Abstract
Soil salinization poses a significant threat to tea plant (Camellia sinensis) production by compromising its bioactive compounds, such as polyphenols, L-theanine, and caffeine, which are key contributors to the plant’s health benefits and economic value. This study investigates the Salt Overly Sensitive 1 (SOS1) gene family, a critical salt-tolerance regulator in tea plants, to elucidate its role in maintaining quality under environmental stress. Genome-wide analysis identified 51 CsSOS1 genes, with phylogenetic and synteny analyses revealing strong evolutionary conservation with Populus trichocarpa and Arabidopsis thaliana. Promoter analysis detected stress- and hormone-responsive cis-elements, indicating adaptive functions in abiotic stress. Expression profiling demonstrated tissue-specific patterns, highlighting significant upregulation of CsSOS1-15 and CsSOS1-41 under salt and drought stress. Co-expression network analysis further linked CsSOS1 genes to carbohydrate metabolism, implicating their roles in stress resilience and secondary metabolite synthesis. Our findings provide molecular insights into CsSOS1-mediated salt tolerance, proposing potential targets for preserving bioactive compounds. This work facilitates developing salt-resistant tea plant cultivars to ensure sustainable production and quality stability amid environmental challenges.
1. Introduction
Soil salinization represents an escalating global threat to irrigated agricultural lands, negatively impacting plant growth and development, and consequently reducing crop yield and quality [1,2]. Salt stress primarily affects plants through osmotic stress, ion toxicity, nutritional imbalance, and oxidative damage, with ion toxicity induced by the accumulation of Na+ and Cl− ions identified as the most significant contributor [3]. Among soluble salts, sodium chloride (NaCl) is the most abundant and problematic in soils [4]. Although sodium (Na+) is not essential for plant growth, it significantly contributes to salt stress by disrupting cellular membranes, destabilizing proteins, and impairing essential cellular and physiological processes [5]. These disruptions include alterations in osmotic balance, ionic homeostasis, nutrient uptake, metabolic activities, and an increase in oxidative stress, ultimately leading to cellular injury [6]. To cope with these stresses, plants have evolved mechanisms to perceive and respond effectively to environmental stimuli [7]. A central component of plant salt-stress response at the cellular level is the Salt Overly Sensitive (SOS) signaling pathway, which has been demonstrated to mediate cellular signaling under saline conditions and maintain ionic homeostasis.
The primary components involved in maintaining ionic homeostasis through the Salt Overly Sensitive (SOS) signaling pathway include the SOS1, SOS2, and SOS3 proteins [8,9]. SOS1, a plasma membrane-localized Na+/H+ antiporter, is pivotal for maintaining low cytosolic Na+ concentrations. SOS2 interacts with SOS3 via the FISL motif located in its C-terminal regulatory domain, facilitating activation [10]. SOS3 encodes a calcium-binding protein with an N-terminal myristoylation site; this interaction activates SOS2 kinase, subsequently phosphorylating SOS1 and enhancing its sodium extrusion activity in yeast [11]. Loss-of-function mutants of SOS genes exhibit hypersensitivity to NaCl stress. Notably, sos1 mutants display the most severe sensitivity observed to date [12].
The activity of plant SOS1 protein was characterized primarily by its sodium efflux capacity. Located at the plasma membrane, SOS1 utilizes energy derived from ATP hydrolysis by H+-ATPase, generating a proton electrochemical gradient. This gradient drives SOS1 to export excess Na+ ions, mitigating sodium toxicity and enhancing plant salt tolerance [13,14]. The SOS1 gene family encodes plasma membrane Na+/H+ antiporters, essential for maintaining ion homeostasis under salt stress. These proteins contain characteristic domains, including the Na+/H+ exchanger (NHX) domain, which facilitates the extrusion of Na+ ions. In Arabidopsis thaliana, SOS1 plays a pivotal role in salt tolerance by mediating Na+ efflux from roots and leaves to the stem [15]. The family has been identified in various plants, including Populus trichocarpa and Oryza sativa, and exhibits evolutionary conservation across monocots and dicots [16]. Physiological analyses of sos1 mutants and gene overexpression studies in Arabidopsis thaliana have further demonstrated SOS1′s crucial role in ion efflux from epidermal cell cytosol to external solutions and from parenchyma to vascular tissues, effectively maintaining low Na+ concentrations in root cells [17]. SOS1 is conserved across monocot and dicot species, underscoring its importance in plant evolution [18]. Interestingly, SOS1 activity is indispensable for the halophyte Thellungiella salsuginea, a close relative of Arabidopsis thaliana; a 50% reduction in SOS1 expression by RNA interference results in significant ion accumulation and loss of halophytic properties [5]. Guo et al. (2012) reported that under high salt stress, Suaeda salsa exhibits excessive Na+ accumulation in shoots, highlighting SOS1 and HKT proteins as central to sodium management when the compartmentalization capacity of NHX1 is limited [19]. Silencing SlSOS1 in tomato results in altered Na+ distribution patterns, notably reduced Na+ content in stems, and significantly elevated levels in roots and leaves compared to wild-type plants [20]. Similarly, heterologous expression of CcSOS1 from Chrysanthemum crassum in yeast mutants deficient in Na+ transport enabled normal growth under 70 mM NaCl, confirming its role in effective sodium efflux [21]. Collectively, these findings indicate that SOS1 proteins facilitate long-distance transport of harmful Na+ ions from roots and leaves to stems, thereby protecting vital photosynthetic and root tissues from salt-induced damage.
Tea plant (Camellia sinensis) is an economically important beverage crop, holding substantial economic and health importance [22]. Its derivatives support an industry valued at over one hundred billion US dollars annually, primarily due to bioactive compounds beneficial to human health [23]. Tea leaves contain abundant active substances, including tea polyphenols (EGCG), L-theanine, and caffeine, which clinical studies have consistently demonstrated to possess antioxidant, anti-inflammatory, lipid metabolism regulatory, and neuroprotective properties [24]. Tea plants prefer acidic soils, thriving optimally within a pH range of 4.5 to 5.5, and struggle to achieve economically viable yields in neutral or alkaline conditions [25]. Under salt stress, the photosynthetic pigment content in tea leaves decreases significantly, suppressing both photosynthesis and respiration [26]. Initial fluorescence efficiency increases notably, whereas maximum fluorescence, variable fluorescence, potential photochemical efficiency, and maximum photochemical efficiency all decline considerably [27]. Additionally, transcription factors and functional genes related to salt-stress adaptation, such as CsWRKY57 and CsWRKY40, are significantly upregulated under salt stress [28]. Overexpression of the tea plant gene CsbZIP4 in Arabidopsis thaliana markedly enhances salt tolerance, indicating its crucial role in salt-stress response [29]. Transcriptome analysis further reveals significant downregulation of genes associated with caffeine and tea polyphenol biosynthesis under prolonged salt stress [30].
Salinity is a critical environmental stress that can profoundly affect crop quality, influencing both nutritional value and flavor of food products [31]. In tea plant, recent studies indicate that salt stress elicits complex biochemical responses with dual effects on quality [32]. On one hand, moderate NaCl exposure can stimulate the accumulation of desirable metabolites; for example, salt treatment significantly increased the levels of L-theanine—a unique amino acid responsible for tea plant umami flavor—in young tea plant shoots. Salt-induced stress may also elevate certain polyphenolic compounds in tea plant leaves (key contributors to astringency and antioxidant capacity) as part of a general plant defense response [32]. Taken together, these findings illustrate that salt stress is a double-edged sword for food quality: mild-to-moderate salinity can trigger beneficial biochemical changes that enhance specific quality traits, whereas higher salt concentrations introduce undesirable effects—including excessive sodium build-up in edible tissues and stunted growth or yield reduction—which ultimately diminish crop quality [31,33]. However, current research on salt tolerance in tea plants primarily focuses on physiological responses, with limited molecular-level investigations.
Therefore, identifying and characterizing salt-stress resistance genes is essential for enhancing tea plant resilience, improving adaptability, and boosting tea quality and economic value. Despite this, studies on the SOS1 gene family in tea plants remain unreported, with their expression patterns and regulatory mechanisms poorly understood. To address this lack of information, this study conducted a genome-wide analysis to identify the tea plant SOS1 gene family, characterize conserved motifs, and elucidate SOS1 genes structures. Expression patterns of tea plant SOS1 genes were further analyzed across various environmental conditions and developmental stages. These findings provide a crucial biological foundation for future studies on salt tolerance in tea plants, offering theoretical support for breeding salt-tolerant tea cultivars.
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
Cuttings of tea plant (Camellia sinensis L. cv. ‘Shuchazao’) were cultivated hydroponically in nutrient solution ((NH4)2SO4 (0.75 mM), Ca(NO3)2·4(H2O)3 (0.25 mM), KH2PO4 (0.05 mM), K2SO4 (0.35 mM), CaCl2 (0.395 mM), MgSO4 (0.21 mM), NaFeEDTA (35.0 μM), H3BO3 (46.1 μM), MnSO4 (2.0 μM), CuSO4 (0.3 μM), ZnSO4 (2.0 μM), and Na2MoSO4 (0.5 μM)) and grown in a controlled growth chamber. The environmental conditions were set to 28/25 °C (day/night), 75% relative humidity, a 16/8 h light/dark photoperiod, and a light intensity of 300 μmol·m−2·s−1. The nutrient solution was replenished every five days [34]. For salt-stress treatment, cuttings were transferred to hydroponic solution supplemented with 200 mM NaCl. The first fully expanded leaf was harvested at 24, 48, and 72 h post-treatment. For drought-stress treatment, cuttings were subjected to 20% (w/v) PEG-6000 solution, with the first fully expanded leaf collected at identical time points [35]. For auxin treatment, cuttings were incubated in hydroponic solution containing 200 μM NAA. The first fully expanded leaf was sampled at 1, 8, and 15 days post-treatment [36]. All harvested leaves were flash-frozen in liquid nitrogen and stored at −80 °C for RNA extraction. Three biological replicates were performed per treatment.
2.2. Identification of the SOS1 Gene Family in Tea Plant
The reference genome of Camellia sinensis cv. ‘Shuchazao’ was obtained from publicly available data [37]. Whole-genome protein sequences (FASTA format) were retrieved from the Genome Warehouse database [38]. SOS1 protein sequences from Arabidopsis thaliana (At), Oryza sativa (Os), and Populus trichocarpa (Pt) were acquired from respective databases: TAIR (https://www.arabidopsis.org, accessed on 14 January 2025), Rice Genome Annotation Project (http://rice.plantbiology.msu.edu, accessed on 14 January 2025), and Ensembl Plants (https://plants.ensembl.org/Populus_trichocarpa, accessed on 14 January 2025). Candidate CsSOS1 genes were screened using the SOS1 domain HMM profile (PF00999) via HMMER v3.0, with domain validation performed using the Pfam database.
2.3. Phylogenetic Analysis of the Tea Plant SOS1 Protein Family
SOS1 protein sequences from tea plant (Cs) and reference species (At, Os, and Pt,) were aligned using MAFFT software (aligned using MAFFT v7.475, and then a maximum-likelihood (ML)). phylogenetic tree was constructed using FastTree version 2.1.10 with a Generalized Time-Reversible (GTR) model and Shimodaira–Hasegawa (SH) test [39,40]. Protein physicochemical properties (molecular weight, theoretical pI) were predicted using ExPASy ProtParam, and subcellular localization was inferred via CELLO v2.5.
2.4. Chromosomal Localization and Collinearity Analysis
Chromosomal coordinates of CsSOS1 genes were extracted from genome annotation (GFF3) files using TBtools-II v1.120. Intra-genomic collinear blocks were identified with OrthoFinder v2.2.6 (default parameters) and visualized as synteny maps [41].
2.5. Protein Sequence Analysis and Gene Nomenclature
Based on protein sequence characteristics and phylogenetic relationships, CsSOS1 genes were classified into subfamilies. Genes were systematically named according to their chromosomal positions from smallest to largest. Physicochemical properties, including amino acid length, molecular weight, and theoretical pI, were analyzed using ExPASy (https://web.expasy.org/compute_pi, accessed on 24 January 2025). TMHMM Server v2.0 (https://services.healthtech.dtu.dk/services/TMHMM-2.0, accessed on 24 January 2025) was employed to predict transmembrane helices.
