Characterization of Germin-like Proteins (GLPs) and Their Expression in Response to Abiotic and Biotic Stresses in Cucumber
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification and Property Characterization of GLPs in Cucumber
2.2. Phylogenetic, Conserved Motif and Gene Structure Analyses
2.3. Chromosomal Distribution and Gene Duplication Analysis
2.4. RNA-Seq Expression Analysis of the CsGLP Genes
2.5. qRT-PCR Analysis of CsGLP Genes in Response to Abiotic Stress
3. Results
3.1. Identification of the CsGLP Genes in Cucumber
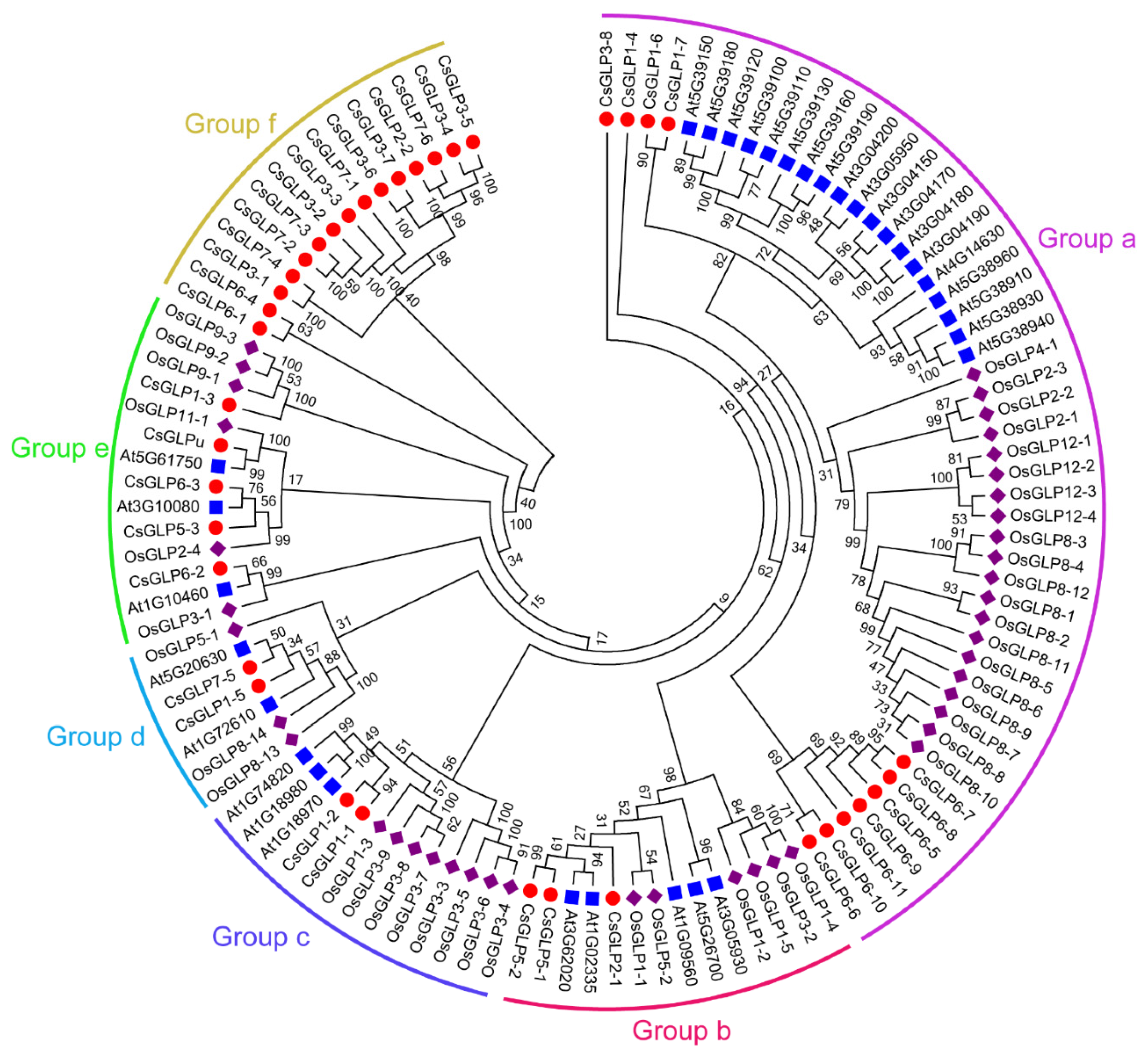
3.2. Phylogenetic Analysis of GLP Proteins from Cucumber and Other Plant Species
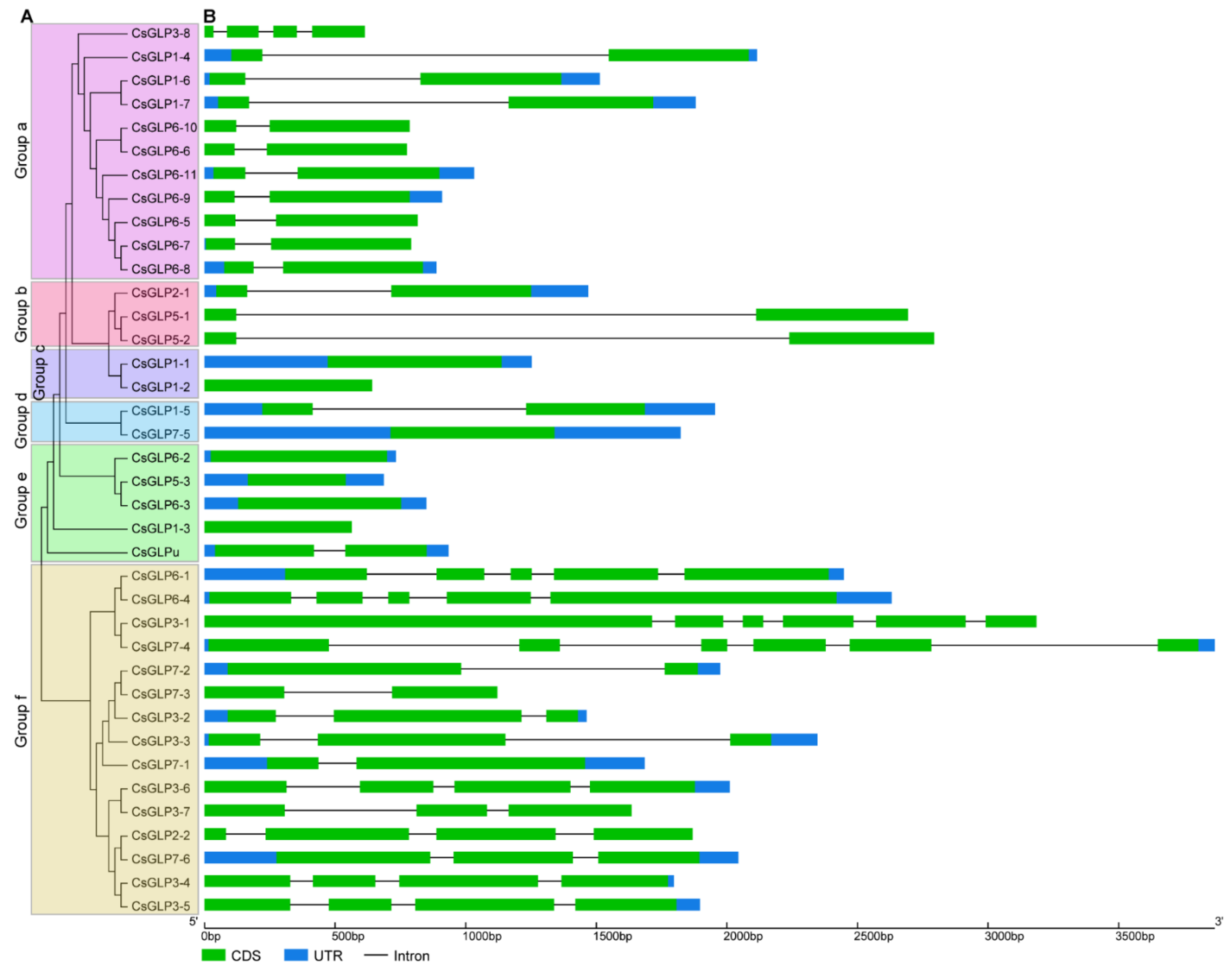
3.3. Conserved Motif Analysis of Cucumber GLP Proteins
3.4. Structure Analysis of CsGLP Genes
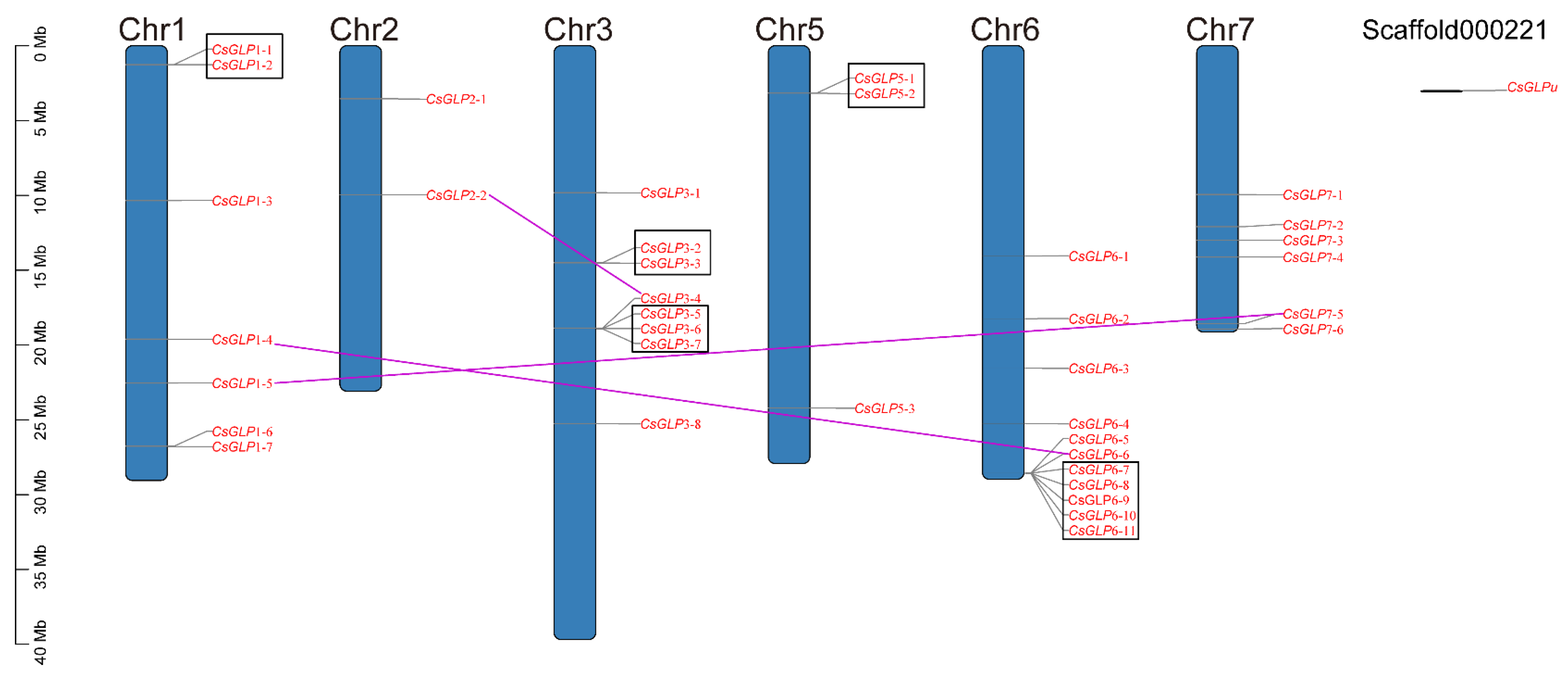
3.5. Chromosomal Location and Gene Duplication of CsGLP genes
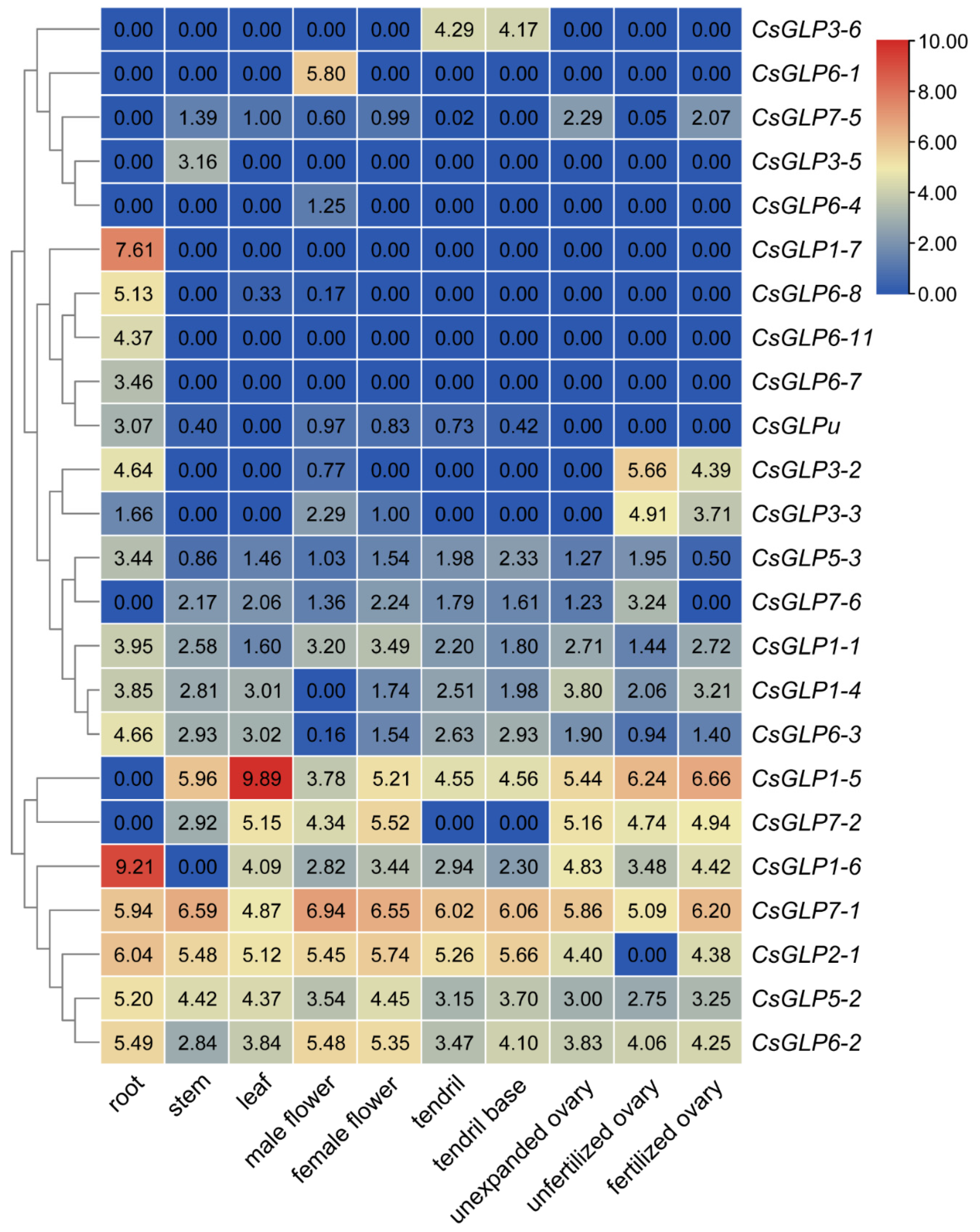
3.6. Expression Profiles of CsGLP Genes in Various Tissues
3.7. Expression Patterns of CsGLP Genes in Response to DM Treatment
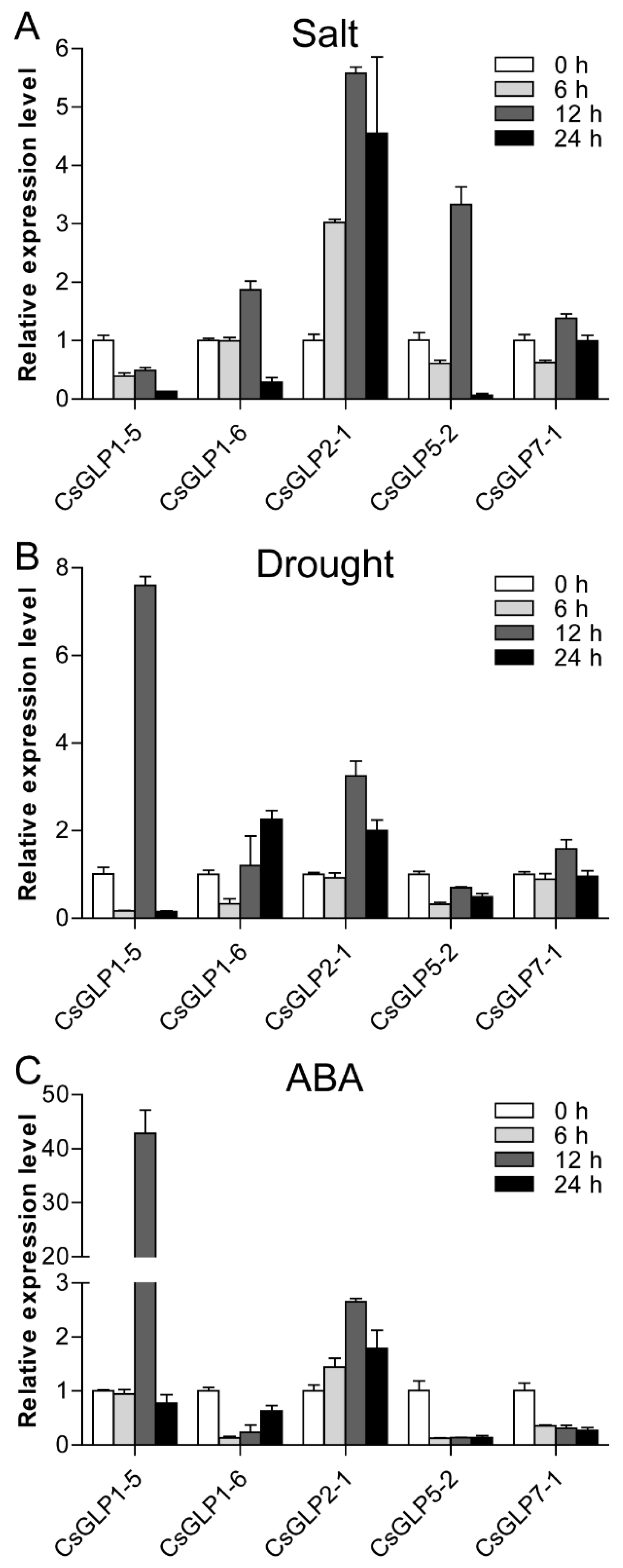
3.8. Expression Patterns of Selected CsGLP Genes in Response to Salt, Drought and ABA Treatments
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lane, B.G.; Cuming, A.C.; Frégeau, J.; Carpita, N.C.; Hurkman, W.J.; Bernier, F.; Dratewka-Kos, E.; Kennedy, T.D. Germin isoforms are discrete temporal markers of wheat development. Eur. J. Biochem. 1992, 209, 961–969. [Google Scholar] [CrossRef] [PubMed]
- Thompson, E.W.; Lane, B.G. Relation of protein synthesis in imbibing wheat embryos to the cell-free translational capacities of bulk mRNA from dry and imbibing embryos. J. Biol. Chem. 1980, 255, 5965–5970. [Google Scholar] [CrossRef]
- Patnaik, D.; Khurana, P. Germins and germin like proteins: An overview. Indian J. Exp. Biol. 2001, 39, 191–200. [Google Scholar] [PubMed]
- Dunwell, J.M.; Gibbings, J.G.; Mahmood, T.; Saqlan Naqvi, S.M. Germin and germin-like proteins: Evolution, structure, and function. Crit. Rev. Plant Sci. 2008, 27, 342–375. [Google Scholar] [CrossRef]
