Analysis of Light-Independent Anthocyanin Accumulation in Mango (Mangifera indica L.)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fruit Materials and Experimental Treatment
2.2. Metabolomic Profiling
2.3. DNA Isolation, RNA Extraction and cDNA Synthesis
2.4. Q-PCR Analysis
2.5. Cloning of Promoter Regions of MiMYB1
2.6. Statistical Analysis
3. Results
3.1. Fruit Color and Metabolomic Profiling
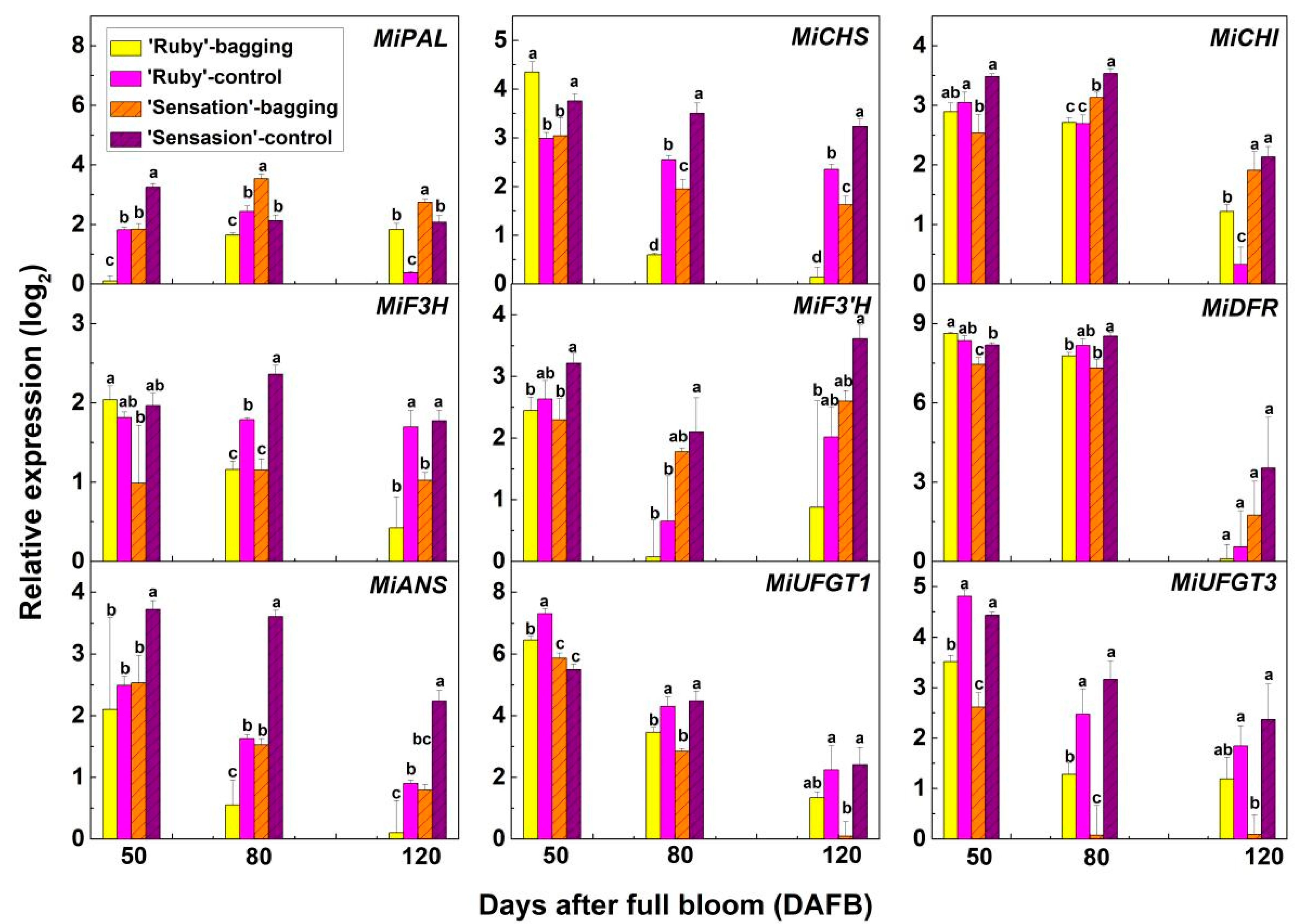
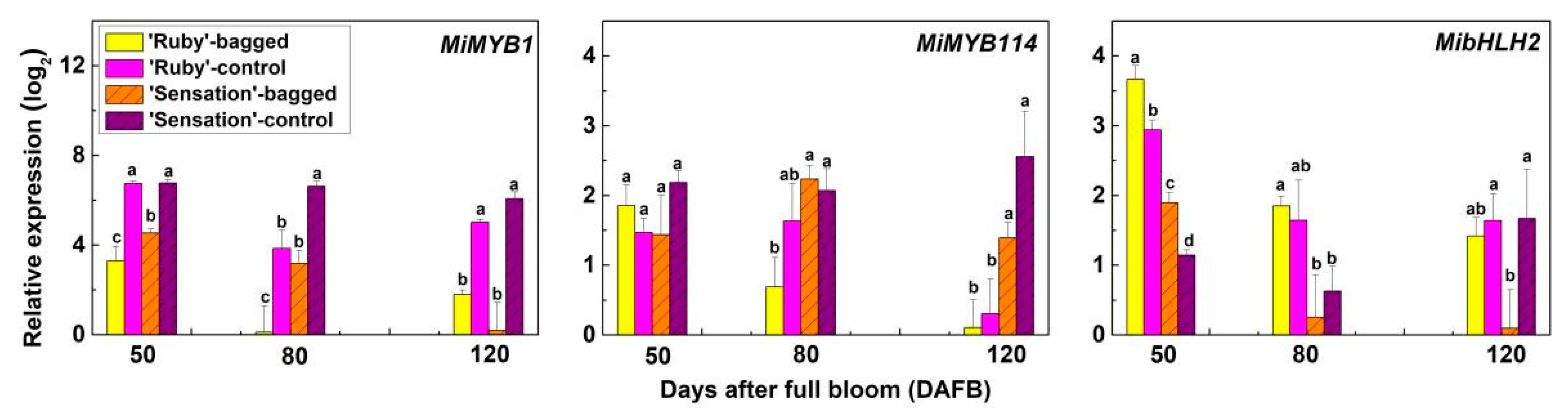
3.2. Expression of Anthocyanin Biosynthetic and Regulatory Genes in Fruit Skin
3.3. MiMYB1 Promoter Sequence Isolation and Comparison in ’Ruby’ and ‘Sensation’
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tharanathan, R.N.; Yashoda, H.M.; Prabha, T.N. Mango (Mangifera indica L.), “the king of fruits”—An overview. Food Rev. Int. 2006, 22, 95–123. [Google Scholar] [CrossRef]
- Medlicott, A.P.; Bhogal, M.; Reynolds, S.B. Changes in peel pigmentation during ripening of mango fruit (Mangifera indica var. Tommy Atkins). Ann. Appl. Biol. 1986, 109, 651–656. [Google Scholar] [CrossRef]
- Berardini, N.; Fezer, R.; Conrad, J.; Beifuss, U.; Carle, R.; Schieber, A. Screening of mango (Mangifera indica L.) cul-tivars for their contents of flavonol O- and xanthone C-glycosides, anthocyanins, and pectin. J. Agric. Food Chem. 2005, 53, 1563–1570. [Google Scholar] [CrossRef]
- Berardini, N.; Schieber, A.; Klaiber, I.; Beifuss, U.; Carle, R.; Conrad, J. 7-O-Methylcyanidin 3-O-β-D-Galactopyranoside, a novel anthocyanin from mango (Mangifera indica L. cv. ‘Tommy Atkins’) peels. Z. Nat. B 2005, 60, 801–804. [Google Scholar] [CrossRef]
- Bajpai, A.; Khan, K.; Muthukumar, M.; Rajan, S.; Singh, N.K. Molecular analysis of anthocyanin biosynthesis pathway genes and their differential expression in mango peel. Genome 2018, 61, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Kanzaki, S.; Kamikawa, S.; Ichihi, A.; Tanaka, Y.; Shimizu, K.; Koeda, S.; Utsunomiya, N. Isolation of UDP:flavonoid 3-O-glycosyltransferase (UFGT)-like genes and expression analysis of genes associated with anthocyanin accumulation in mango ‘Irwin’ skin. Hortic. J. 2019, 88, 435–443. [Google Scholar] [CrossRef] [Green Version]
- Allan, A.C.; Hellens, R.; Laing, W. MYB transcription factors that colour our fruit. Trends Plant Sci. 2008, 13, 99–102. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Dubos, C.; Lepiniec, L. Transcriptional control of flavonoid biosynthesis by MYB–bHLH–WDR complexes. Trends Plant Sci. 2015, 20, 176–185. [Google Scholar] [CrossRef]