2.6. Analysis of Conserved Motifs and Gene Structures
Conserved motifs of CsSOS1 proteins were identified using MEME (https://meme-suite.org/meme/tools/meme, accessed on 25 January 2025). Gene structural features were extracted from genome annotation files, and integrated phylogenetic, motif, and gene structure diagrams were visualized using TBtools.
2.7. Prediction of Cis-Acting Regulatory Elements in Promoters
Cis-acting regulatory elements in the promoter regions (2000 bp upstream of the start codon) of CsSOS1 genes were identified using PlantCARE and PLACE databases (http://www.dna.affrc.go.jp/PLACE, accessed on 25 January 2025). Potential transcription factor binding sites were predicted using the Plant Reg Map tool, and their physical locations were visualized [39,42].
2.8. Expression Analysis of SOS1 in Various Tea Plant Tissues
Transcriptomic data (TPM values) of identified CsSOS1 genes across different tea plant tissues (flowers, stems, roots, buds, young leaves, mature leaves, and old leaves) were downloaded from the TPIA database (https://tpia.teaplants.cn, accessed on 25 January 2025). The data were normalized, and expression heatmaps were generated using TBtools software.
2.9. RNA-seq-Based Expression Analysis of CsSOS1 Genes
The transcriptomic data from treatments with melatonin, gibberellic acid, and NAA were used to determine the expression patterns of the tea plant SOS1 gene family under different hormone treatments [35,36].
2.10. RT-qPCR Analysis of CsSOS1 Gene Expression
Total RNA from leaves (the first leaf of two-year-old cutting seedlings) of tea plant was extracted using the modified CTAB method [43]. First-strand cDNA was synthesized from 1 μg of total RNA using Prime Script RT Reagent Kit (Takara, Otsu, Shiga, Japan). Real-time PCR was performed using the Bio-Rad Real-time thermal cycler CFX96 with SYBR Premix ExTaq™ Kit (Takara, Otsu, Shiga, Japan). The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and β-actin were used as controls. Candidate gene-specific primers were designed with Primer Premier v6.0 software (Table S1). RT-qPCR was carried out using a CFX100 Real-time PCR System (Bio-Rad, Hercules, CA, USA) with the following cycling conditions: initial denaturation at 95 °C for 30 s, followed by 40 cycles of 95 °C for 15 s, 60 °C for 30 s, and 72 °C for 30 s. Melting curves ranged from 55 °C to 95 °C for 5 s, with samples stored at 4 °C. SYBR Green Real-time PCR Mastermix was used as per the manufacturer’s protocol, and relative expression levels were calculated using the 2−ΔΔCT method [44].
3. Results
3.1. Identification of the CsSOS1 Gene Family in Tea Plant
A total of 51 SOS1 genes were identified in the tea plant genome and systematically designated CsSOS1-1 to CsSOS1-51 according to their chromosomal positions (short arm to long arm), adhering to standard nomenclature conventions (Table S2). To further characterize the physicochemical properties of the CsSOS1 genes, we predicted their protein lengths (in amino acids), molecular weights (MWs), and isoelectric points (pI). The analysis revealed that protein lengths varied from 119 amino acids (CsSOS1-14) to 1231 amino acids (CsSOS1-18 and CsSOS1-40), with molecular weights ranging from 13.20 kDa (CsSOS1-14) to 132.43 kDa (CsSOS1-18) and isoelectric points ranging from 4.98 (CsSOS1-38) to 9.71 (CsSOS1-27) (Table S3).
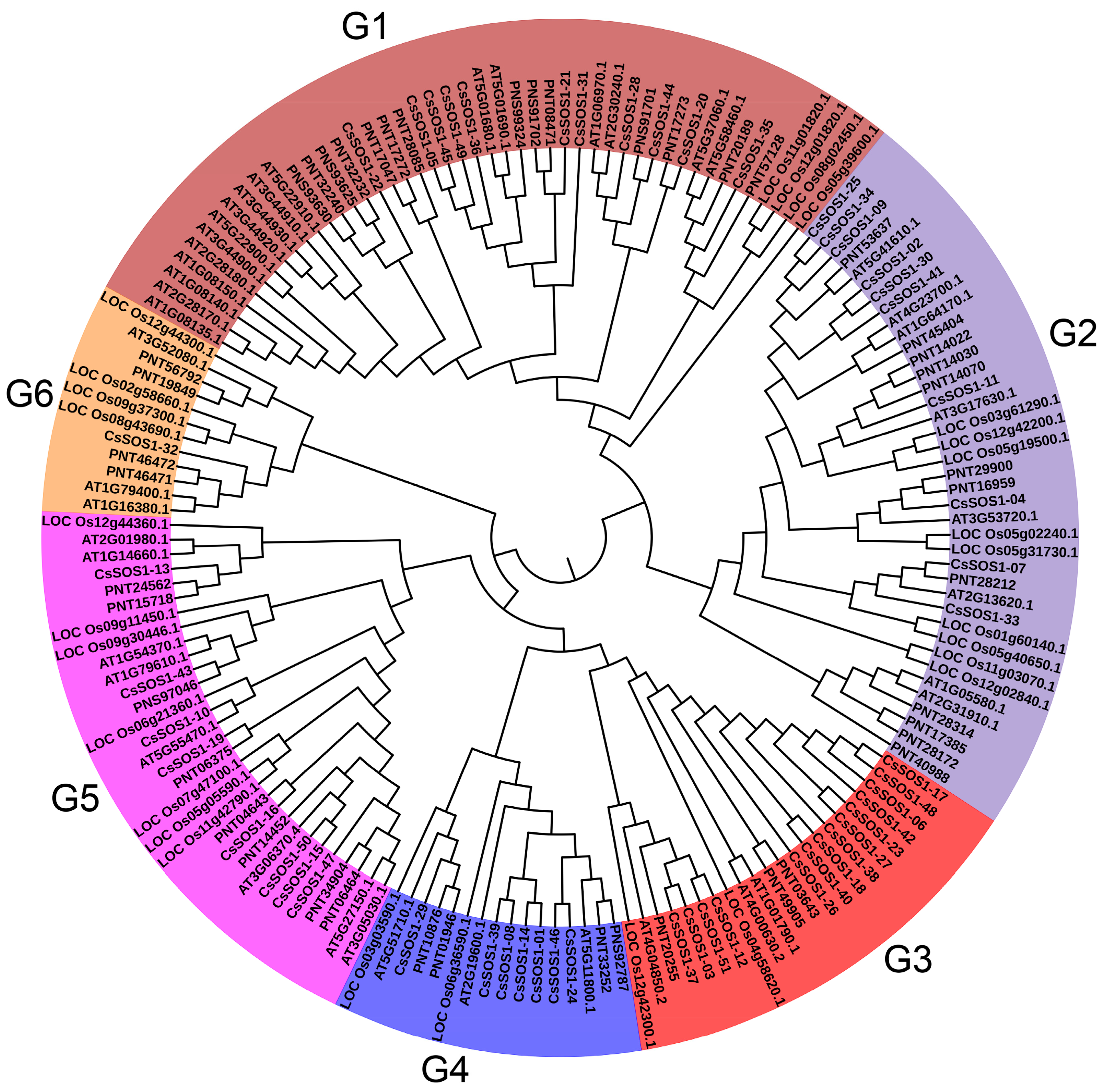
Subcellular localization predictions using the Cell-PLoc 2.0 tool indicated that most CsSOS1 proteins were localized to the plasma membrane, with the exception of CsSOS1-26, which was located in the cytoplasm. These findings provided preliminary insights into the molecular functions of the CsSOS1 proteins (Table S4). To investigate the evolutionary relationships of the CsSOS1 protein family and their counterparts in other species, we constructed a phylogenetic tree of SOS1 proteins from Camellia sinensis, Populus trichocarp, Oryza sativa, and Arabidopsis thaliana. The phylogenetic analysis revealed that CsSOS1 proteins fall into six subgroups, with the largest subgroup containing 46 SOS1 members, 11 of which were CsSOS1 proteins. This subgroup was the most abundant across all six groups. Phylogenetic comparisons further indicated that tea plant SOS1 proteins shared strong homology with those in Populus trichocarp and Arabidopsis thaliana (Figure 1).
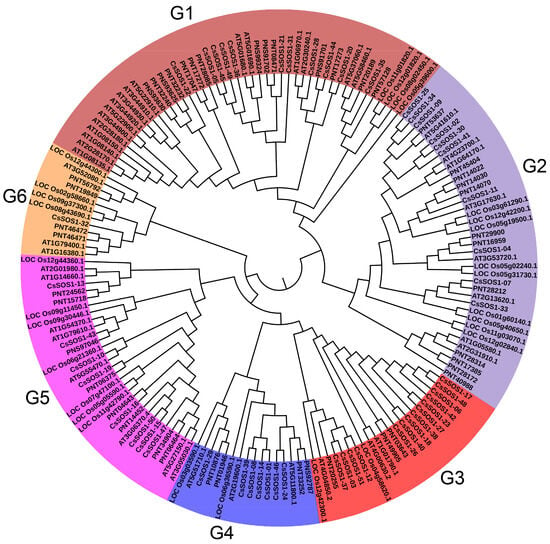
Figure 1.
Phylogenetic analysis of SOS1s proteins among Camellia sinensis, Populus trichocarp, Oryza sativa, and Arabidopsis thaliana. Different colors correspond to different groups.
3.2. Chromosomal Localization and Synteny Analysis of the CsSOS1 Gene Family in Tea Plant
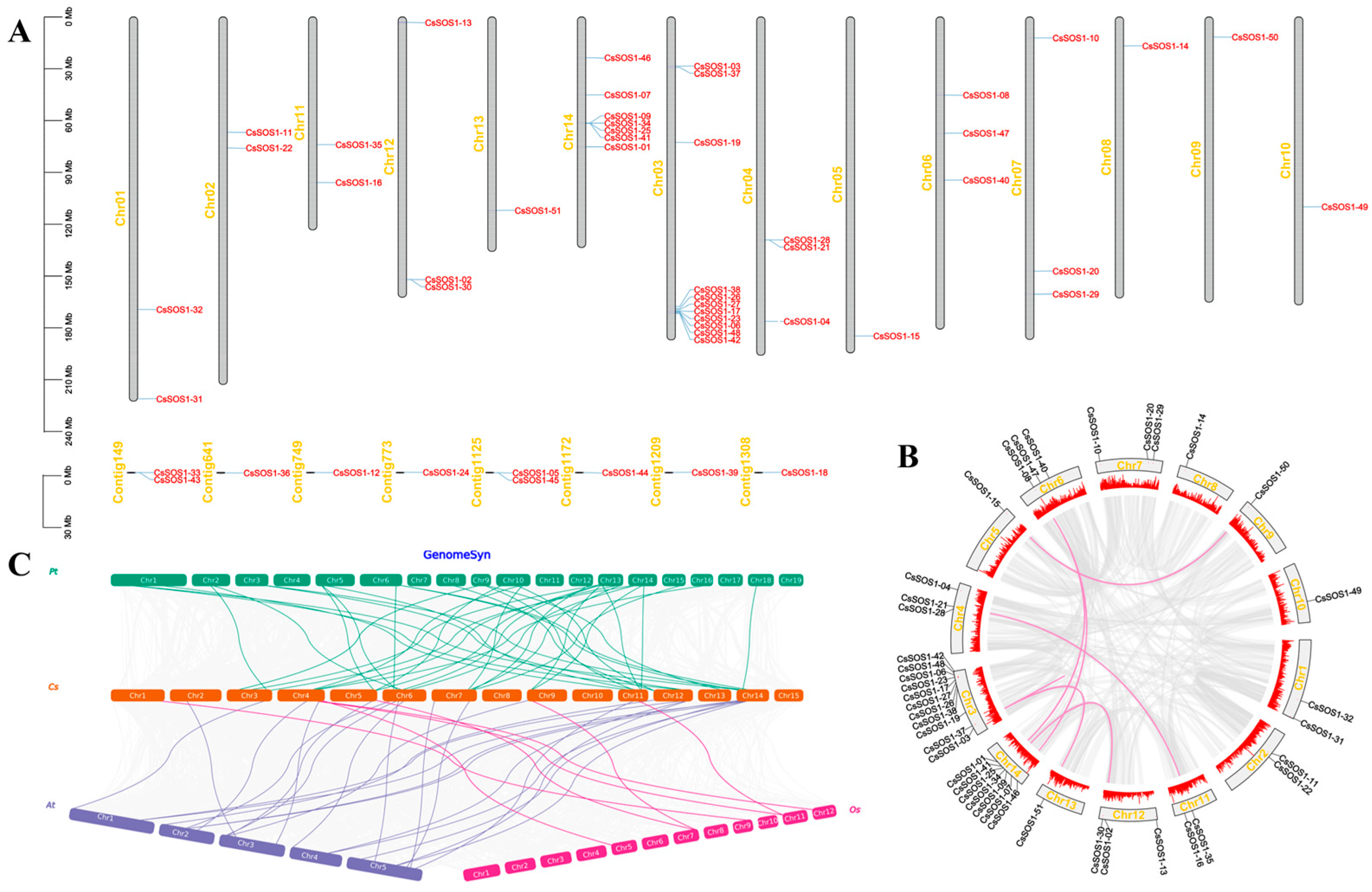
Genomic localization analysis revealed that the 51 CsSOS1 genes are distributed across 14 chromosomes. Chromosome 3 harbored the highest density (11 genes), followed by chromosome 14 (7 genes) (Figure 2A). Tandem duplication analysis identified 11 clusters encompassing 13 CsSOS1 genes (Figure 2B; Table S5). Comparative synteny analysis between Camellia sinensis, Arabidopsis thaliana, Oryza sativa, and Populus trichocarpa revealed collinear ortholog pairs: 23 with A. thaliana, 7 with O. sativa, and 41 with P. trichocarpa (Figure 2C; Table S6). These results demonstrate stronger collinearity between tea plant SOS1 genes and eudicot models (P. trichocarpa and A. thaliana) than with monocots (O. sativa), highlighting evolutionary conservation within dicotyledonous plants.