- Ayub, S.; Hayat, R.; Zainab, Z.; Akhtar, W.; Mahmood, T. OsRGLP2 promoter derived GUS expression in transgenic tobacco in response to salicylic acid, H2O2, PEG, NaCl and auxins. Plant Gene 2019, 19, 100190. [Google Scholar] [CrossRef]
- Li, L.; Xu, X.; Chen, C.; Shen, Z. Genome-wide characterization and expression analysis of the germin-like protein family in rice and Arabidopsis. Int. J. Mol. Sci. 2016, 17, 1622. [Google Scholar] [CrossRef] [Green Version]
- Breen, J.; Bellgard, M. Germin-like proteins (GLPs) in cereal genomes: Gene clustering and dynamic roles in plant defence. Funct. Integr. Genom. 2010, 10, 463–476. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, A.; Ahmed, A.; Essa, R.; Baber, S.; Jamshed, A.; Shoukat, A.; Younas, T.; Saleem, M.A.; Naveed, M. In-silico analysis of grapevine germin like protein (VvGLP3) and its probable role in defense against powdery mildew disease. Life Sci. J. Pak. 2019, 1, 17–23. [Google Scholar]
- Barman, A.R.; Banerjee, J. Versatility of germin-like proteins in their sequences, expressions, and functions. Funct. Integr. Genom. 2015, 15, 533–548. [Google Scholar] [CrossRef] [PubMed]
- Pei, Y.; Zhu, Y.; Jia, Y.; Ge, X.; Li, X.; Li, F.; Hou, Y. Molecular evidence for the involvement of cotton GhGLP2, in enhanced resistance to Verticillium and Fusarium Wilts and oxidative stress. Sci. Rep. 2020, 10, 12510. [Google Scholar] [CrossRef] [PubMed]
- Beracochea, V.C.; Almasia, N.I.; Peluffo, L.; Nahirñak, V.; Hopp, E.H.; Paniego, N.; Heinz, R.A.; Vazquez-Rovere, C.; Lia, V.V. Sunflower germin-like protein HaGLP1 promotes ROS accumulation and enhances protection against fungal pathogens in transgenic Arabidopsis thaliana. Plant Cell Rep. 2015, 34, 1717–1733. [Google Scholar] [CrossRef]
- Rodríguez-López, M.; Baroja-Fernández, E.; Zandueta-Criado, A.; Moreno-Bruna, B.; Muñoz, F.J.; Akazawa, T.; Pozueta-Romero, J. Two isoforms of a nucleotide-sugar pyrophosphatase/phosphodiesterase from barley leaves (Hordeum vulgare L.) are distinct oligomers of HvGLP1, a germin-like protein. FEBS Lett. 2001, 490, 44–48. [Google Scholar] [CrossRef] [Green Version]
