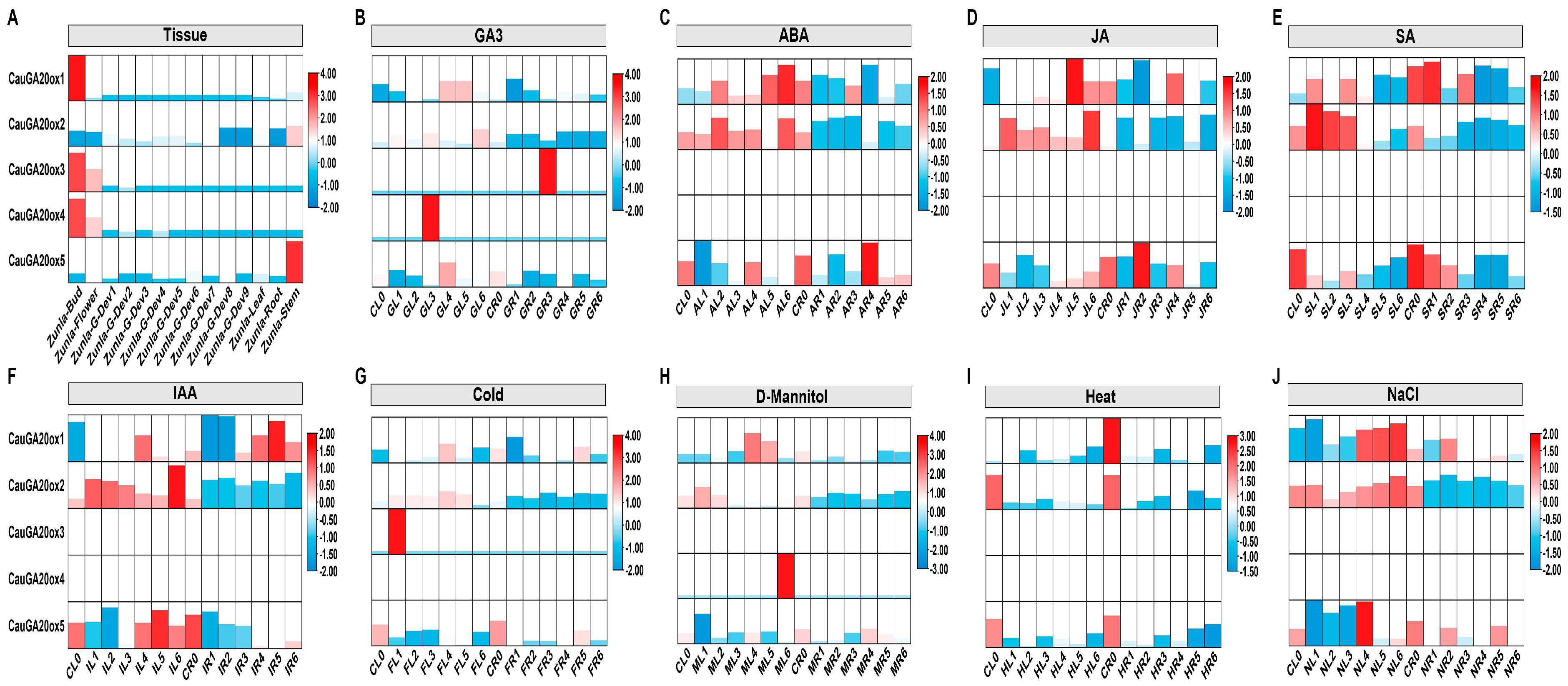
2.5. Characterization of CauGA20oxs Gene Expression in Pepper
2.6. RNA Extraction and cDNA Synthesis
2.7. RNA-Seq Analysis
2.8. Real-Time Fluorescence Quantitative PCR (qPCR) Analysis of Gene Expression
2.9. Pearson Correlation Coefficient Analysis of GA Metabolism Pathway and Flower Development Genes
2.10. Pollen Viability and Germination Assays
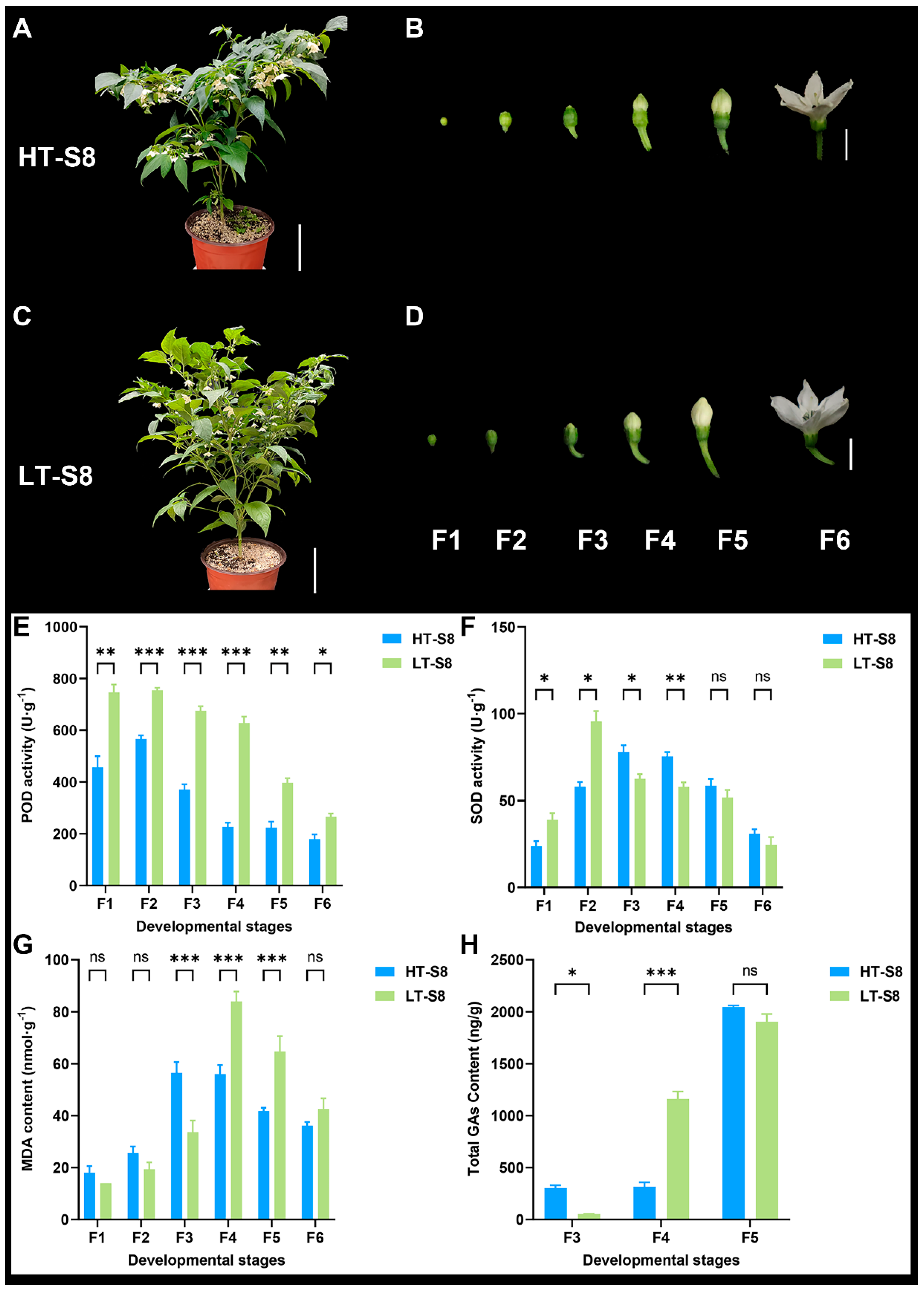
2.11. Evaluation of Antioxidant Enzyme Activities and Oxidative Stress in S8 Flowers at Different Temperatures
2.12. Determination of GA Content in Pepper Flowers
2.13. Subcellular Localization in N. benthamiana
2.14. Statistical Analysis
3. Results
3.1. Identification and Characterization of CaGA20ox Genes
3.2. Phylogenetic, Chromosomal Location, and Collinearity Analyses of CaGA20ox Genes
3.3. Conserved Motif, Cis-Element, and Gene Structure Analyses of CazGA20ox Genes in ‘Zhangshugang’ Pepper
3.4. CauGa20ox Genes Exhibit Distinct Tissue- and Developmental Stage-Specific Expression Profiles
3.5. Pepper Flower Development at Different Temperatures
3.6. RNA-Seq Analysis of Pepper Flowers at Different Temperatures
3.7. Four Candidate CazGA20ox Genes May Be Involved in Temperature-Driven Flower Development
4. Discussion
4.1. Characteristics of the CaGA20ox Gene Family
4.2. Temperature Significantly Affects Peppers’ Floral Organ Development and GA Content
4.3. Four Candidate CazGA20ox Genes May Be Involved in Floral Organ Development
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Lyu, J.; Aiwaili, P.; Gu, Z.; Xu, Y.; Zhang, Y.; Wang, Z.; Huang, H.; Zeng, R.; Ma, C.; Gao, J.; et al. Chrysanthemum MAF2 regulates flowering by repressing gibberellin biosynthesis in response to low temperature. Plant J. 2022, 112, 1159–1175. [Google Scholar] [CrossRef]
- Lantzouni, O.; Alkofer, A.; Falter-Braun, P.; Schwechheimer, C. GROWTH-REGULATING FACTORS Interact with DELLAs and Regulate Growth in Cold Stress. Plant Cell 2020, 32, 1018–1034. [Google Scholar] [CrossRef] [PubMed]
- Busi, M.V.; Bustamante, C.; D’Angelo, C.; Hidalgo-Cuevas, M.; Boggio, S.B.; Valle, E.M.; Zabaleta, E. MADS-box genes expressed during tomato seed and fruit development. Plant Mol. Biol. 2003, 52, 801–815. [Google Scholar] [CrossRef]