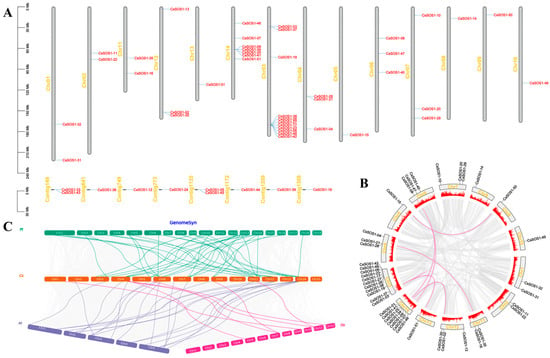
Figure 2.
Chromosomal localization and gene collinearity analysis for CsSOS1s. (A): The localization of CsSOS1s on 14 chromosomes. (B): Intraspecific collinearity analysis of CsSOS1s. (C): Interspecific collinearity analysis of CsSOS1 genes in Camellia sinensis (Cs), populus trichocarp (Pt), Oryza sativa (Os), and Arabidopsis thaliana (At).
3.3. Prediction of Cis-Elements in the Promoter Sequences of the CsSOS1 Gene Family in Tea Plant
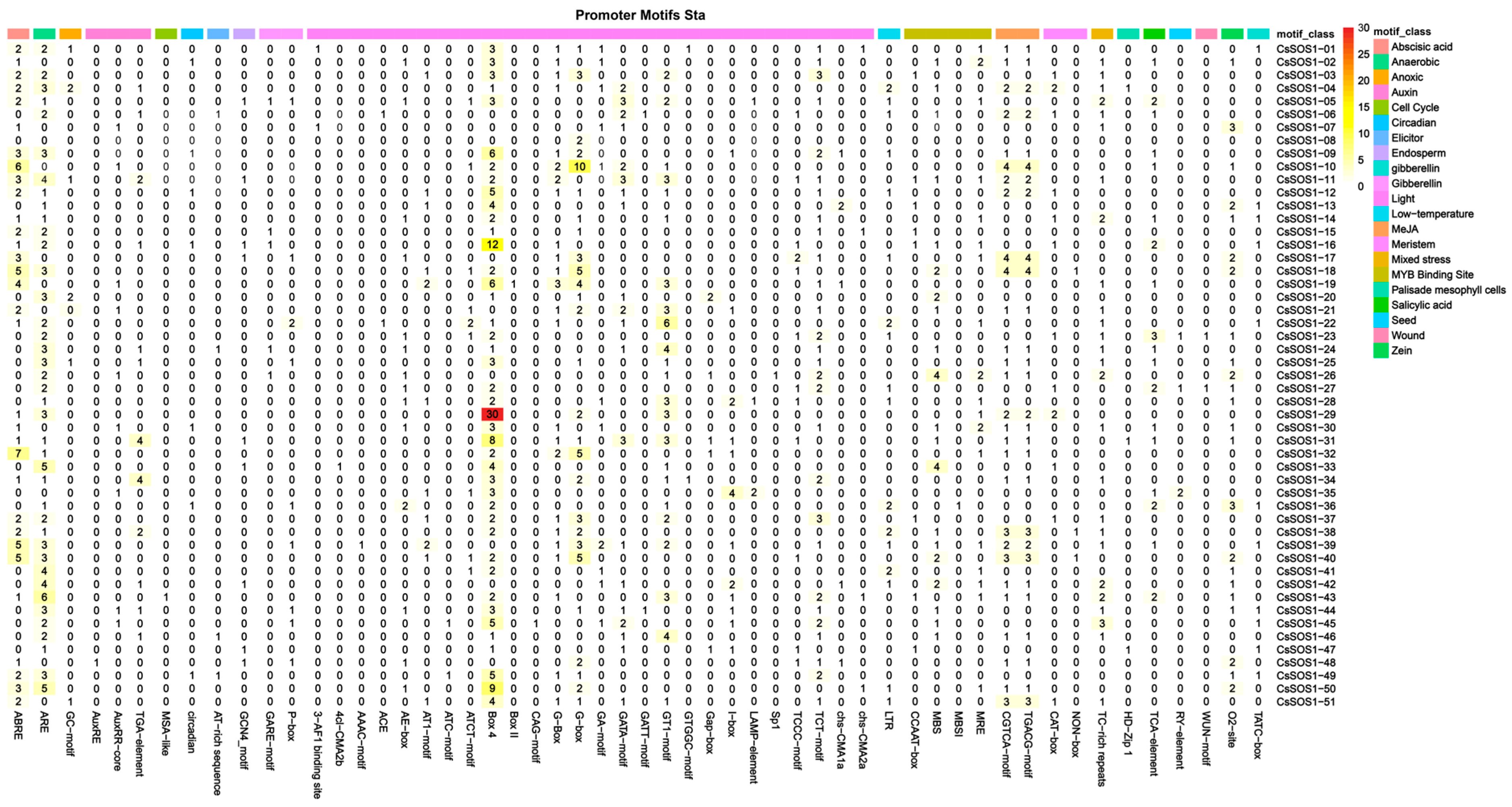
To investigate hormonal, environmental stress, and developmental signals associated with the CsSOS1 gene family, promoter regions (2000 bp upstream) of all 51 genes were analyzed using PlantCARE. The analysis identified diverse core cis-regulatory elements, including hormone-responsive motifs: methyl jasmonate (MeJA; CGTCA- and TGACG-motifs), abscisic acid (ABA; ABRE), gibberellin (GA; P-box, TATC-box, GARE-motif), auxin (IAA; TGA-element, AuxRR-core, TGA-box, AuxRE), and salicylic acid (SA; TCA-element); alongside stress-related elements: ARE (anaerobic induction), MBS (drought-responsive MYB binding), LTR (low-temperature response), GC-motif (hypoxia-specific), and TC-rich repeats (defense/stress response) (Figure 3). Quantitatively, MeJA-responsive elements were most abundant, followed by ABA-responsive elements, while among stress elements, light-response motifs predominated with anaerobic induction elements ranking second. These findings demonstrate that CsSOS1 genes play integral roles in mediating hormone signaling and coordinating responses to diverse biotic/abiotic stresses.
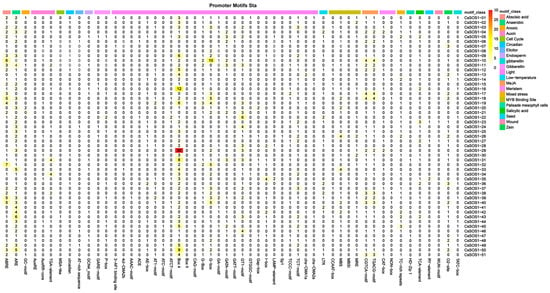
Figure 3.
Analysis of cis-acting elements upstream of the CsSOS1s. Different CAREs are displayed in different colors.
3.4. Analysis of Conserved Protein Motifs and Gene Structure in the CsSOS1 Gene Family of Tea Plant
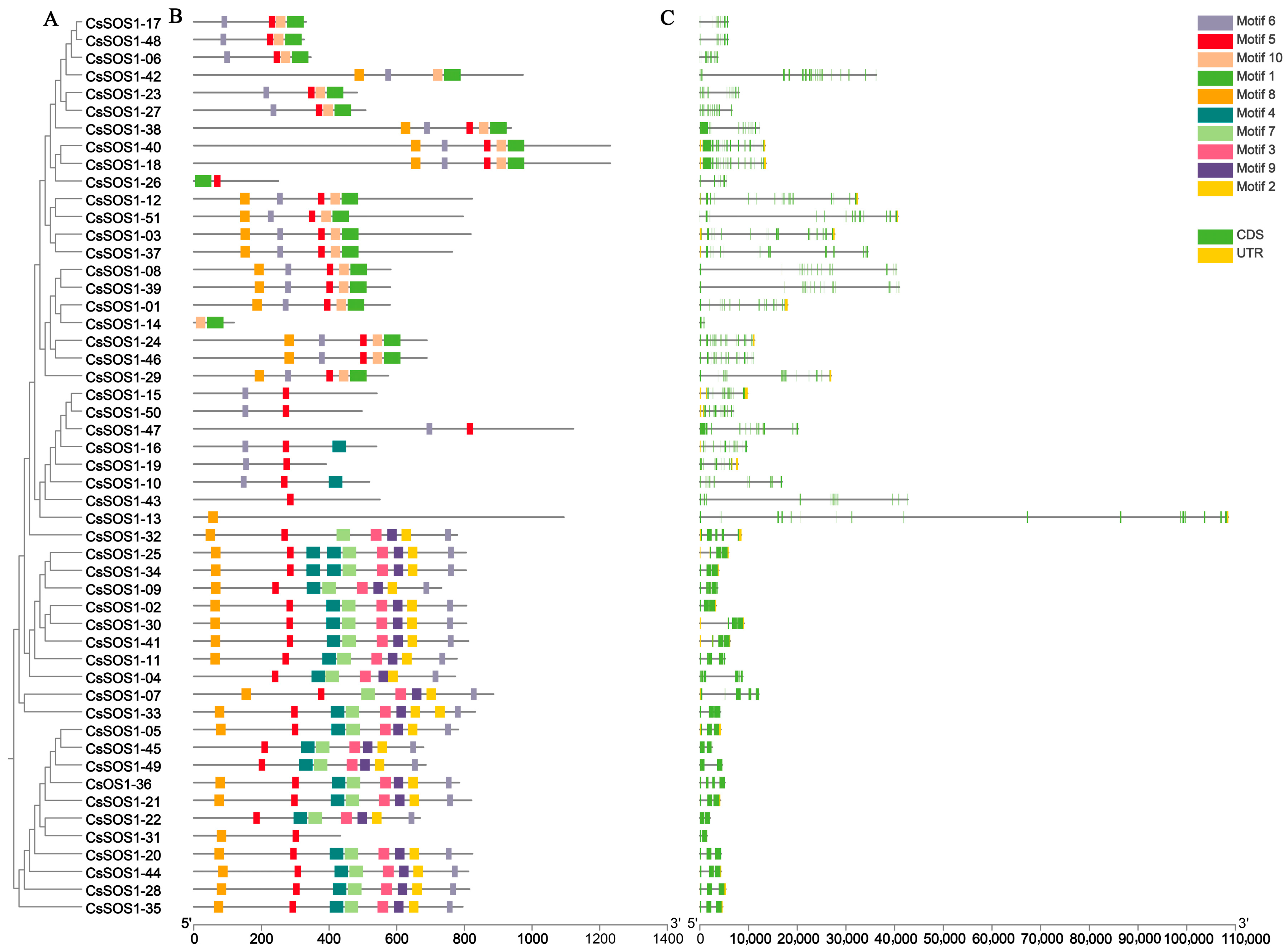
The exon–intron structure is a key evolutionary feature of genes and provides important clues about gene functional diversification. Therefore, we analyzed the conserved protein motifs and gene structure of the CsSOS1 gene family. Phylogenetic analysis of the CsSOS1 genes in tea plant revealed that the motif architecture within the same evolutionary branch was largely consistent, suggesting that these genes might share similar functions (Figure 4A). Exon–intron mapping demonstrated variable exon counts ranging from 2 (CsSOS1-49, CsSOS1-22, and CsSOS1-31) to 25 (CsSOS1-42), with phylogenetically proximate genes exhibiting analogous structures primarily differing in exon/intron lengths (Figure 4B). Gene lengths spanned from 360 bp (CsSOS1-14) to 3693 bp (CsSOS1-18). Analysis of the functional sites within the conserved motifs using the Eukaryotic Linear Motif (ELM) resource server revealed significant functional diversity among these sites. Most of the functional sites were associated with phosphorylation, kinase phosphorylation, protein interactions, and sorting signals (Figure 4C).