- Huang, N.; Cheng, X.; Hu, W.; Pan, S. Inactivation, aggregation, secondary and tertiary structural changes of germin-like protein in Satsuma mandarine with high polyphenol oxidase activity induced by ultrasonic processing. Biophys. Chem. 2015, 197, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Huang, X.; Liu, S.; Tang, M.; Hu, W.; Pan, S. Characterization of germin-like protein with polyphenol oxidase activity from Satsuma mandarine. Biochem. Biophys. Res. Commun. 2014, 449, 313–318. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, J.; Maiti, M.K. Functional role of rice germin-like protein1 in regulation of plant height and disease resistance. Biochem. Biophys. Res. Commun. 2010, 394, 178–183. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, Y.; Xiao, N.; Zhang, G.; Wang, F.; Chen, X.; Fang, R. Rice GERMIN-LIKE PROTEIN 2-1 functions in seed dormancy under the control of abscisic acid and gibberellic acid signaling pathways. Plant Physiol. 2020, 183, 1157–1170. [Google Scholar] [CrossRef]
- Perveen, S.; Khan, T.A.; Shaheen, H.; Naz, R.; Hyder, M.Z.; Ijaz, B.; Naqvi, S.M.S.; Yasmin, T. Morpho-physiological investigations in transgenic tomato (Solanum lycopersicum L.) over expressing OsRGLP1 gene. Vitr. Cell Dev. Biol.-Plant 2021. [Google Scholar] [CrossRef]
- Fu, J.Y.; Wang, X.C.; Mao, T.F.; Cheng, H.; Chen, F.; Yang, Y.J. Identification and functional analysis of germin-like protein gene family in tea plant (Camellia sinensis). Sci. Hortic. 2018, 234, 166–175. [Google Scholar] [CrossRef]
- Das, A.; Pramanik, K.; Sharma, R.; Gantait, S.; Banerjee, J. In-silico study of biotic and abiotic stress-related transcription factor binding sites in the promoter regions of rice germin-like protein genes. PLoS ONE 2019, 14, e0211887. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, J.; Gantait, S.; Maiti, M.K. Physiological role of rice germin-like protein 1 (OsGLP1) at early stages of growth and development in indica rice cultivar under salt stress condition. Plant Cell Tissue Organ Cult. 2017, 131, 127–137. [Google Scholar] [CrossRef]