- Lohmann, J.U.; Weigel, D. Building beauty: The genetic control of floral patterning. Dev. Cell 2002, 2, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Krizek, B.A.; Fletcher, J.C. Molecular mechanisms of flower development: An armchair guide. Nat. Rev. Genet. 2005, 6, 688–698. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yang, Y.; Zhang, Y.; Zhao, T.; Jiang, J.; Li, J.; Xu, X.; Yang, H. Transcriptome Analysis of Flower Development and Mining of Genes Related to Flowering Time in Tomato (Solanum lycopersicum). Int. J. Mol. Sci. 2021, 22, 8128. [Google Scholar] [CrossRef]
- Tang, B.; Yang, H.; Zhang, X.; Du, J.; Xie, L.; Dai, X.; Zou, X.; Liu, F. A global view of transcriptome dynamics during flower development in Capsicum annuum L. Hortic. Plant J. 2023, 9, 999–1012. [Google Scholar] [CrossRef]
- Sandoval-Oliveros, R.; Guevara-Olvera, L.; Beltrán, J.P.; Gómez-Mena, C.; Acosta-García, G. Developmental landmarks during floral ontogeny of jalapeño chili pepper (Capsicum annuum L.) and the effect of gibberellin on ovary growth. Plant Reprod. 2017, 30, 119–129. [Google Scholar] [CrossRef]
- Fukazawa, J.; Ohashi, Y.; Takahashi, R.; Nakai, K.; Takahashi, Y. DELLA degradation by gibberellin promotes flowering via GAF1-TPR-dependent repression of floral repressors in Arabidopsis. Plant Cell 2021, 33, 2258–2272. [Google Scholar] [CrossRef]
- Yamaguchi, S.; Kamiya, Y. Gibberellins and Light-Stimulated Seed Germination. J. Plant Growth Regul. 2001, 20, 369–376. [Google Scholar] [CrossRef]
- Arnaud, N.; Girin, T.; Sorefan, K.; Fuentes, S.; Wood, T.A.; Lawrenson, T.; Sablowski, R.; Østergaard, L. Gibberellins control fruit patterning in Arabidopsis thaliana. Genes Dev. 2010, 24, 2127–2132. [Google Scholar] [CrossRef]
- Waadt, R.; Seller, C.A.; Hsu, P.K.; Takahashi, Y.; Munemasa, S.; Schroeder, J.I. Plant hormone regulation of abiotic stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 680–694. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Qin, L.; Lee, S.; Fu, X.; Richards, D.E.; Cao, D.; Luo, D.; Harberd, N.P.; Peng, J. Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development 2004, 131, 1055–1064. [Google Scholar] [CrossRef] [PubMed]
- Plackett, A.R.; Thomas, S.G.; Wilson, Z.A.; Hedden, P. Gibberellin control of stamen development: A fertile field. Trends Plant Sci. 2011, 16, 568–578. [Google Scholar] [CrossRef]