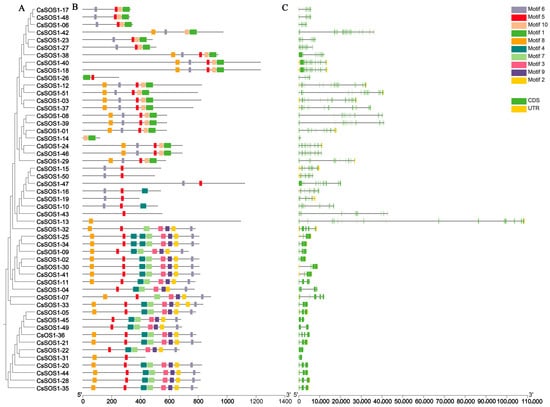
Figure 4.
The phylogenetic tree, conserved motifs, and gene structure of the CsSOS1 gene family members. (A): A phylogenetic tree was constructed using the maximum likelihood method, incorporating 51 CsSOS1 proteins. (B): The conserved motifs of CsSOS1 proteins were depicted, with various patterns represented by colored boxes. (C): The coding sequences (CDS) and untranslated regions (UTR) were highlighted with yellow and green boxes, respectively, while introns were indicated by black lines. Phylogenetic tree, motif, and gene structure predictions were performed using TBtools, with the scale bar located at the bottom.
3.5. Gene Ontology (GO) Enrichment Analysis of the CsSOS1 Gene Family in Tea Plant
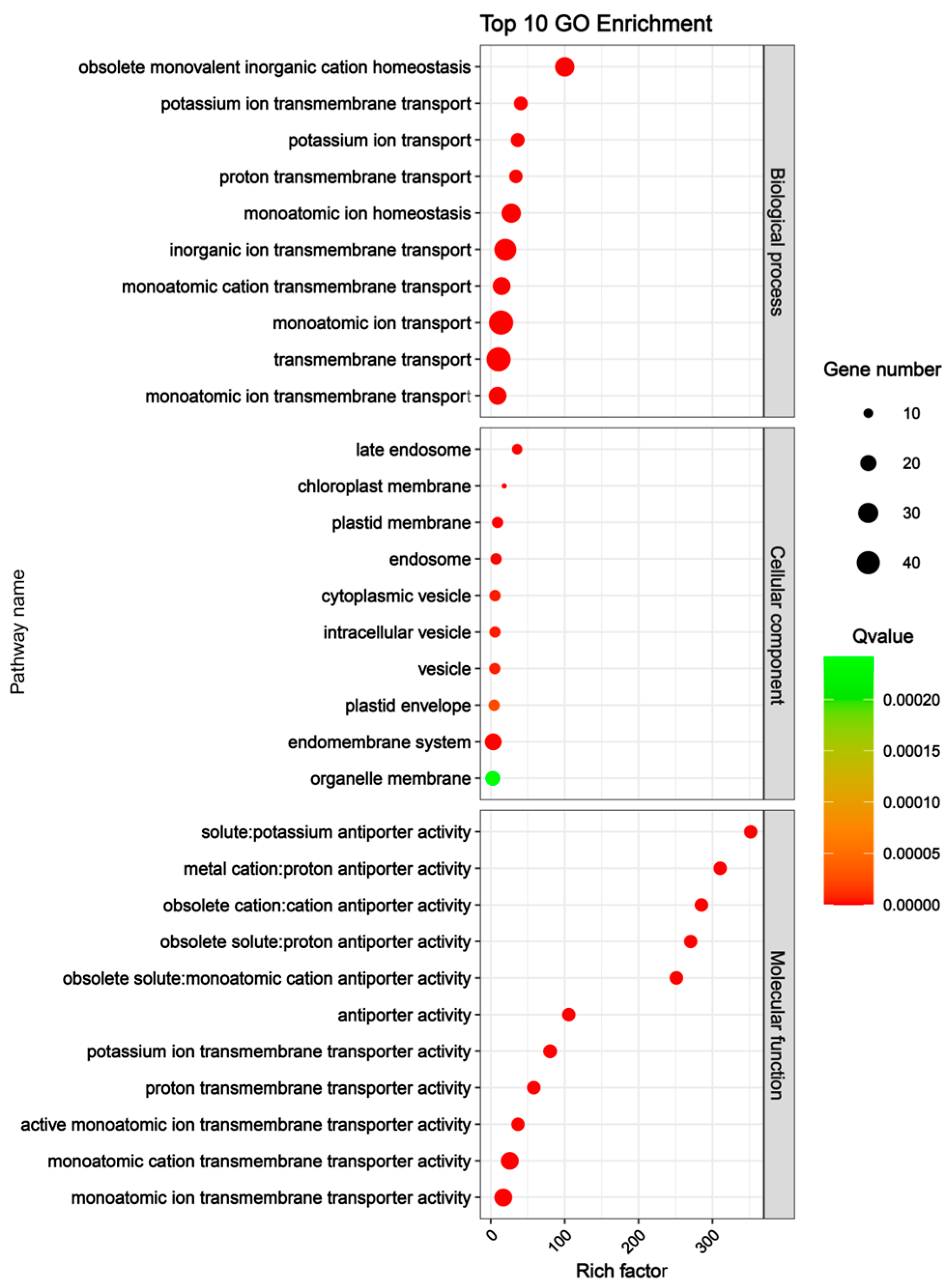
To explore the potential biological functions of the CsSOS1 gene family, we performed Gene Ontology (GO) enrichment analysis on the 51 CsSOS1 genes, categorizing their involvement across three biological processes: biological process, molecular function, and cellular component. The results revealed that, within the biological process category, CsSOS1 genes were predominantly enriched in the pathways of monoatomic ion transport and transmembrane transport. In terms of molecular function, these genes were primarily associated with monoatomic cation transmembrane transporter activity and monoatomic ion transmembrane transporter activity. Regarding cellular component, CsSOS1 genes were mainly enriched in the endomembrane system and organelle membrane pathways (Figure 5). These GO enrichment results indicated that CsSOS1 genes were primarily involved in transmembrane transport and cellular membrane composition, supporting their functional role in maintaining cytosolic ionic homeostasis.
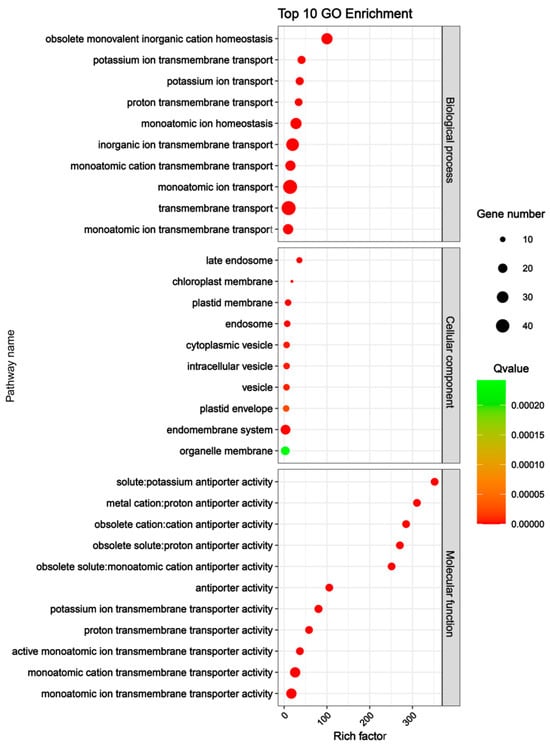
Figure 5.
GO enrichment analysis of CsSOS1 genes in tea plant.
3.6. Co-Expression Analysis of the CsSOS1 Gene Family with Other Genes in Tea Plant
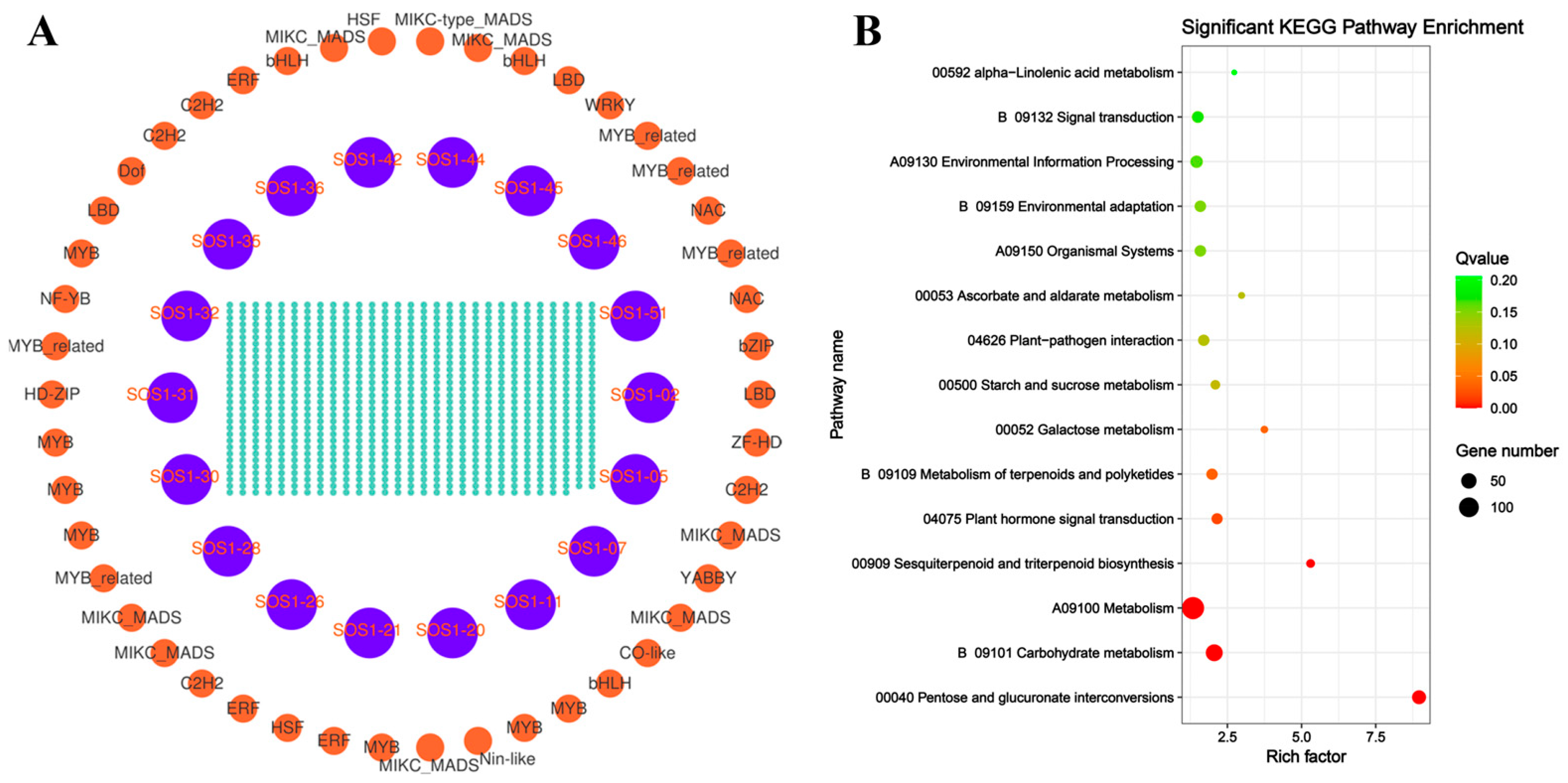
To further investigate the functional relationships and regulatory networks between CsSOS1 genes and other genes in tea plant, we calculated the Pearson correlation coefficients and constructed a co-expression network for CsSOS1 genes using Cytoscape software (version 3.10.1). KEGG enrichment analysis was then performed on the co-expressed genes to explore the association between CsSOS1 and other tea plant genes. The results revealed that a total of 197 genes were co-expressed with CsSOS1s. KEGG enrichment analysis showed that the most enriched pathway was the metabolism pathway, which included 135 genes, followed by the carbohydrate metabolism pathway, with 66 genes. These findings suggest that these gene sets played a central role in metabolic regulation, with carbohydrate metabolism serving as a crucial hub for energy supply, signal transduction, and stress adaptation (Figure 6, Table S7).