- Borevitz, J.O.; Xia, Y.; Blount, J.; Dixon, R.A.; Lamb, C. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 2000, 12, 2383–2393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ban, Y.; Honda, C.; Hatsuyama, Y.; Igarashi, M.; Bessho, H.; Moriguchi, T. Isolation and Functional Analysis of a MYB Transcription Factor Gene that is a Key Regulator for the Development of Red Coloration in Apple Skin. Plant Cell Physiol. 2007, 48, 958–970. [Google Scholar] [CrossRef]
- Espley, R.V.; Hellens, R.P.; Putterill, J.; Stevenson, D.E.; Kutty-Amma, S.; Allan, A.C. Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 2007, 49, 414–427. [Google Scholar] [CrossRef] [Green Version]
- Takos, A.M.; Jaffé, F.W.; Jacob, S.R.; Bogs, J.; Robinson, S.; Walker, A.R. Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 2006, 142, 1216–1232. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, S.; Goto-Yamamoto, N.; Hirochika, H. Retrotransposon-induced mutations in grape skin color. Science 2004, 304, 982. [Google Scholar] [CrossRef]
- Butelli, E.; Licciardello, C.; Zhang, Y.; Liu, J.; Mackay, S.; Bailey, P.; Reforgiato-Recupero, G.; Martin, C. Retrotransposons control fruit-specific, cold-dependent accumulation of anthocyanins in blood oranges. Plant Cell 2012, 24, 1242–1255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, S.; Wang, Y.; Yang, S.; Xu, Y.; Chen, X. Anthocyanin biosynthesis in pears is regulated by a R2R3-MYB transcription factor PyMYB10. Planta 2010, 232, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.L.; Bolitho, K.; Grafton, K.; Kortstee, A.; Karunairetnam, S.; McGhie, T.K.; Espley, R.V.; Hellens, R.P.; Allan, A.C. An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol. 2010, 10, 50. [Google Scholar] [CrossRef] [Green Version]
- Rahim, A.; Busatto, N.; Trainotti, L. Regulation of anthocyanin biosynthesis in peach fruits. Planta 2014, 240, 913–929. [Google Scholar] [CrossRef] [PubMed]
- Yao, G.; Ming, M.; Allan, A.C.; Gu, C.; Li, L.; Wu, X.; Wang, R.; Chang, Y.; Qi, K.; Zhang, S.; et al. Map-based cloning of the pear gene MYB114 identifies an interaction with other transcription factors to coordinately regulate fruit anthocyanin biosynthesis. Plant J. 2017, 92, 437–451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanzaki, S.; Ichihi, A.; Tanaka, Y.; Fujishige, S.; Koeda, S.; Shimizu, K. The R2R3-MYB transcription factor MiMYB1 regulates light dependent red coloration of ‘Irwin’ mango fruit skin. Sci. Hortic. 2020, 272, 109567. [Google Scholar] [CrossRef]