- Ueguchi-Tanaka, M.; Ashikari, M.; Nakajima, M.; Itoh, H.; Katoh, E.; Kobayashi, M.; Chow, T.Y.; Hsing, Y.I.; Kitano, H.; Yamaguchi, I.; et al. GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 2005, 437, 693–698. [Google Scholar] [CrossRef]
- Cheng, H.; Song, S.; Xiao, L.; Soo, H.M.; Cheng, Z.; Xie, D.; Peng, J. Gibberellin acts through jasmonate to control the expression of MYB21, MYB24, and MYB57 to promote stamen filament growth in Arabidopsis. PLoS Genet. 2009, 5, e1000440. [Google Scholar] [CrossRef] [PubMed]
- Sakata, T.; Oda, S.; Tsunaga, Y.; Shomura, H.; Kawagishi-Kobayashi, M.; Aya, K.; Saeki, K.; Endo, T.; Nagano, K.; Kojima, M.; et al. Reduction of gibberellin by low temperature disrupts pollen development in rice. Plant Physiol. 2014, 164, 2011–2019. [Google Scholar] [CrossRef]
- Li, P.; Tian, J.; Guo, C.; Luo, S.; Li, J. Interaction of gibberellin and other hormones in almond anthers: Phenotypic and physiological changes and transcriptomic reprogramming. Hortic. Res. 2021, 8, 94. [Google Scholar] [CrossRef]
- Tang, X.; Hao, Y.J.; Lu, J.X.; Lu, G.; Zhang, T. Transcriptomic analysis reveals the mechanism of thermosensitive genic male sterility (TGMS) of Brassica napus under the high temperature inducement. BMC Genom. 2019, 20, 644. [Google Scholar] [CrossRef]
- Murase, K.; Hirano, Y.; Sun, T.P.; Hakoshima, T. Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 2008, 456, 459–463. [Google Scholar] [CrossRef]
- Wuddineh, W.A.; Mazarei, M.; Zhang, J.; Poovaiah, C.R.; Mann, D.G.; Ziebell, A.; Sykes, R.W.; Davis, M.F.; Udvardi, M.K.; Stewart, C.N., Jr. Identification and overexpression of gibberellin 2-oxidase (GA2ox) in switchgrass (Panicum virgatum L.) for improved plant architecture and reduced biomass recalcitrance. Plant Biotechnol. J. 2015, 13, 636–647. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, T.; Miura, K.; Itoh, H.; Tatsumi, T.; Ueguchi-Tanaka, M.; Ishiyama, K.; Kobayashi, M.; Agrawal, G.K.; Takeda, S.; Abe, K.; et al. An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol. 2004, 134, 1642–1653. [Google Scholar] [CrossRef]
- Fukazawa, J.; Mori, M.; Watanabe, S.; Miyamoto, C.; Ito, T.; Takahashi, Y. DELLA-GAF1 Complex Is a Main Component in Gibberellin Feedback Regulation of GA20 Oxidase 2. Plant Physiol. 2017, 175, 1395–1406. [Google Scholar] [CrossRef]