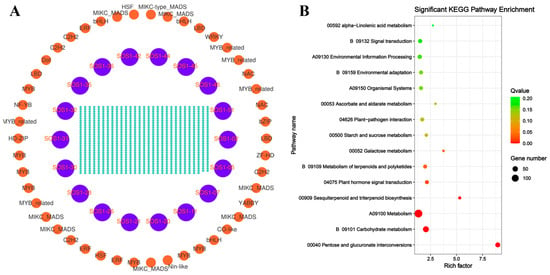
Figure 6.
Co-expression analysis of tea plant genes. (A): Relationship of co-expressed genes in tea plants. (B): KEGG pathway enrichment analysis of the co-expressed genes. The criteria for co-expression of the CsSOS1s gene family were |R| ≥ 0.8 and p ≤ 0.001.
3.7. Expression Profiling of CsSOS1s Under Different Tissues and Environmental Stress
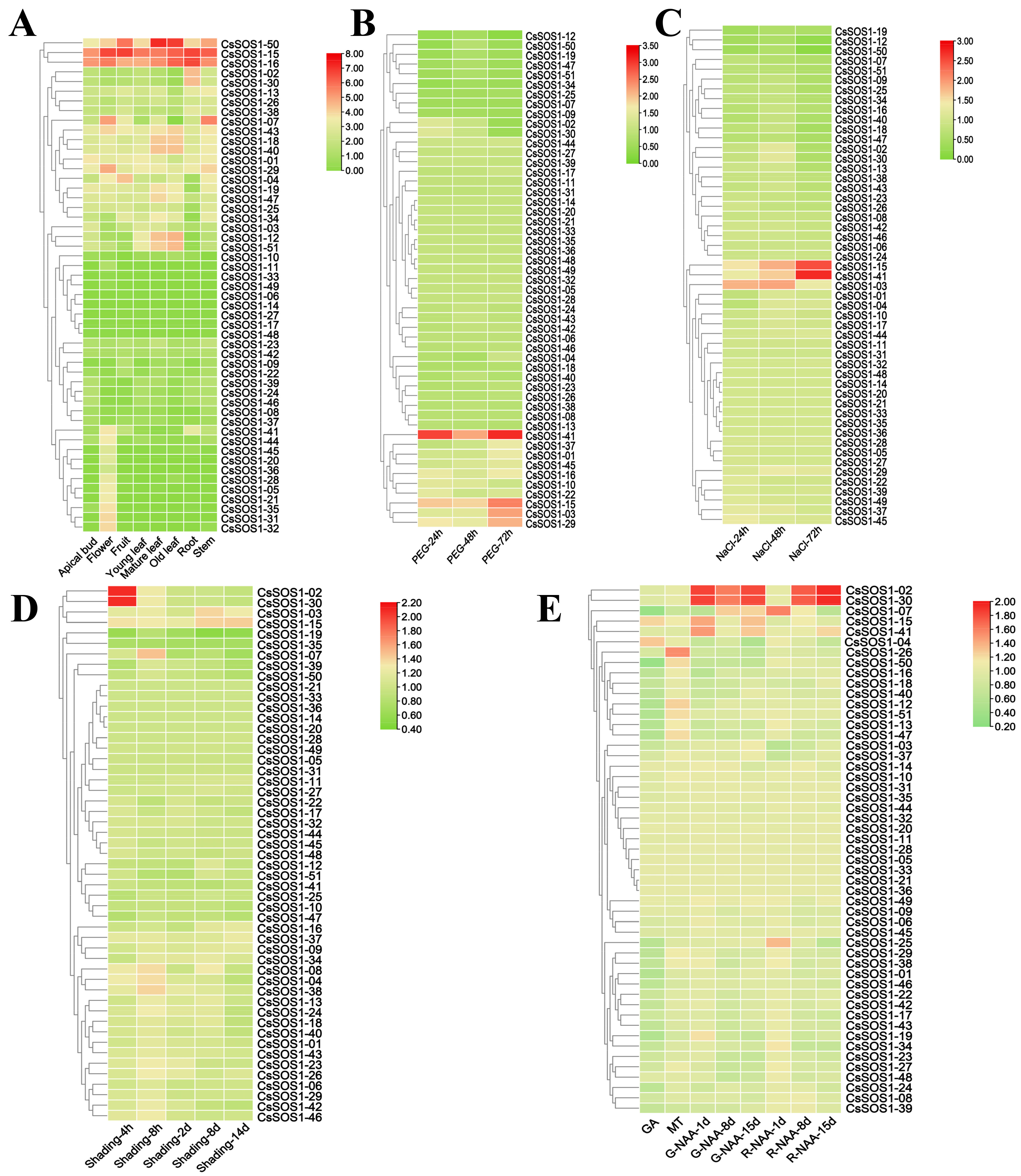
To investigate the biological functions of the CsSOS1 gene family across different tissues, we analyzed the expression profiles of all identified CsSOS1 genes in eight distinct tissue types: apical bud, flower, fruit, young leaf, mature leaf, old leaf, root, and stem. The results revealed that CsSOS1-15 and CsSOS1-16 exhibited high expression levels across all tissues, while CsSOS1-50 was highly expressed in the fruit, mature leaf, and old leaf. Several members of the CsSOS1 family showed tissue-specific expression patterns; for example, CsSOS1-07 was exclusively expressed in the flower and stem, while CsSOS1-02 and CsSOS1-30 were expressed only in the root (Figure 7A).
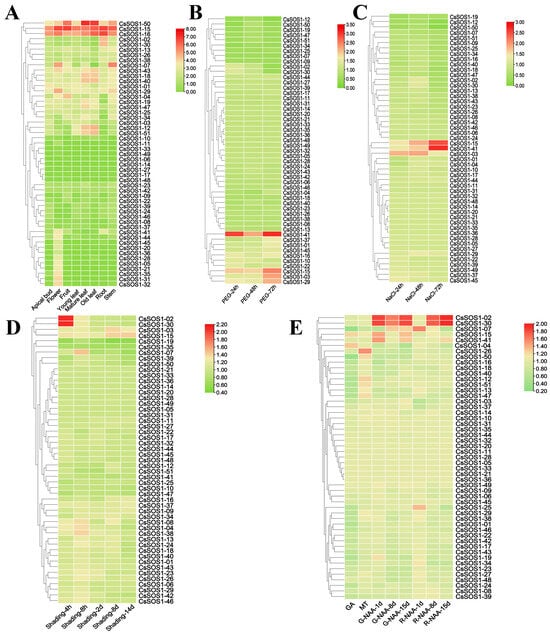
Figure 7.
Expression profile of CsSOS1 genes across different tissues and under various environmental stresses. (A): Expression pattern of CsSOS1 genes in different tissues. (B): Expression profile of CsSOS1 genes under drought stress. (C): Expression pattern of CsSOS1 genes under salt stress. (D): Expression profile of CsSOS1 genes under shade stress. (E): Expression of CsSOS1 genes following hormone treatments (GA: Gibberellin; MT: Melatonin; NAA: Naphthalene acetic acid; h: hours; d: days).
We further investigated CsSOS1 transcriptional responses to abiotic stresses (drought, salt, and shading) and plant hormones (auxin (NAA), gibberellin (GA), and melatonin (MT)) (Figure 7, Table S7). Under drought stress, the expression of CsSOS1-41 was significantly upregulated, while CsSOS1-15, CsSOS1-03, and CsSOS1-29 showed moderate upregulation (Figure 7B). In response to salt stress, CsSOS1-03 was highly expressed during early stages of treatment, while CsSOS1-15 and CsSOS1-41 showed significantly increased expression at later stages (Figure 7C). These results indicated that CsSOS1-15 and CsSOS1-41 played a prominent role in the tea plant’s response to environmental stress. Furthermore, CsSOS1-02 and CsSOS1-30 exhibited a significant upregulation in expression during early shading treatments, and their expression remained unique among all CsSOS1 genes, suggesting their potential involvement in the shading response of tea plant (Figure 7D). Regarding hormone treatments, the transcriptional profiles revealed no significant change in CsSOS1s expression under GA treatment. However, CsSOS1-26 showed specific upregulation under MT treatment. NAA treatment elicited distinct induction of CsSOS1-02 and CsSOS1-30 peaking significantly at 8 and 15 days post-treatment (Figure 7E).
Research across various plant species has demonstrated that SOS1 was a conserved protein present in both monocots and dicots. The SOS1 protein facilitates the long-distance transport of Na+, a harmful ion to plant growth, from roots and leaves to the stem, thereby protecting root and photosynthetic tissues. Additionally, SOS1 plays a crucial role in regulating intracellular K+ homeostasis. Oh et al. (2010) found that in Arabidopsis thaliana plants silenced for AtSOS1, the activity of K+-transporting proteins was significantly suppressed, along with a corresponding reduction in gene expression levels [45]. This suggests that under salt stress, SOS1 not only helps in Na+ efflux but also ensures K+ uptake, maintaining ionic balance. To investigate the potential regulatory functions of the CsSOS1 gene family in tea plant, we analyzed transcriptomic data to map the expression patterns of SOS1 genes under drought, salt, shading stress, and hormone treatments. Our results revealed that CsSOS1-03, CsSOS1-15, and CsSOS1-41 were specifically upregulated under both salt and drought stress, demonstrating that the SOS1 family was functionally active not only in salt-stress adaptation but also in drought-stress response mechanisms. Hormonal treatment analyses showed that CsSOS1-02 and CsSOS1-30 were upregulated in response to auxin treatment. Previous studies have demonstrated that auxin regulated plant responses to abiotic stress by modulating root architecture, antioxidant systems, ion homeostasis, and interactions with other hormones. Previous studies demonstrate that auxin regulates plant responses to abiotic stress by modulating root architecture, antioxidant systems, ion homeostasis, and hormonal crosstalk. Building on this, we propose that CsSOS1-02 and CsSOS1-30 in tea plant enhance abiotic stress tolerance through auxin-mediated maintenance of ion homeostasis.