- Qian, M.; Zhang, D.; Yue, X.; Wang, S.; Li, X.; Teng, Y. Analysis of different pigmentation patterns in ‘Mantianhong’ (Pyrus pyrifolia Nakai) and ‘Cascade’ (Pyrus communis L.) under bagging treatment and postharvest UV-B/visible irradiation conditions. Sci. Hortic. 2013, 151, 75–82. [Google Scholar] [CrossRef]
- Bai, S.; Tuan, P.A.; Saito, T.; Honda, C.; Hatsuyama, Y.; Ito, A.; Moriguchi, T. Epigenetic regulation of MdMYB1 is associated with paper bagging-induced red pigmentation of apples. Planta 2016, 244, 573–586. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Li, J.H.; Xin, H.P.; Wang, N.; Guan, L.; Wu, B.H.; Li, S.H. Anthocyanin profile and gene expression in berry skin of two red Vitis vinifera grape cultivars that are sunlight dependent versus sunlight independent. Aust. J. Grape Wine R. 2013, 19, 238–248. [Google Scholar] [CrossRef]
- Karanjalker, G.R.; Ravishankar, K.V.; Shivashankara, K.S.; Dinesh, M.R. Influence of bagging on color, anthocyanin and anthocyanin biosynthetic genes in peel of red colored mango cv. ‘Lily’. Erwerbs-Obstbau 2018, 60, 281–287. [Google Scholar] [CrossRef]
- Downey, M.O.; Harvey, J.S.; Robinson, S.P. The effect of bunch shading on berry development and flavonoid accumulation in Shiraz grapes. Aust. J. Grape Wine R. 2004, 10, 55–73. [Google Scholar] [CrossRef]
- Human, M.; Bindon, K. Interactive effect of ethephon and shading on the anthocyanin composition of Vitis vinifera L. cv. Crimson Seedless. S. Afr. J. Enol. Vitic. 2016, 28, 50–58. [Google Scholar] [CrossRef] [Green Version]
- Ristic, R.; Downey, M.O.; Iland, P.G.; Bindon, K.; Francis, I.L.; Herderich, M.; Robinson, S.P. Exclusion of sunlight from Shiraz grapes alters wine colour, tannin and sensory properties. Aust. J. Grape Wine Res. 2007, 13, 53–65. [Google Scholar] [CrossRef]
- Espley, R.V.; Brendolise, C.; Chagné, D.; Kutty-Amma, S.; Green, S.; Volz, R.; Putterill, J.; Schouten, H.J.; Gardiner, S.E.; Hellens, R.P.; et al. Multiple repeats of a promoter segment causes transcription factor autoregulation in red apples. Plant Cell 2009, 21, 168–183. [Google Scholar] [CrossRef] [Green Version]
- Zhou, H.; Lin-Wang, K.; Wang, H.; Gu, C.; Dare, A.P.; Espley, R.; He, H.; Allan, A.C.; Han, Y. Molecular genetics of blood-fleshed peach reveals activation of anthocyanin biosynthesis by NAC transcription factors. Plant J. 2015, 82, 105–121. [Google Scholar] [CrossRef]