- Lee, D.H.; Lee, I.C.; Kim, K.J.; Kim, D.S.; Na, H.J.; Lee, I.J.; Kang, S.M.; Jeon, H.W.; Le, P.Y.; Ko, J.H. Expression of gibberellin 2-oxidase 4 from Arabidopsis under the control of a senescence-associated promoter results in a dominant semi-dwarf plant with normal flowering. J. Plant Biol. 2014, 57, 106–116. [Google Scholar] [CrossRef]
- El-Sharkawy, I.; El Kayal, W.; Prasath, D.; Fernández, H.; Bouzayen, M.; Svircev, A.M.; Jayasankar, S. Identification and genetic characterization of a gibberellin 2-oxidase gene that controls tree stature and reproductive growth in plum. J. Exp. Bot. 2012, 63, 1225–1239. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, A.; Ashikari, M.; Ueguchi-Tanaka, M.; Itoh, H.; Nishimura, A.; Swapan, D.; Ishiyama, K.; Saito, T.; Kobayashi, M.; Khush, G.S.; et al. Green revolution: A mutant gibberellin-synthesis gene in rice. Nature 2002, 416, 701–702. [Google Scholar] [CrossRef]
- Rieu, I.; Ruiz-Rivero, O.; Fernandez-Garcia, N.; Griffiths, J.; Powers, S.J.; Gong, F.; Linhartova, T.; Eriksson, S.; Nilsson, O.; Thomas, S.G.; et al. The gibberellin biosynthetic genes AtGA20ox1 and AtGA20ox2 act, partially redundantly, to promote growth and development throughout the Arabidopsis life cycle. Plant J. 2008, 53, 488–504. [Google Scholar] [CrossRef]
- Plackett, A.R.; Powers, S.J.; Fernandez-Garcia, N.; Urbanova, T.; Takebayashi, Y.; Seo, M.; Jikumaru, Y.; Benlloch, R.; Nilsson, O.; Ruiz-Rivero, O.; et al. Analysis of the developmental roles of the Arabidopsis gibberellin 20-oxidases demonstrates that GA20ox1, -2, and -3 are the dominant paralogs. Plant Cell 2012, 24, 941–960. [Google Scholar] [CrossRef]
- Qin, C.; Yu, C.; Shen, Y.; Fang, X.; Chen, L.; Min, J.; Cheng, J.; Zhao, S.; Xu, M.; Luo, Y.; et al. Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proc. Natl. Acad. Sci. USA 2014, 111, 5135–5140. [Google Scholar] [CrossRef]
- Kim, S.; Park, M.; Yeom, S.I.; Kim, Y.M.; Lee, J.M.; Lee, H.A.; Seo, E.; Choi, J.; Cheong, K.; Kim, K.T.; et al. Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat. Genet. 2014, 46, 270–278. [Google Scholar] [CrossRef]