3.8. RT-qPCR Validation of CsSOS1s Under Drought, Salt, and Auxin Treatments
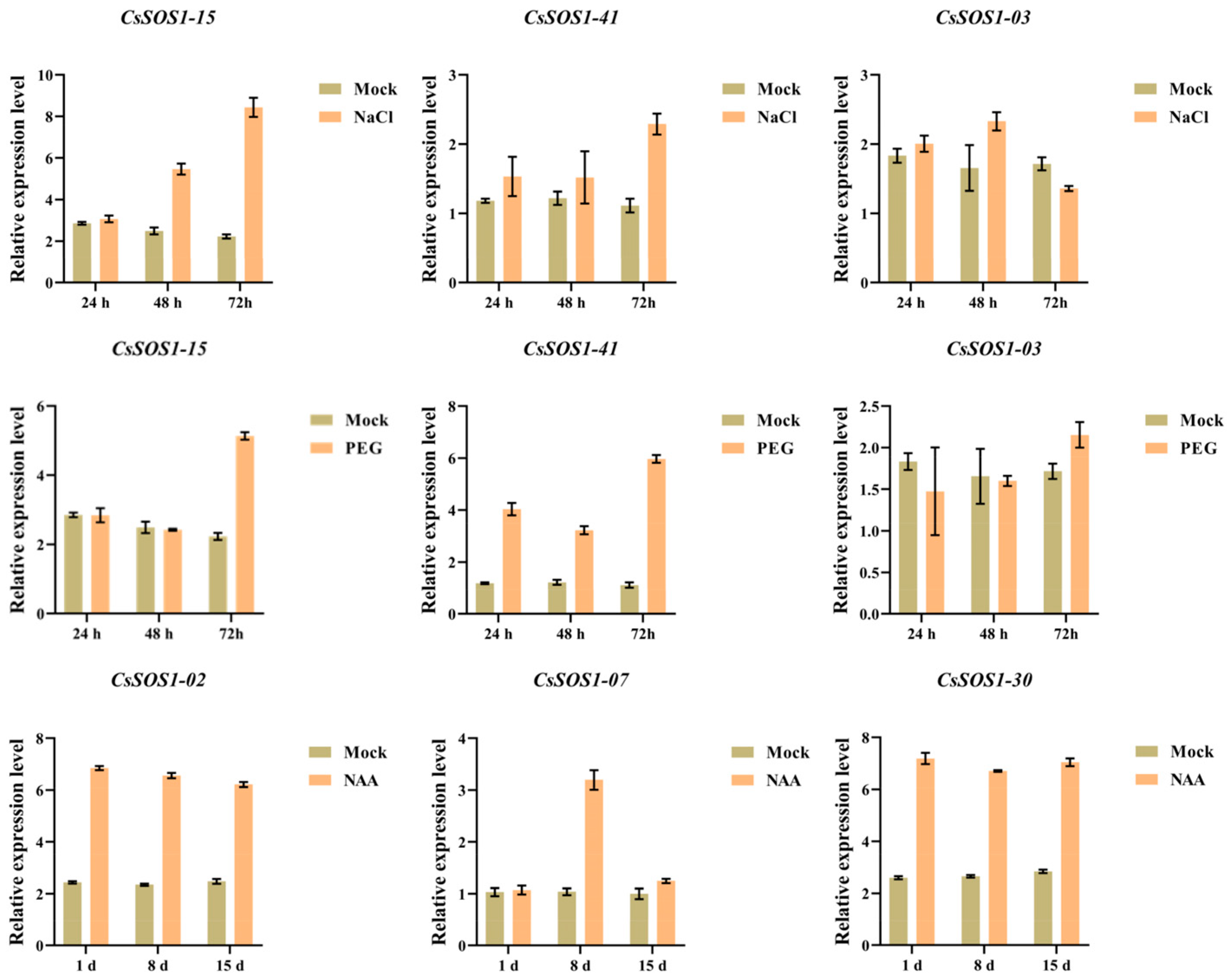
Transcriptomic analysis under abiotic stress and hormone treatments revealed that CsSOS1-03, CsSOS1-15, and CsSOS1-41 were transcriptionally upregulated in response to both drought and salt stress, while CsSOS1-02, CsSOS1-07, and CsSOS1-30 showed enhanced expression following auxin treatment. To validate these expression patterns, RT-qPCR was performed on selected genes under corresponding stress and hormonal conditions (Figure 8). The RT-qPCR analysis revealed that under salt stress, CsSOS1-15 expression was significantly elevated compared to the control group at 48 h and 72 h post-treatment (2.2- and 3.8-fold higher than the Mock, respectively), while CsSOS1-41 exhibited substantially higher expression at 72 h (2.1-fold higher than Mock). CsSOS1-03 expression significantly increased at 48 h following salt exposure (1.5-fold higher than Mock). Under drought stress, all three genes (CsSOS1-15, CsSOS1-41, and CsSOS1-03) showed significantly upregulated expression at 72 h (2.3, 5.5, and 1.3-fold higher than the Mock, respectively) with CsSOS1-41 demonstrating heightened sensitivity to drought (3.5, 2.8, and 5.5-fold higher than the Mock, respectively). For NAA treatment, CsSOS1-02 and CsSOS1-30 displayed significantly elevated expression levels versus the control at 1 d, 8 d, and 15 d (CsSOS1-02: 2.8, 2.8, and 2.6-fold higher than the Mock; CsSOS1-30: 2.7, 2.7, and 2.5-fold higher than the Mock), whereas CsSOS1-07 expression was significantly higher at 8 d (2.9-fold higher than the Mock). The RT-qPCR results corroborated the transcriptome data, confirming the consistent upregulation of these CsSOS1 genes under the respective treatments.
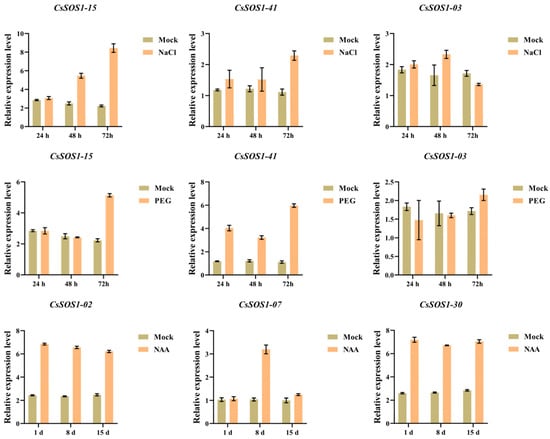
Figure 8.
RT-qPCR profiles of CsSOS1 genes under salt, drought, and auxin treatments. h stands for hour, and d stands for day (Mock represents the control group, and error bars represent SD).
4. Discussion
High soil salinity is one of the most pervasive abiotic stresses limiting crop productivity worldwide [46]. To cope with high external Na+, plants have evolved the Salt Overly Sensitive (SOS) pathway—comprising SOS1, SOS2, and SOS3—which regulates cellular signaling and maintains ion homeostasis under salt stress [47]. SOS1, a plasma membrane Na+/H+ antiporter energized by H+-ATPase activity, expels cytosolic Na+ in exchange for H+ and thereby mediates Na+ efflux, K+ homeostasis, Ca2+ transport, pH regulation, oxidative-stress balance, and long-distance Na+ translocation [48]. Functional complementation studies show that overexpression of rice OsSOS1 or poplar PtSOS1 rescues the salt-sensitive phenotype of Arabidopsis thaliana sos1 mutants, and co-expression of OsSOS1 or AtSOS2 in Na+-export-deficient yeast dramatically enhances Na+/H+ exchange activity and restores salt tolerance [45,49].
As sessile perennials, tea plants face inevitable exposure to drought, low temperature, salinity, and other environmental extremes [50]. With intensifying soil salinization, salt stress has become a major constraint on tea cultivation, reducing photosynthetic pigment content, impairing photosynthetic efficiency, and consequently diminishing growth parameters (plant height and stem diameter), biomass accumulation, yield, and leaf quality [51]. Recent mechanistic advances reveal that exogenous salicylic acid and hydrogen sulfide alleviate salt sensitivity; arbuscular mycorrhizal fungi inoculation reduces malondialdehyde accumulation and membrane lipid peroxidation; exogenous spermidine mitigates ROS-induced oxidative damage [32,52]. Transcriptomic analyses under salt stress reveal significant differential expression of stress-related proteins and transcription factors such as HSP, LEA, WRKY, and bHLH [53]. Despite SOS1′s established role in salt tolerance across species, its functional characterization in tea plants remains unexplored. This study, therefore, aims to identify abiotic stress-responsive CsSOS1 homologs in the tea genome, establishing a molecular foundation for elucidating their regulatory mechanisms in salt tolerance.
This study identified 51 SOS1 gene family members in the tea plant genome, distributed across all 14 chromosomes (Figure 1; Table S2). This gene count substantially exceeds those of Arabidopsis thaliana and Populus trichocarpa, suggesting extensive lineage-specific gene duplications likely driven by selective pressures to adapt to environmental stresses. Phylogenetic analysis resolved the 51 CsSOS1 genes into six well-supported clades (Figure 1), reflecting evolutionary trajectories and domain architecture conservation. All clades retain conserved Na+/H+ exchanger (NHX) domains critical for ion transport yet exhibit structural divergence indicative of functional specialization during evolution. This divergence may underpin distinct roles in stress response and metabolic regulation. The clade formation highlights both deep evolutionary conservation and potential functional redundancy conferred by shared domain architecture. Collectively, the CsSOS1 family expansion implies functional divergence among paralogs and/or acquisition of novel functions through species-specific adaptive evolution.
Comparative synteny analysis among Camellia sinensis, Arabidopsis thaliana, Oryza sativa, and Populus trichocarpa revealed lineage-specific evolutionary dynamics in the SOS1 gene family. Tea and poplar, both long-lived woody perennials, are exposed to prolonged and complex environmental challenges such as drought and salinity, possibly driving the retention of structurally conserved and functionally relevant SOS1 genes. Although Arabidopsis thaliana is herbaceous, its phylogenetic proximity to tea may explain the moderate synteny observed. In contrast, rice (a monocot with distinct genome architecture and ecological niche) exhibits significantly reduced synteny with tea, likely due to gene loss, genomic rearrangements, or neofunctionalization during diversification. These results underscore how ecological strategies, life history traits, and evolutionary trajectories shape SOS1 family divergence
Motif analysis revealed that all CsSOS1 proteins shared ten conserved motifs (Figure 4), indicating functional coherence within the family. Furthermore, phylogenetically proximate members exhibit highly conserved exon–intron architectures and motif compositions. Notably, substantial variations in gene length (360 bp to 3693 bp) and exon number (2 to 25) suggest evolutionary exon gain/loss events, reflecting functional specialization of duplicated genes—a pattern consistent with other plant lineages [54,55].
To explore the regulatory potential of the CsSOS1 gene family, we analyzed cis-acting elements in their promoter regions. Similar to findings in Brassica juncea, potato, and Arabidopsis thaliana, the promoters of CsSOS1 genes harbored abundant hormone-responsive elements (including SA, MeJA, ABA, auxin, and gibberellin) and stress-related elements responsive to cold, dehydration, and defense signaling [56,57]. Remarkably, light-responsive elements were the most enriched, consistent with the photophilic nature of tea plants and the central role of light signaling in their development and stress response. The interaction between light signaling and salt-stress pathways may enable SOS1 to participate in a coordinated light-salt regulatory network, enhancing the environmental adaptability of tea.
Previous research in salt-tolerant tobacco demonstrated that vesicle-mediated vacuolar Na+ sequestration complements SOS1 antiporter function, collectively enhancing cellular salt tolerance [58]. Upon activation of the SOS pathway, SOS1 efficiently extrudes Na+ through Na+/H+ exchange at the plasma membrane, thereby restoring ionic balance under salt stress [9]. To functionally characterize CsSOS1 genes, Gene Ontology (GO) enrichment analysis revealed significant enrichment in cellular processes, transmembrane transport, and membrane components. These results confirm their essential role in ion exchange and support broader functions in membrane stabilization and intracellular signaling. Co-expression network analysis further implicated CsSOS1 genes in metabolic regulation—particularly carbohydrate metabolism—indicating functional crosstalk between ionic homeostasis and energy dynamics. Collectively, CsSOS1 genes not only maintain ion balance but also integrate metabolic coordination and stress adaptation mechanisms, expanding our understanding of their regulatory roles in tea plant resilience.