- He, Z.D.; Tao, M.L.; Leung, D.W.M.; Yan, X.Y.; Chen, L.; Peng, X.X.; Liu, E.E. The rice germin-like protein OsGLP1 participates in acclimation to UV-B radiation. Plant Physiol. 2021, 186, 1254–1268. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Guan, R.; Yang, Y.; Bai, Z.; Ge, F.; Liu, D. Isolation and characterization of a Fusarium oxysporum-resistant gene LrGLP1 from Lilium regale Wilson. In Vitro Cell Dev. Biol.-Plant 2017, 53, 461–468. [Google Scholar] [CrossRef]
- Majeed, N.; Hussain, I.; Abbas, Z.; Iqbal, U.; Sultana, T.; Naqvi, S.M.S. Heterologous expression of OsRGLP1 gene induces resistance against fusarium infection in transgenic potato. Pak. J. Phytopathol. 2016, 28, 45–55. [Google Scholar]
- Sultana, T.; Deeba, F.; Naz, F.; Rose, R.J.; Saqlan Naqvi, S.M. Expression of a rice GLP in Medicago truncatula exerting pleiotropic effects on resistance against Fusarium oxysporum through enhancing FeSOD-like activity. Acta Physiol. Plant. 2016, 38, 255. [Google Scholar] [CrossRef]
- Liu, Q.; Yang, J.; Yan, S.; Zhang, S.; Zhao, J.; Wang, W.; Yang, T.; Wang, X.; Mao, X.; Dong, J.; et al. The germin-like protein OsGLP2-1 enhances resistance to fungal blast and bacterial blight in rice. Plant Mol. Biol. 2016, 92, 411–423. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, X.; Chang, X.; Sun, M.; Zhang, Y.; Li, W.; Li, Y. Overexpression of germin-like protein GmGLP10 enhances resistance to Sclerotinia sclerotiorum in transgenic tobacco. Biochem. Biophys. Res. Commun. 2018, 497, 160–166. [Google Scholar] [CrossRef] [PubMed]
- Pei, Y.; Li, X.; Zhu, Y.; Ge, X.; Sun, Y.; Liu, N.; Jia, Y.; Li, F.; Hou, Y. GhABP19, a novel germin-like protein from Gossypium hirsutum, plays an important role in the regulation of resistance to verticillium and fusarium wilt pathogens. Front. Plant Sci. 2019, 10, 583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakata, M.; Watanabe, Y.; Sakurai, Y.; Hashimoto, Y.; Matsuzaki, M.; Takahashi, Y.; Satoh, T. Germin-like protein gene family of a moss, Physcomitrella patens, phylogenetically falls into two characteristic new clades. Plant Mol. Biol. 2004, 56, 381–395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, M.; Han, Y.P.; Gao, J.G.; Wang, X.J.; Li, W.B. Identification and analysis of the germin-like gene family in soybean. BMC Genom. 2010, 11, 620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Zhang, H.; Gao, Y.; Sun, G.; Zhang, W.; Qiu, L. A comprehensive analysis of the Cupin gene family in soybean (Glycine max). PLoS ONE 2014, 9, e110092. [Google Scholar] [CrossRef] [Green Version]