- Mano, H.; Ogasawara, F.; Sato, K.; Higo, H.; Minobe, Y. Isolation of a regulatory gene of anthocyanin biosynthesis in tuberous roots of purple-fleshed sweet potato. Plant Physiol. 2007, 143, 1252–1268. [Google Scholar] [CrossRef] [Green Version]
- He, Q.; Wu, J.; Xue, Y.; Zhao, W.; Li, R.; Zhang, L. The novel gene BrMYB2, located on chromosome A07, with a short intron 1 controls the purple-head trait of Chinese cabbage (Brassica rapa L.). Hortic. Res. 2020, 7, 1–19. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhu, Q.; Hu, M.; Gao, Z.; An, F.; Li, M.; Jiang, Y. Low-temperature conditioning induces chilling tolerance in stored mango fruit. Food Chem. 2017, 219, 76–84. [Google Scholar] [CrossRef]
- Boss, P.; Davies, C.; Robinson, S. Analysis of the expression of anthocyanin pathway genes in developing Vitis vinifera L. cv Shiraz grape berries and the implications for pathway regulation. Plant Physiol. 1996, 111, 1059–1066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaakola, L.; Maatta, K.; Pirttila, A.M.; Torronen, R.; Karenlampi, S.; Hohtola, A. Expression of genes involved in anthocyanin biosynthesis in relation to anthocyanin, proanthocyanidin, and flavonol levels during bilberry fruit development. Plant Physiol. 2002, 130, 729–739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harborne, J.B. Phytochemistry of fruit and vegetables: An ecological overview. In Phytochemistry of Fruits and Vegetables; Tomas-Barberan, F., Ed.; Oxford University Press: New York, NY, USA, 1996; pp. 353–367. [Google Scholar]
- Harborne, J.B.; Williams, C.A. Advances in flavonoid research since 1992. Phytochemistry 2000, 55, 481–504. [Google Scholar] [CrossRef]
- Tohge, T.; Nishiyama, Y.; Hirai, M.Y.; Yano, M.; Nakajima, J.; Awazuhara, M.; Inoue, E.; Takahashi, H.; Goodenowe, D.B.; Kitayama, M.; et al. Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J. 2005, 42, 218–235. [Google Scholar] [CrossRef] [PubMed]
- Xie, D.Y.; Sharma, S.B.; Paiva, N.L.; Ferreira, D.; Dixon, R.A. Role of anthocyanidin reductase, encoded by BANYULS in plant flavonoid biosynthesis. Science 2003, 299, 396–399. [Google Scholar] [CrossRef] [Green Version]
- Han, Y.; Vimolmangkang, S.; Soria-Guerra, R.E.; Korban, S.S. Introduction of apple ANR genes into tobacco inhibits expression of both CHI and DFR genes in flowers, leading to loss of anthocyanin. J. Exp. Bot. 2012, 63, 2437–2447. [Google Scholar] [CrossRef] [Green Version]
- Ali, M.; Anwar, R.; Yousef, A.; Li, B.; Luvisi, A.; Bellis, L.; Aprile, A.; Chen, F. Influence of bagging on the development and quality of fruits. Plants 2021, 10, 358. [Google Scholar] [CrossRef]