Studies have demonstrated that SOS1 genes are expressed across diverse plant tissues, suggesting their broad functional roles in growth and development. In wheat, SOS1 homologs are expressed in buds, leaves, spikes, and caryopses; in Arabidopsis thaliana, their expression has been detected in roots, flowers, and leaves; and in potato, SOS1 transcripts are particularly enriched in flowers and leaves [55,59,60]. Consistent with these findings, transcriptomic analysis in the present study revealed that CsSOS1 genes in tea plants are predominantly expressed in leaves and flowers. Notably, CsSOS1-15, CsSOS1-16, and CsSOS1-50 showed consistently high expression levels across multiple tissue types, indicating their potential roles in the developmental regulation of diverse organs in Camellia sinensis (Figure 7). The observed tissue-specific expression patterns of CsSOS1s resemble those reported in Arabidopsis thaliana and potato, suggesting that the spatial expression and possibly the functional roles of SOS1 genes are evolutionarily conserved across species.
Research across various plant species has demonstrated that SOS1 is a conserved protein present in both monocots and dicots. The SOS1 protein facilitates the long-distance transport of Na+, a harmful ion to plant growth, from roots and leaves to the stem, thereby protecting root and photosynthetic tissues [61]. Additionally, SOS1 plays a crucial role in regulating intracellular K+ homeostasis. Oh et al. (2010) found that in Arabidopsis thaliana plants silenced for AtSOS1, the activity of K+-transporting proteins was significantly suppressed, along with a corresponding reduction in gene expression levels. This suggests that under salt stress, SOS1 not only helps in Na+ efflux but also ensures K+ uptake, maintaining ionic balance [45]. To investigate the potential regulatory functions of the CsSOS1 gene family in tea plant, we analyzed transcriptomic data to map the expression patterns of SOS1 genes under drought, salt, shading stress, and hormone treatments. Our results revealed that CsSOS1-03, CsSOS1-15, and CsSOS1-41 were specifically upregulated under salt and drought stress. To validate these findings, we performed RT-qPCR and confirmed that the expression levels of these genes under both stress conditions were consistent with the transcriptomic data. These results further indicated that the SOS1 family not only played a biological role under salt stress but also possessed regulatory potential under drought stress. Hormonal treatment analyses showed that CsSOS1-02 and CsSOS1-30 were upregulated in response to auxin treatment. Previous studies have demonstrated that auxin regulates plant responses to abiotic stress by modulating root architecture, antioxidant systems, ion homeostasis, and interactions with other hormones [62]. Therefore, it is likely that CsSOS1-02 and CsSOS1-30 in tea plant contribute to ion homeostasis regulation through auxin-mediated mechanisms, thereby enhancing the plant’s stress tolerance.
The potential candidate CsSOS1 genes identified in this study, including CsSOS1-03, CsSOS1-15, and CsSOS1-41, play critical roles in tea plant adaptation to salt and drought stress. These genes were significantly upregulated in response to both stresses, as confirmed by our transcriptomic analysis and RT-qPCR validation. Notably, CsSOS1-15 and CsSOS1-41 exhibited consistent expression patterns across different environmental conditions, highlighting their central role in ion homeostasis and stress mitigation. The co-expression network analysis further revealed their interaction with key metabolic pathways, such as carbohydrate metabolism, which may contribute to energy allocation and secondary metabolite synthesis under stress. These findings underscore the multifaceted functions of CsSOS1 genes not only in ion regulation but also in metabolic processes critical for tea quality. Future functional validation of these genes will be pivotal in developing salt-tolerant tea plant cultivars, enhancing both yield and the preservation of bioactive compounds essential for tea’s nutritional and medicinal properties.
5. Conclusions
This study elucidates the critical role of the CsSOS1 gene family in tea plants’ adaptation to salt and drought stress. The identification of 51 CsSOS1 genes, their conserved motifs, and stress-responsive cis-elements underscores their evolutionary and functional significance. Tissue-specific expression patterns and stress-induced upregulation of key genes (CsSOS1-03, CsSOS1-15, and CsSOS1-41) highlight their contribution to ionic homeostasis and stress mitigation. The interaction between CsSOS1s and auxin signaling further suggests a hormone-mediated regulatory mechanism for stress tolerance. These findings bridge the gap in molecular understanding of tea plant salt-stress responses and provide actionable targets for genetic engineering. By leveraging CsSOS1 genes, breeders can develop tea plant cultivars with enhanced resilience to soil salinization and drought, ensuring stable yields and preserving bioactive compounds critical for tea quality and health benefits. Future research should focus on functional validation of candidate genes and their integration into agronomic practices, advancing sustainable tea plant production in the face of climate change and environmental degradation.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/horticulturae11070855/s1, Table S1. Primer sequences; Table S2. Identification of CsSOS1 gene family members; Table S3. Physicochemical properties of CsSOS1 gene family members; Table S4. Subcellular localization prediction of CsSOS1 gene family members; Table S5. Collinearity analysis among CsSOS1 gene family members; Table S6. Collinearity analysis between the CsSOS1 gene family and the SOS1 gene families of Arabidopsis thaliana and Populus trichocarpa; Table S7. Co-expression analysis of the CsSOS1 gene family.
Author Contributions
S.H.: Project administration, Investigation, Funding Acquisition, Preparation, Writing, Formal Analysis, and Data Curation; P.J.: Data Curation; Q.G.: Visualization and Funding Acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by project for the Jiangsu Funding Program for Excellent Postdoctoral Talent (JB23024), National Natural Science Foundation of China (32402624).
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Yadav, N.S.; Shukla, P.S.; Jha, A.; Agarwal, P.K.; Jha, B. The SbSOS1 gene from the extreme halophyte Salicornia brachiata enhances Na+ loading in xylem and confers salt tolerance in transgenic tobacco. BMC Plant Biol. 2012, 12, 188. [Google Scholar] [CrossRef] [PubMed]
- Volkov, V. Salinity tolerance in plants. Quantitative approach to ion transport starting from halophytes and stepping to genetic and protein engineering for manipulating ion fluxes. Front. Plant Sci. 2015, 6, 873. [Google Scholar] [CrossRef] [PubMed]
- Adem, G.D.; Roy, S.J.; Zhou, M.; Bowman, J.P.; Shabala, S. Evaluating contribution of ionic, osmotic and oxidative stress components towards salinity tolerance in barley. BMC Plant Biol. 2014, 14, 113. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, P.M.; Bressan, R.A.; Zhu, J.K.; Bohnert, H.J. Plant Cellular and Molecular Responses to High Salinity. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2000, 51, 463–499. [Google Scholar] [CrossRef] [PubMed]
- Oh, D.H.; Leidi, E.; Zhang, Q.; Hwang, S.M.; Li, Y.; Quintero, F.J.; Jiang, X.; D’Urzo, M.P.; Lee, S.Y.; Zhao, Y.; et al. Loss of halophytism by interference with SOS1 expression. Plant Physiol. 2009, 151, 210–222. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.M.; WR, W.X.; Li, H.W.; Jin, F.X.; Guo, L.N.; Wang, J.; Da, H.J.; Xu, X. Co-expression of the Arabidopsis SOS genes enhances salt tolerance in transgenic tall fescue (Festuca arundinacea Schreb.). Protoplasma 2014, 251, 219–231. [Google Scholar] [CrossRef] [PubMed]
- Heydarian, Z.; Yu, M.; Gruber, M.; Coutu, C.; Robinson, S.J.; Hegedus, D.D. Changes in gene expression in Camelina sativa roots and vegetative tissues in response to salinity stress. Sci. Rep. 2018, 8, 9804. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.; Pardo, J.M.; Batelli, G.; Van Oosten, M.J.; Bressan, R.A.; Li, X. The Salt Overly Sensitive (SOS) pathway: Established and emerging roles. Mol. Plant 2013, 6, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Xie, Q.; Zhou, Y.; Jiang, X. Structure, Function, and Regulation of the Plasma Membrane Na+/H+ Antiporter Salt Overly Sensitive 1 in Plants. Front. Plant Sci. 2022, 13, 866265. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.F.; Wang, X.Z. [Research progress in salt tolerance genes of SOS in Arabidopsis thaliana]. Yi Chuan 2002, 24, 190–192. [Google Scholar] [PubMed]
- Guo, Y.; Qiu, Q.S.; Quintero, F.J.; Pardo, J.M.; Ohta, M.; Zhang, C.; Schumaker, K.S.; Zhu, J.K. Transgenic evaluation of activated mutant alleles of SOS2 reveals a critical requirement for its kinase activity and C-terminal regulatory domain for salt tolerance in Arabidopsis thaliana. Plant Cell 2004, 16, 435–449. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.J.; Ding, L.; Zhu, J.K. SOS1, a Genetic Locus Essential for Salt Tolerance and Potassium Acquisition. Plant Cell 1996, 8, 617–627. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Q.S.; Guo, Y.; Dietrich, M.A.; Schumaker, K.S.; Zhu, J.K. Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. USA 2002, 99, 8436–8441. [Google Scholar] [CrossRef] [PubMed]
- Bose, J.; Xie, Y.; Shen, W.; Shabala, S. Haem oxygenase modifies salinity tolerance in Arabidopsis by controlling K+ retention via regulation of the plasma membrane H+-ATPase and by altering SOS1 transcript levels in roots. J. Exp. Bot. 2013, 64, 471–481. [Google Scholar] [CrossRef] [PubMed]
- Song, R.F.; Liao, C.Y.; Wang, L.F.; Lu, K.K.; Zhang, C.; Wu, R.X.; Wu, J.X.; Ma, Y.Q.; Kuang, L.; Guo, N.; et al. SORTING NEXIN1 facilitates SALT OVERLY SENSITIVE1 protein accumulation to enhance salt tolerance in Arabidopsis. Plant Physiol. 2024, 197, kiae633. [Google Scholar] [CrossRef] [PubMed]
- Meng, K.; Wu, Y. Footprints of divergent evolution in two Na+/H+ type antiporter gene families (NHX and SOS1) in the genus Populus. Tree Physiol. 2018, 38, 813–824. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Lee, B.H.; Wu, S.J.; Zhu, J.K. Overexpression of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana. Nat. Biotechnol. 2003, 21, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Pan, C.; Chen, Q.; Xie, Q.; Gao, Y.; He, L.; Li, Y.; Dong, Y.; Jiang, X.; Zhao, Y. Architecture and autoinhibitory mechanism of the plasma membrane Na+/H+ antiporter SOS1 in Arabidopsis. Nat. Commun. 2023, 14, 4487. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Wang, P.; Ma, Q.; Zhang, J.L.; Bao, A.K.; Wang, S.M. Selective transport capacity for K+ over Na+ is linked to the expression levels of PtSOS1 in halophyte Puccinellia tenuiflora. Funct. Plant Biol. 2012, 39, 1047–1057. [Google Scholar] [CrossRef] [PubMed]