- Yuan, B.; Yang, Y.; Fan, P.; Liu, J.; Xing, H.; Liu, Y.; Feng, D. Genome-wide identification and characterization of germin and germin-like proteins (GLPs) and their response under powdery mildew stress in wheat (Triticum aestivum L.). Plant Mol. Biol. Rep. 2021. [Google Scholar] [CrossRef]
- Lai, W.; Zhou, Y.; Pan, R.; Liao, L.; He, J.; Liu, H.; Yang, Y.; Liu, S. Identification and expression analysis of stress-associated proteins (SAPs) containing A20/AN1 zinc finger in cucumber. Plants 2020, 9, 400. [Google Scholar] [CrossRef] [Green Version]
- Lai, W.; Zhu, C.; Hu, Z.; Liu, S.; Wu, H.; Zhou, Y. Identification and transcriptional analysis of zinc finger-homeodomain (ZF-HD) family genes in cucumber. Biochem. Genet. 2021, 59, 884–901. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, Z.; Yan, P.; Huang, S.; Fei, Z.; Lin, K. RNA-Seq improves annotation of protein-coding genes in the cucumber genome. BMC Genom. 2011, 12, 540. [Google Scholar] [CrossRef] [Green Version]
- Adhikari, B.N.; Savory, E.A.; Vaillancourt, B.; Childs, K.L.; Hamilton, J.P.; Day, B.; Buell, C.R. Expression profiling of Cucumis sativus in response to infection by Pseudoperonospora cubensis. PLoS ONE 2012, 7, e34954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Gu, R.; Song, X.; Liu, X.; Yan, L.; Zhou, Z.; Zhang, X. Genome-wide analysis of CsWOX transcription factor gene family in cucumber (Cucumis sativus L.). Sci. Rep. 2020, 10, 6216. [Google Scholar] [CrossRef]
- Zhou, Y.; Li, G.; Zhang, L.; Xu, J.; Hu, L.; Jiang, L.; Liu, S. Comprehensive genomic analysis and expression profiling of the BTB and TAZ (BT) genes in cucumber (Cucumis sativus L.). Czech J. Genet. Plant Breed. 2020, 56, 15–23. [Google Scholar] [CrossRef] [Green Version]
- Muhammad, I.; Muhammad, I.; Tariq, M.; Hazrat, H.; Latif-ur, R.; Ijaz, N.; Khaliq-ur, R. Analysis of germin-like protein genes (OsGLPs) family in rice using various in silico approaches. Curr. Bioinform. 2020, 15, 17–33. [Google Scholar]