- Liu, F.; Zhao, J.; Sun, H.; Xiong, C.; Sun, X.; Wang, X.; Wang, Z.; Jarret, R.; Wang, J.; Tang, B.; et al. Genomes of cultivated and wild Capsicum species provide insights into pepper domestication and population differentiation. Nat. Commun. 2023, 14, 5487. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Wang, J.; Zhu, Z.; Liu, Y.; Chen, J.; Zhou, Y.; Liu, F.; Lei, J.; Gaut, B.S.; Cao, B.; et al. The 3D architecture of the pepper genome and its relationship to function and evolution. Nat. Commun. 2022, 13, 3479. [Google Scholar] [CrossRef]
- Chen, W.; Wang, X.; Sun, J.; Wang, X.; Zhu, Z.; Ayhan, D.H.; Yi, S.; Yan, M.; Zhang, L.; Meng, T.; et al. Two telomere-to-telomere gapless genomes reveal insights into Capsicum evolution and capsaicinoid biosynthesis. Nat. Commun. 2024, 15, 4295. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; de Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, J.; Paterson, A.H. MCScanX-transposed: Detecting transposed gene duplications based on multiple colinearity scans. Bioinformatics 2013, 29, 1458–1460. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Yu, H.; Deng, Y.; Zheng, J.; Liu, M.; Ou, L.; Yang, B.; Dai, X.; Ma, Y.; Feng, S.; et al. PepperHub, an Informatics Hub for the Chili Pepper Research Community. Mol. Plant 2017, 10, 1129–1132. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Wan, H.; Yuan, W.; Ruan, M.; Ye, Q.; Wang, R.; Li, Z.; Zhou, G.; Yao, Z.; Zhao, J.; Liu, S.; et al. Identification of reference genes for reverse transcription quantitative real-time PCR normalization in pepper (Capsicum annuum L.). Biochem. Biophys. Res. Commun. 2011, 416, 24–30. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Cai, B.D.; Zhu, J.X.; Gao, Q.; Luo, D.; Yuan, B.F.; Feng, Y.Q. Rapid and high-throughput determination of endogenous cytokinins in Oryza sativa by bare Fe3O4 nanoparticles-based magnetic solid-phase extraction. J. Chromatogr. A 2014, 1340, 146–150. [Google Scholar] [CrossRef]
- Niu, Q.; Zong, Y.; Qian, M.; Yang, F.; Teng, Y. Simultaneous quantitative determination of major plant hormones in pear flowers and fruit by UPLC/ESI-MS/MS. Anal. Methods 2014, 6, 1766–1773. [Google Scholar] [CrossRef]