- Olias, R.; Eljakaoui, Z.; Li, J.; De Morales, P.A.; Marin-Manzano, M.C.; Pardo, J.M.; Belver, A. The plasma membrane Na+/H+ antiporter SOS1 is essential for salt tolerance in tomato and affects the partitioning of Na+ between plant organs. Plant Cell Environ. 2009, 32, 904–916. [Google Scholar] [CrossRef] [PubMed]
- Song, A.; Lu, J.; Jiang, J.; Chen, S.; Guan, Z.; Fang, W.; Chen, F. Isolation and characterisation of Chrysanthemum crassum SOS1, encoding a putative plasma membrane Na+/H+ antiporter. Plant Biol. 2012, 14, 706–713. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Tong, W.; Li, F.; Wang, Y.; Wu, Q.; Wan, X.; Xia, E. TPIA2: An updated tea plant information archive for Camellia genomics. Nucleic Acids Res. 2024, 52, D1661–D1667. [Google Scholar] [CrossRef] [PubMed]
- Xing, L.; Zhang, H.; Qi, R.; Tsao, R.; Mine, Y. Recent Advances in the Understanding of the Health Benefits and Molecular Mechanisms Associated with Green Tea Polyphenols. J. Agric. Food Chem. 2019, 67, 1029–1043. [Google Scholar] [CrossRef] [PubMed]
- Luo, Q.; Luo, L.; Zhao, J.; Wang, Y.; Luo, H. Biological potential and mechanisms of Tea’s bioactive compounds: An Updated review. J. Adv. Res. 2024, 65, 345–363. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Cui, C.; Cao, Y.; Dai, J.; Cheng, X.; Hua, S.; Wang, W.; Duan, Y.; Petropoulos, E.; Wang, H.; et al. Tea plant-legume intercropping simultaneously improves soil fertility and tea quality by changing Bacillus species composition. Hortic. Res. 2022, 9, uhac046. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yang, Y.; Sun, K.; Chen, Y.; Chen, X.; Li, X. Exogenous Melatonin Enhances Cold, Salt and Drought Stress Tolerance by Improving Antioxidant Defense in Tea Plant (Camellia sinensis (L.) O. Kuntze). Molecules 2019, 24, 1826. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Ma, X.; Tang, D.; Chen, Y.; Chen, G.; Zou, J.; Tan, L.; Tang, Q.; Chen, W. Effects of Brassinosteroid on the Physiological Changes on Two Varieties of Tea Plants Under Salt Stress. Int. J. Mol. Sci. 2024, 25, 13445. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Li, C.; Wu, F.; Yang, Y.; Yu, A.; Wang, Z.; Zhao, L.; Zhang, X.; Qu, F.; Gao, L.; et al. Genome-wide identification and expression pattern analysis of WRKY transcription factors in response to biotic and abiotic stresses in tea plants (Camellia sinensis). Plant Physiol. Biochem. 2024, 211, 108670. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Liu, Y.; Li, J.; Zhou, B.; Chen, Y.; Tang, H.; Cui, Y.; Liu, J.; Tang, J. Evolutionary and Expression Analyses of the bZIP Family in Tea Plants (Camellia sinensis) and Functional Characterization of CsbZIP3/42/6 in Response to Environmental Stresses. J. Agric. Food Chem. 2024, 72, 24989–25000. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Ye, Z.; Wang, Y.; Zhang, X.; Kong, W. Haplotype-Resolution Transcriptome Analysis Reveals Important Responsive Gene Modules and Allele-Specific Expression Contributions under Continuous Salt and Drought in Camellia sinensis. Genes 2023, 14, 1417. [Google Scholar] [CrossRef] [PubMed]
- Moles, T.M.; de Brito Francisco, R.; Mariotti, L.; Pompeiano, A.; Lupini, A.; Incrocci, L.; Carmassi, G.; Scartazza, A.; Pistelli, L.; Guglielminetti, L.; et al. Salinity in Autumn-Winter Season and Fruit Quality of Tomato Landraces. Front. Plant Sci. 2019, 10, 1078. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Lin, S.; Li, J.; Chen, T.; Gu, Q.; Yang, T.; Zhang, Z. Theanine Improves Salt Stress Tolerance via Modulating Redox Homeostasis in Tea Plants (Camellia sinensis L.). Front. Plant Sci. 2021, 12, 770398. [Google Scholar] [CrossRef] [PubMed]
- Qiu, N.; Liu, Q.; Li, J.; Zhang, Y.; Wang, F.; Gao, J. Physiological and Transcriptomic Responses of Chinese Cabbage (Brassica rapa L. ssp. Pekinensis) to Salt Stress. Int. J. Mol. Sci. 2017, 18, 1953. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Liu, S.; Wang, Y.; Zhuang, J.; Chen, X.; Li, X. The combined analysis of transcriptome and phytohormone provides new insights into signaling mechanism for lateral root formation of tea plant (Camellia sinensis). Sci. Hortic. 2024, 338, 113758. [Google Scholar] [CrossRef]
- Di, T.; Zhao, L.; Chen, H.; Qian, W.; Wang, P.; Zhang, X.; Xia, T. Transcriptomic and Metabolic Insights into the Distinctive Effects of Exogenous Melatonin and Gibberellin on Terpenoid Synthesis and Plant Hormone Signal Transduction Pathway in Camellia sinensis. J. Agric. Food Chem. 2019, 67, 4689–4699. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Pang, D.; Ruan, L.; Liang, J.; Zhang, Q.; Qian, Y.; Zhang, Y.; Bai, P.; Wu, L.; Cheng, H.; et al. Integrated transcriptome and hormonal analysis of naphthalene acetic acid-induced adventitious root formation of tea cuttings (Camellia sinensis). BMC Plant Biol. 2022, 22, 319. [Google Scholar] [CrossRef] [PubMed]
- Xia, E.; Tong, W.; Hou, Y.; An, Y.; Chen, L.; Wu, Q.; Liu, Y.; Yu, J.; Li, F.; Li, R.; et al. The Reference Genome of Tea Plant and Resequencing of 81 Diverse Accessions Provide Insights into Its Genome Evolution and Adaptation. Mol. Plant 2020, 13, 1013–1026. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Feng, H.; Chang, Y.; Ma, C.; Wang, L.; Hao, X.; Li, A.; Cheng, H.; Wang, L.; Cui, P.; et al. Population sequencing enhances understanding of tea plant evolution. Nat. Commun. 2020, 11, 4447. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Tian, F.; Yang, D.C.; Meng, Y.Q.; Jin, J.; Gao, G. PlantRegMap: Charting functional regulatory maps in plants. Nucleic Acids Res. 2020, 48, D1104–D1113. [Google Scholar] [CrossRef] [PubMed]
- Kiss, T.; Karacsony, Z.; Gomba-Toth, A.; Szabadi, K.L.; Spitzmuller, Z.; Hegyi-Kalo, J.; Cels, T.; Otto, M.; Golen, R.; Hegyi, A.I.; et al. A modified CTAB method for the extraction of high-quality RNA from mono-and dicotyledonous plants rich in secondary metabolites. Plant Methods 2024, 20, 62. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Oh, D.H.; Lee, S.Y.; Bressan, R.A.; Yun, D.J.; Bohnert, H.J. Intracellular consequences of SOS1 deficiency during salt stress. J. Exp. Bot. 2010, 61, 1205–1213. [Google Scholar] [CrossRef] [PubMed]
- Rolly, N.K.; Imran, Q.M.; Lee, I.J.; Yun, B.W. Salinity Stress-Mediated Suppression of Expression of Salt Overly Sensitive Signaling Pathway Genes Suggests Negative Regulation by AtbZIP62 Transcription Factor in Arabidopsis thaliana. Int. J. Mol. Sci. 2020, 21, 1726. [Google Scholar] [CrossRef] [PubMed]
- Cha, J.Y.; Kim, J.; Jeong, S.Y.; Shin, G.I.; Ji, M.G.; Hwang, J.W.; Khaleda, L.; Liao, X.; Ahn, G.; Park, H.J.; et al. The Na+/H+ antiporter SALT OVERLY SENSITIVE 1 regulates salt compensation of circadian rhythms by stabilizing GIGANTEA in Arabidopsis. Proc. Natl. Acad. Sci. USA 2022, 119, e2207275119. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.Y.; Tang, L.H.; Nie, J.W.; Zhang, C.R.; Han, X.; Li, Q.Y.; Qin, L.; Wang, M.H.; Huang, X.; Yu, F.; et al. Structure and activation mechanism of the rice Salt Overly Sensitive 1 (SOS1) Na+/H+ antiporter. Nat. Plants 2023, 9, 1924–1936. [Google Scholar] [CrossRef] [PubMed]
- Shivani, S.; Ghosh, R.; Mitra, A.; Das, A.; Banerjee, J. Typical tetra-mediated signaling and plant architectural changes regulate salt-stress tolerance in indica rice genotypes. Protoplasma 2025, 262, 837–855. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.K.; Zhang, M.; Yang, Y.Q.; Xuan, W.; Zou, Z.W.; Arkorful, E.; Chen, Y.; Ma, Q.P.; Jeyaraj, A.; Chen, X.; et al. A novel insight into nitrogen and auxin signaling in lateral root formation in tea plant [Camellia sinensis (L.) O. Kuntze]. BMC Plant Biol. 2020, 20, 232. [Google Scholar] [CrossRef] [PubMed]
- Borah, A.; Hazarika, S.N.; Thakur, D. Potentiality of actinobacteria to combat against biotic and abiotic stresses in tea [Camellia sinensis (L) O. Kuntze]. J. Appl. Microbiol. 2022, 133, 2314–2330. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Tang, X.; Li, F.; Zhu, J.; Wu, M.; Wei, X.; Wang, Y. Green and Oolong Tea Extracts With Different Phytochemical Compositions Prevent Hypertension and Modulate the Intestinal Flora in a High-Salt Diet Fed Wistar Rats. Front. Nutr. 2022, 9, 892801. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Ye, X.; Xing, A.; Wu, Z.; Li, X.; Shu, Z.; Wang, Y. Camellia sinensis small GTPase gene (CsRAC1) involves in response to salt stress, drought stress and ABA signaling pathway. Gene 2022, 821, 146318. [Google Scholar] [CrossRef] [PubMed]
- Shang, C.; Sihui, L.; Li, C.; Hussain, Q.; Chen, P.; Hussain, M.A.; Nkoh Nkoh, J. SOS1 gene family in mangrove (Kandelia obovata): Genome-wide identification, characterization, and expression analyses under salt and copper stress. BMC Plant Biol. 2024, 24, 805. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Guo, L.; Zhai, Y.; Hou, Z.; Wu, W.; Zhang, X.; Wu, Y.; Liu, X.; Guo, S.; Gao, G.; et al. Genome-wide characterization of SOS1 gene family in potato (Solanum tuberosum) and expression analyses under salt and hormone stress. Front. Plant Sci. 2023, 14, 1201730. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Zhong, Y.; Wang, Q.; Cai, Z.; Wang, D.; Li, C. Genome-wide identification and gene expression analysis of SOS family genes in tuber mustard (Brassica juncea var. tumida). PLoS ONE 2019, 14, e0224672. [Google Scholar] [CrossRef] [PubMed]
- Maghraby, A.; Alzalaty, M. Genome-wide identification, characterization, and functional analysis of the CHX, SOS, and RLK genes in Solanum lycopersicum under salt stress. Sci. Rep. 2025, 15, 1142. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Wang, D.; Zhang, X.; Lv, X.; Li, B. Current progress in deciphering the molecular mechanisms underlying plant salt tolerance. Curr. Opin. Plant Biol. 2025, 83, 102671. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Shabala, L.; Cuin, T.A.; Huang, X.; Zhou, M.; Munns, R.; Shabala, S. Nax loci affect SOS1-like Na+/H+ exchanger expression and activity in wheat. J. Exp. Bot. 2016, 67, 835–844. [Google Scholar] [CrossRef] [PubMed]
- Ullah, A.; Dutta, D.; Fliegel, L. Expression and characterization of the SOS1 Arabidopsis salt tolerance protein. Mol. Cell. Biochem. 2016, 415, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.K.; Song, R.F.; Guo, J.X.; Zhang, Y.; Zuo, J.X.; Chen, H.H.; Liao, C.Y.; Hu, X.Y.; Ren, F.; Lu, Y.T.; et al. CycC1;1-WRKY75 complex-mediated transcriptional regulation of SOS1 controls salt stress tolerance in Arabidopsis. Plant Cell 2023, 35, 2570–2591. [Google Scholar] [CrossRef] [PubMed]
- Jing, H.; Wilkinson, E.G.; Sageman-Furnas, K.; Strader, L.C. Auxin and abiotic stress responses. J. Exp. Bot. 2023, 74, 7000–7014. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).