- Sun, M.; Ye, Z.; Tan, J.; Chen, S.; Zhang, X.; Tu, L. A cotton germin-like protein GbGLP2 controls fiber length via regulating genes involved in secondary cell wall synthesis. Mol. Breed. 2020, 40, 98. [Google Scholar] [CrossRef]
- Sun, C.; Song, X.; Zheng, J.; Li, X.; Feng, Z.; Yan, L. Comparative proteomic profiles of resistant/susceptible cucumber leaves in response to downy mildew infection. Hortic. Plant J. 2021, 7, 327–340. [Google Scholar] [CrossRef]
- Gao, X.; Guo, P.; Wang, Z.; Chen, C.; Ren, Z. Transcriptome profiling reveals response genes for downy mildew resistance in cucumber. Planta 2021, 253, 112. [Google Scholar] [CrossRef] [PubMed]
- Manosalva, P.M.; Davidson, R.M.; Liu, B.; Zhu, X.; Hulbert, S.H.; Leung, H.; Leach, J.E. A germin-like protein gene family functions as a complex quantitative trait locus conferring broad-spectrum disease resistance in rice. Plant Physiol. 2009, 149, 286–296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giarola, V.; Chen, P.; Dulitz, S.J.; König, M.; Manduzio, S.; Bartels, D. The dehydration- and ABA-inducible germin-like protein CpGLP1 from Craterostigma plantagineum has SOD activity and may contribute to cell wall integrity during desiccation. Planta 2020, 252, 84. [Google Scholar] [CrossRef] [PubMed]


| Gene | Locus | Chromosome | Chromosomal Position | gDNA (bp) | CDS (bp) | Protein | |||
|---|---|---|---|---|---|---|---|---|---|
| Length (aa) | MW (Da) | pI | Subcellular Prediction | ||||||
| CsGLP1-1 | Csa1G007890.1 | 1 | 1255048-1256300 | 1253 | 666 | 221 | 23,666.44 | 7.73 | Cell wall |
| CsGLP1-2 | Csa1G007900.1 | 1 | 1258859-1259500 | 642 | 564 | 213 | 22,984.84 | 7.81 | Cell wall |
| CsGLP1-3 | Csa1G166250.1 | 1 | 10344063-10344626 | 564 | 675 | 187 | 19,869.54 | 5.16 | Cell wall |
| CsGLP1-4 | Csa1G537570.1 | 1 | 19610111-19612225 | 2115 | 672 | 217 | 23,108.48 | 5.79 | Cell wall |
| CsGLP1-5 | Csa1G596420.1 | 1 | 22537064-22539018 | 1955 | 642 | 215 | 22,364.19 | 6.81 | Cell wall |
| CsGLP1-6 | Csa1G662790.1 | 1 | 26758087-26759600 | 1514 | 648 | 224 | 23,991.83 | 7.82 | Cell wall |