- Xiao, H.M.; Cai, W.J.; Ye, T.T.; Ding, J.; Feng, Y.Q. Spatio-temporal profiling of abscisic acid, indoleacetic acid and jasmonic acid in single rice seed during seed germination. Anal. Chim. Acta 2018, 1031, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Šimura, J.; Antoniadi, I.; Široká, J.; Tarkowská, D.; Strnad, M.; Ljung, K.; Novák, O. Plant Hormonomics: Multiple Phytohormone Profiling by Targeted Metabolomics. Plant Physiol. 2018, 177, 476–489. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Welti, R.; Wang, X. Quantitative analysis of major plant hormones in crude plant extracts by high-performance liquid chromatography-mass spectrometry. Nat. Protoc. 2010, 5, 986–992. [Google Scholar] [CrossRef]
- Han, F.; Zhu, B. Evolutionary analysis of three gibberellin oxidase genes in rice, Arabidopsis, and soybean. Gene 2011, 473, 23–35. [Google Scholar] [CrossRef]
- Ci, J.; Wang, X.; Wang, Q.; Zhao, F.; Yang, W.; Cui, X.; Jiang, L.; Ren, X.; Yang, W. Genome-wide analysis of gibberellin-dioxygenases gene family and their responses to GA applications in maize. PLoS ONE 2021, 16, e0250349. [Google Scholar] [CrossRef]
- Sun, J.; Zhang, X.; Fu, C.; Ahmad, N.; Zhao, C.; Hou, L.; Naeem, M.; Pan, J.; Wang, X.; Zhao, S. Genome-wide identification and expression analysis of GA20ox and GA3ox genes during pod development in peanut. PeerJ 2023, 11, e16279. [Google Scholar] [CrossRef]
- He, H.; Liang, G.; Lu, S.; Wang, P.; Liu, T.; Ma, Z.; Zuo, C.; Sun, X.; Chen, B.; Mao, J. Genome-Wide Identification and Expression Analysis of GA2ox, GA3ox, and GA20ox Are Related to Gibberellin Oxidase Genes in Grape (Vitis vinifera L.). Genes 2019, 10, 680. [Google Scholar] [CrossRef]
- Oliver, S.N.; Dennis, E.S.; Dolferus, R. ABA regulates apoplastic sugar transport and is a potential signal for cold-induced pollen sterility in rice. Plant Cell Physiol. 2007, 48, 1319–1330. [Google Scholar] [CrossRef]
- Khan, T.A.; Fariduddin, Q.; Yusuf, M. Lycopersicon esculentum under low temperature stress: An approach toward enhanced antioxidants and yield. Environ. Sci. Pollut. Res. Int. 2015, 22, 14178–14188. [Google Scholar] [CrossRef]
- Xu, H.; Xu, L.; Hassan, M.A. Mitigating low-temperature stress in alfalfa by postponing phosphorus application and remodeling of antioxidant activities and carbon-nitrogen metabolism. Front. Plant Sci. 2025, 16, 1550026. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Cai, X.; Tang, Y. Outcomes of Low-Temperature Stress on Biological Alterations within Pothos (Epipremnum aureum) Leaves. Life 2022, 12, 1432. [Google Scholar] [CrossRef] [PubMed]