- Bai, S.; Sun, Y.; Qian, M.; Yang, F.; Ni, J.; Tao, R.; Li, L.; Shu, Q.; Zhang, D.; Teng, Y. Transcriptome analysis of bagging-treated red Chinese sand pear peels reveals light-responsive pathway functions in anthocyanin accumulation. Sci. Rep. 2017, 7, 63. [Google Scholar] [CrossRef] [Green Version]
- Qian, M.; Sun, Y.; Allan, A.C.; Teng, Y.; Zhang, D. The red sport of ‘Zaosu’ pear and its red-striped pigmentation pattern are associated with demethylation of the PyMYB10 promoter. Phytochemistry 2014, 107, 16–23. [Google Scholar] [CrossRef]
- Li, Y.Y.; Mao, K.; Zhao, C.; Zhao, X.Y.; Zhang, H.L.; Shu, H.R.; Hao, Y.J. MdCOP1 ubiquitin E3 ligases interact with MdMYB1 to regulate light-induced anthocyanin biosynthesis and red fruit coloration in apple. Plant Physiol. 2012, 160, 1011–1022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giovannoni, J.J. Genetic Regulation of fruit development and ripening. Plant Cell 2004, 16, S170–S180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- An, J.P.; Wang, X.F.; Li, Y.Y.; Song, L.Q.; Zhao, L.L.; You, C.X.; Hao, Y.J. EIN3-LIKE1, MYB1, and ETHYLENE RE-SPONSE FACTOR3 act in a regulatory loop that synergistically modulates ethylene biosynthesis and anthocyanin accumulation. Plant Physiol. 2018, 178, 808–823. [Google Scholar] [CrossRef]
- An, J.P.; Yao, J.F.; Xu, R.R.; You, C.X.; Wang, X.F.; Hao, Y.J. Apple bZIP transcription factor MdbZIP44 regulates abscisic acid-promoted anthocyanin accumulation. Plant Cell Environ. 2018, 41, 2678–2692. [Google Scholar] [CrossRef]
- Rudell, D.; Mattheis, J.; Fan, X.; Fellman, J. Methyl jasmonate enhances anthocyanin accumulation and modifies production of phenolics and pigments in `Fuji’ apples. J. Am. Soc. Hortic. Sci. 2002, 127, 435–441. [Google Scholar] [CrossRef]
- Qian, M.; Yu, B.; Li, X.; Sun, Y.; Zhang, N.; Teng, Y. Isolation and expression analysis of anthocyanin biosynthesis genes from the red Chinese sand pear, Pyrus pyrifolia Nakai cv. Mantianhong, in response to methyl jasmonate treatment and UV-B/VIS conditions. Plant Mol. Biol. Rep. 2014, 32, 428–437. [Google Scholar] [CrossRef]
- Kumar, S.P.; Maurer, D.; Feygenberg, O.; Love, C.; Alkan, N. Improving the red color and fruit quality of ‘Kent’ mango fruit by pruning and preharvest spraying of prohydrojasmon or abscisic acid. Agronomy 2020, 10, 944. [Google Scholar] [CrossRef]