| CsGLP1-7 | Csa1G662810.1 | 1 | 26786634-26788514 | 1881 | 654 | 223 | 24,103.81 | 6.82 | Cell wall |
| CsGLP2-1 | Csa2G035370.1 | 2 | 3544666-3546134 | 1469 | 654 | 217 | 22,892.41 | 9.30 | Cell wall |
| CsGLP2-2 | Csa2G174130.1 | 2 | 9968726-9970593 | 1868 | 1464 | 487 | 55,464.56 | 8.83 | Vacuole |
| CsGLP3-1 | Csa3G146460.1 | 3 | 9816841-9820025 | 3185 | 1440 | 927 | 112,934.56 | 8.43 | Nucleus |
| CsGLP3-2 | Csa3G218160.1 | 3 | 14499431-14500893 | 1463 | 1023 | 340 | 37,226.71 | 5.83 | Cell wall, Vacuole |
| CsGLP3-3 | Csa3G218170.1 | 3 | 14512631-14514977 | 2347 | 1071 | 356 | 38,684.19 | 5.19 | Vacuole |
| CsGLP3-4 | Csa3G384800.1 | 3 | 18869164-18870960 | 1797 | 2784 | 501 | 57,611.85 | 7.20 | Vacuole |
| CsGLP3-5 | Csa3G386310.1 | 3 | 18896546-18898442 | 1897 | 1485 | 494 | 56,770.06 | 8.60 | Vacuole |
| CsGLP3-6 | Csa3G386810.1 | 3 | 18901334-18903344 | 2011 | 447 | 479 | 54,416.18 | 5.71 | Vacuole |
| CsGLP3-7 | Csa3G386820.1 | 3 | 18920239-18921873 | 1635 | 1047 | 348 | 40,180.34 | 5.51 | Vacuole |
| CsGLP3-8 | Csa3G644800.1 | 3 | 25260870-25261483 | 614 | 1506 | 148 | 16,322.28 | 6.63 | Cell wall |
| CsGLP5-1 | Csa5G128780.1 | 5 | 3162233-3164925 | 2693 | 675 | 233 | 25,061.75 | 6.06 | Cell wall |
| CsGLP5-2 | Csa5G129280.1 | 5 | 3171528-3174320 | 2793 | 702 | 224 | 23,714.30 | 6.96 | Cell wall |
| CsGLP5-3 | Csa5G614670.1 | 5 | 24218204-24218890 | 687 | 375 | 124 | 13,540.72 | 6.51 | Cell wall |
| CsGLP6-1 | Csa6G290870.1 | 6 | 14050717-14053164 | 2448 | 1527 | 508 | 58,057.85 | 5.74 | Vacuole |
| CsGLP6-2 | Csa6G404270.1 | 6 | 18237396-18238128 | 733 | 663 | 224 | 24,344.08 | 8.37 | Cell wall |
| CsGLP6-3 | Csa6G452110.1 | 6 | 21556627-21557475 | 849 | 657 | 207 | 22,359.95 | 8.77 | Cell wall |
| CsGLP6-4 | Csa6G502040.1 | 6 | 25250904-25253534 | 2631 | 648 | 661 | 78,191.37 | 6.23 | Nucleus |
| CsGLP6-5 | Csa6G525540.1 | 6 | 28560994-28561809 | 816 | 648 | 219 | 23,425.11 | 7.80 | Cell wall |
| CsGLP6-6 | Csa6G525550.1 | 6 | 28563130-28563904 | 775 | 651 | 216 | 22,819.23 | 6.69 | Cell wall |
| CsGLP6-7 | Csa6G525580.1 | 6 | 28567264-28568054 | 791 | 660 | 215 | 22,597.97 | 5.47 | Cell wall |
| CsGLP6-8 | Csa6G525590.1 | 6 | 28570081-28570968 | 888 | 651 | 215 | 22,664.07 | 5.84 | Cell wall |