- Shang, M.; Wang, X.; Zhang, J.; Qi, X.; Ping, A.; Hou, L.; Xing, G.; Li, G.; Li, M. Genetic Regulation of GA Metabolism during Vernalization, Floral Bud Initiation and Development in Pak Choi (Brassica rapa ssp. chinensis Makino). Front. Plant Sci. 2017, 8, 1533. [Google Scholar] [CrossRef] [PubMed]
- Barro-Trastoy, D.; Carrera, E.; Baños, J.; Palau-Rodríguez, J.; Ruiz-Rivero, O.; Tornero, P.; Alonso, J.M.; López-Díaz, I.; Gómez, M.D.; Pérez-Amador, M.A. Regulation of ovule initiation by gibberellins and brassinosteroids in tomato and Arabidopsis: Two plant species, two molecular mechanisms. Plant J. 2020, 102, 1026–1041. [Google Scholar] [CrossRef]



| Genome | Name | Gene ID | Amino Acid Length (aa) | pI | Molecular Weight (kDa) | Instability Index | GRAVY |
|---|---|---|---|---|---|---|---|
| Zhangshugang | CazGA20ox1 | Caz01g13200 | 379 | 6.34 | 43.41 | 37.31 | −0.354 |
| CazGA20ox2 | Caz03g21260 | 375 | 6.01 | 42.39 | 40.92 | −0.369 | |
| CazGA20ox3 | Caz03g36510 | 375 | 5.63 | 43.11 | 38.11 | −0.388 | |
| CazGA20ox4 | Caz03g36530 | 294 | 6.75 | 33.60 | 35.83 | −0.254 | |
| CazGA20ox5 | Caz03g36550 | 262 | 4.97 | 29.98 | 42.28 | −0.315 | |
| CazGA20ox6 | Caz11g05440 | 364 | 5.90 | 41.46 | 31.11 | −0.373 | |
| Zunla | CauGA20ox1 | Capana01g001332 | 365 | 6.34 | 41.95 | 36.65 | −0.381 |
| CauGA20ox2 | Capana03g002435 | 375 | 6.01 | 42.39 | 40.92 | −0.369 | |
| CauGA20ox3 | Capana03g003972 | 321 | 6.20 | 36.54 | 34.42 | −0.269 | |
| CauGA20ox4 | Capana03g003973 | 179 | 6.19 | 20.62 | 38.96 | −0.425 | |
| CauGA20ox5 | Capana11g000564 | 267 | 6.24 | 30.62 | 28.69 | −0.36 | |
| Chiltepin | CacGA20ox1 | Capang01g001151 | 365 | 6.34 | 41.95 | 36.65 | −0.381 |
| CacGA20ox2 | Capang02g001466 | 376 | 8.23 | 43.47 | 43.61 | −0.387 | |
| CacGA20ox3 | Capang03g002357 | 375 | 6.11 | 42.43 | 40.26 | −0.379 | |
| CacGA20ox4 | Capang11g000521 | 384 | 5.86 | 43.70 | 30.28 | −0.361 | |
| CacGA20ox5 | Capang00g003932 | 375 | 5.64 | 43.16 | 38.95 | −0.393 | |
| CM334 | CamGA20ox1 | PHT93425 | 377 | 6.34 | 43.20 | 37.45 | −0.352 |
| CamGA20ox2 | PHT87225 | 375 | 6.01 | 42.39 | 40.92 | −0.369 | |
| CamGA20ox3 | PHT78363 | 367 | 8.05 | 42.41 | 39.05 | −0.320 | |