- Qi, T.; Song, S.; Ren, Q.; Wu, D.; Huang, H.; Chen, Y.; Fan, M.; Peng, W.; Ren, C.; Xie, D. The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 2011, 23, 1795–1814. [Google Scholar] [CrossRef] [Green Version]
- Shan, X.; Zhang, Y.; Peng, W.; Wang, Z.; Xie, D. Molecular mechanism for jasmonate-induction of anthocyanin accumulation in Arabidopsis. J. Exp. Bot. 2009, 60, 3849–3860. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Motif | Sequence | Distance from ATG | Strand | Function |
|---|---|---|---|---|
| ABRE | ACGTG | 1030 | − | cis-acting element involved in the abscisic acid responsiveness |
| AE-box | AGAAACAA | 756 | + | part of a module for light response |
| Box 4 | ATTAAT | 1311 | + | part of a conserved DNA module involved in light responsiveness |
| ATTAAT | 697 | − | ||
| CGTCA-motif | CGTCA | 149 | + | cis-acting regulatory element involved in the MeJA-responsiveness |
| CGTCA | 136 | + | ||
| GATA-motif | GATAGGA | 1407 | + | part of a light responsive element |
| GATAGGA | 433 | + | ||
| G-box | CACGAC | 783 | − | cis-acting regulatory element involved in light responsiveness |
| CACGTT | 1030 | + | ||
| GT1-motif | GGTTAA | 1333 | − | light responsive element |
| LTR | CCGAAA | 876 | + | cis-acting element involved in low-temperature responsiveness |
| MBS | CAACTG | 1340 | + | MYB binding site involved in drought-inducibility |
| CAACTG | 244 | − | ||
| CAACAG | 991 | + | ||
| TCA-element | CCATCTTTTT | 1074 | − | cis-acting element involved in salicylic acid responsiveness |
| TGA-element | AACGAC | 731 | + | auxin-responsive element |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, B.; Wu, H.; Zheng, B.; Qian, M.; Gao, A.; Zhou, K. Analysis of Light-Independent Anthocyanin Accumulation in Mango (Mangifera indica L.). Horticulturae 2021, 7, 423. https://doi.org/10.3390/horticulturae7110423
Shi B, Wu H, Zheng B, Qian M, Gao A, Zhou K. Analysis of Light-Independent Anthocyanin Accumulation in Mango (Mangifera indica L.). Horticulturae. 2021; 7(11):423. https://doi.org/10.3390/horticulturae7110423
Chicago/Turabian StyleShi, Bin, Hongxia Wu, Bin Zheng, Minjie Qian, Aiping Gao, and Kaibing Zhou. 2021. "Analysis of Light-Independent Anthocyanin Accumulation in Mango (Mangifera indica L.)" Horticulturae 7, no. 11: 423. https://doi.org/10.3390/horticulturae7110423
APA StyleShi, B., Wu, H., Zheng, B., Qian, M., Gao, A., & Zhou, K. (2021). Analysis of Light-Independent Anthocyanin Accumulation in Mango (Mangifera indica L.). Horticulturae, 7(11), 423. https://doi.org/10.3390/horticulturae7110423