| CsGLP6-9 | Csa6G525600.1 | 6 | 28572339-28573248 | 910 | 624 | 216 | 23,153.73 | 7.85 | Cell wall |
| CsGLP6-10 | Csa6G525610.1 | 6 | 28574435-28575220 | 786 | 1986 | 218 | 22,902.41 | 6.26 | Cell wall |
| CsGLP6-11 | Csa6G525620.1 | 6 | 28576583-28577614 | 1032 | 675 | 220 | 23,148.55 | 6.95 | Cell wall |
| CsGLP7-1 | Csa7G281380.1 | 7 | 9939437-9941121 | 1685 | 1071 | 356 | 38,300.97 | 5.82 | Vacuole |
| CsGLP7-2 | Csa7G337100.1 | 7 | 12086664-12088637 | 1974 | 708 | 339 | 37,153.37 | 5.37 | Vacuole |
| CsGLP7-3 | Csa7G368140.1 | 7 | 12991068-12992188 | 1121 | 1431 | 235 | 25,649.72 | 9.43 | Vacuole |
| CsGLP7-4 | Csa7G380130.1 | 7 | 14118117-14121984 | 3868 | 1458 | 485 | 55,928.57 | 6.94 | Vacuole |
| CsGLP7-5 | Csa7G450510.1 | 7 | 18555157-18556979 | 1823 | 627 | 208 | 21,395.90 | 6.39 | Cell wall |
| CsGLP7-6 | Csa7G452090.1 | 7 | 18921704-18923747 | 2044 | 1020 | 476 | 54,181.04 | 7.69 | Vacuole |
| CsGLPu | CsaUNG024810 | Scaffold000221 | 4018-4951 | 934 | 690 | 229 | 25,058.56 | 7.00 | Cell wall |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liao, L.; Hu, Z.; Liu, S.; Yang, Y.; Zhou, Y. Characterization of Germin-like Proteins (GLPs) and Their Expression in Response to Abiotic and Biotic Stresses in Cucumber. Horticulturae 2021, 7, 412. https://doi.org/10.3390/horticulturae7100412
Liao L, Hu Z, Liu S, Yang Y, Zhou Y. Characterization of Germin-like Proteins (GLPs) and Their Expression in Response to Abiotic and Biotic Stresses in Cucumber. Horticulturae. 2021; 7(10):412. https://doi.org/10.3390/horticulturae7100412
Chicago/Turabian StyleLiao, Liting, Zhaoyang Hu, Shiqiang Liu, Yingui Yang, and Yong Zhou. 2021. "Characterization of Germin-like Proteins (GLPs) and Their Expression in Response to Abiotic and Biotic Stresses in Cucumber" Horticulturae 7, no. 10: 412. https://doi.org/10.3390/horticulturae7100412
APA StyleLiao, L., Hu, Z., Liu, S., Yang, Y., & Zhou, Y. (2021). Characterization of Germin-like Proteins (GLPs) and Their Expression in Response to Abiotic and Biotic Stresses in Cucumber. Horticulturae, 7(10), 412. https://doi.org/10.3390/horticulturae7100412