| CamGA20ox4 | PHT68898 | 384 | 5.86 | 43.70 | 30.28 | −0.361 | |
| CamGA20ox5 | PHT63838 | 375 | 5.75 | 43.19 | 40.54 | −0.406 | |
| CA59 | CaaGA20ox1 | Capann_59V1aChr01g016000 | 365 | 6.34 | 41.95 | 36.65 | −0.381 |
| CaaGA20ox2 | Capann_59V1aChr03g024430 | 375 | 6.01 | 42.39 | 40.92 | −0.369 | |
| CaaGA20ox3 | Capann_59V1aChr03g043420 | 375 | 5.63 | 43.11 | 38.11 | −0.388 | |
| CaaGA20ox4 | Capann_59V1aChr03g043430 | 321 | 6.20 | 36.54 | 34.42 | −0.269 | |
| CaaGA20ox5 | Capann_59V1aChr11g006860 | 348 | 6.06 | 39.56 | 29.53 | −0.312 | |
| T2T | CatGA20ox1 | CaT2T01g01562 | 379 | 6.34 | 43.41 | 37.31 | −0.354 |
| CatGA20ox2 | CaT2T03g02508 | 334 | 6.41 | 37.63 | 39.66 | −0.342 | |
| CatGA20ox3 | CaT2T03g04651 | 321 | 5.65 | 36.82 | 36.90 | −0.321 | |
| CatGA20ox4 | CaT2T03g04653 | 321 | 6.20 | 36.55 | 35.26 | −0.252 | |
| CatGA20ox5 | CaT2T11g00667 | 372 | 5.85 | 42.34 | 29.62 | −0.366 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, Y.; Wang, J.; Ren, C.; Chen, Y.; Yang, S.; Yin, Q.; Wang, M.; Sui, X.; Tian, H.; Liu, F.; et al. Genome-Wide Identification of CaGA20ox Gene Family Members Related to Floral Organ Development in Pepper (Capsicum annuum) at Different Temperatures. Horticulturae 2025, 11, 469. https://doi.org/10.3390/horticulturae11050469
Luo Y, Wang J, Ren C, Chen Y, Yang S, Yin Q, Wang M, Sui X, Tian H, Liu F, et al. Genome-Wide Identification of CaGA20ox Gene Family Members Related to Floral Organ Development in Pepper (Capsicum annuum) at Different Temperatures. Horticulturae. 2025; 11(5):469. https://doi.org/10.3390/horticulturae11050469
Chicago/Turabian StyleLuo, Yin, Jin Wang, Chaohui Ren, Yaqian Chen, Shimei Yang, Qinbiao Yin, Meiqi Wang, Xiaoyan Sui, Hao Tian, Feng Liu, and et al. 2025. "Genome-Wide Identification of CaGA20ox Gene Family Members Related to Floral Organ Development in Pepper (Capsicum annuum) at Different Temperatures" Horticulturae 11, no. 5: 469. https://doi.org/10.3390/horticulturae11050469
APA StyleLuo, Y., Wang, J., Ren, C., Chen, Y., Yang, S., Yin, Q., Wang, M., Sui, X., Tian, H., Liu, F., & Zou, X. (2025). Genome-Wide Identification of CaGA20ox Gene Family Members Related to Floral Organ Development in Pepper (Capsicum annuum) at Different Temperatures. Horticulturae, 11(5), 469. https://doi.org/10.3390/horticulturae11050469






