Rhizospheric Fungal Diversities and Soil Biochemical Factors of Fritillaria taipaiensis over Five Cultivation Years
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Site Location
2.2. Sample Collection
2.3. Soil Biochemical Analysis
2.4. Soil DNA Extraction and High-Throughput Sequencing
2.5. Data Analysis
3. Results
3.1. Alpha Diversities of the Rhizospheric Soils with Different Cultivation Years
3.2. Fungal Taxonomic Composition
3.3. NMDS Analysis
3.4. Soil Biochemical Analysis
3.5. Correlation Analysis
3.6. Relationship between the Rhizospheric Fungal Community and the Soil Biochemical Factors
4. Discussion
4.1. Cultivation Year Changed the Fungal Alpha Diversity in the F. taipaiensis Rhizosphere
4.2. Pathogenic Fungi Accumulated in the F. taipaiensis Rhizosphere with the Cultivation Year
4.3. Analysis on the Effect of Cultivation Years on the Biochemical Factors in the F. taipaiensis rhizosphere
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, D.D.; Wang, S.; Chen, X.; Xu, X.L.; Zhu, J.Y.; Nie, L.H. Antitussive, expectorant and anti-inflammatory activities of four alkaloids isolated from Bulbus of Fritillaria wabuensis. J. Ethnopharmacol. 2000, 139, 189–193. [Google Scholar] [CrossRef]
- Tan, S.F.; Liu, C.F.; Wang, C.S.; Wang, D.H.; Zhang, Y.Q.; Lin, N. Evaluation on the effect of analgesia and expectorant of Aconiti Radix Cocta in coordination with Fritillaria cirrhosa and Fritillaria thunbergii based on the uniform design method. China J. Chin. Mater. Med. 2013, 38, 2706. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, M. Suitable Technology for Production and Processing of Fritillaria cirrhosa, 1st ed.; China Medical Press: Beijing, China, 2018; p. 8. [Google Scholar]
- Li, P.; Xu, G.J. Studies on resources of Chinese drugs Beimu. J. Plant Resour. Environ. 1993, 2, 12–17. [Google Scholar]
- Chinese Pharmacopoeia Commission. Chuanbeimu. In Pharmacopoeia of the People’s Republic of China, 1st ed.; Zhao, Y.Y., Fan, Z.X., Huang, K., Li, Z., Gao, Y.M., Eds.; China Medical Science Press: Beijing, China, 2015; p. 36. [Google Scholar]
- Duan, B.Z.; Chen, X.L.; Huang, L.F.; Lu, Q.F.; Li, X.W.; Chen, S.L. A survey of resource science of Fritillaria Taipaiensis. Mod. Chin. Med. 2010, 12, 12. [Google Scholar] [CrossRef]
- Jiang, S.Y.; Sun, H.B.; Qin, J.H.; Zhu, W.T.; Sun, H. Functional production regionalization for Fritillariae Cirrhosae bulbs based on growth and quality suitability assessment. China. J. Chin. Mater. Med. 2016, 41, 3194. [Google Scholar] [CrossRef]
- Duan, B.Z.; Huang, L.F.; Yu, Y.L.; Wang, L.Z.; Xie, C.X.; Suo, F.M. Regionalization for growing Fritillaria taipaiensis P. Y. Li by TCMGIS-II. World Sci. Technol./Mod. Tradit. Chin. Medic. Mater. Med. 2010, 12, 486–488. [Google Scholar] [CrossRef]
- Gu, W.C.; Mu, M.J.; Yang, M.; Guo, D.Q.; Zhou, N. Correlation analysis between bulb quality and rhizosphere soil factors of Fritillaria taipaiensis. Chin. J. Exp. Tradit. Med. Form. 2020, 26, 165–177. [Google Scholar] [CrossRef]
- Mu, M.J.; Zhang, D.G.; Zhang, H.; Yang, M.; Guo, D.Q.; Zhou, N. Correlation between rhizospheric microorganisms distribution and alkaloid content of Fritillaria taipaiensis. China J. Chin. Mater. Med. 2019, 44, 2231–2235. [Google Scholar] [CrossRef]
- Peng, R.; Ma, P.; Mo, R.Y.; Sun, N.X. Analysis of the bioactive components from different growth stages of Fritillaria taipaiensis P. Y. Li. Acta Pharm. Sin. 2013, 3, 167–173. [Google Scholar] [CrossRef] [Green Version]
- Nannipieri, P.; Kandeler, E.; Ruggiero, P.; Burns, R.G.; Dick, R.P. Enzymes in the Environment: Activity, Ecology and Applications, 1st ed.; Marcel Dekker: New York, NY, USA, 2002. [Google Scholar]
- Sparling, G.P. Biological Indicators of Soil Health, 1st ed.; CAB International: Wallingford, UK, 1997. [Google Scholar]
- Alkorta, I.; Aizpurua, A.; Riga, P.; Albizu, I.; Amezaga, I.; Garbisu, C. Soil enzyme activities as biological indicators of soil health. Rev. Environ. Health 2003, 18, 65–73. [Google Scholar] [CrossRef]
- Lu, L.H.; Yin, S.X.; Liu, X.; Zhang, W.M.; Gu, T.Y.; Shen, Q.R. Fungal networks in yield-invigorating and debilitating soils induced by prolonged potato monoculture. Soil Biol. Biochem. 2013, 65, 186–194. [Google Scholar] [CrossRef] [Green Version]
- Sun, J.; Zhang, Q.; Zhou, J.; Wei, Q.P. Illumina amplicon sequencing of 16S rRNA Tag reveals bacterial community development in the rhizosphere of apple nurseries at a replant disease site and a new planting site. PLoS ONE 2014, 9, e111744. [Google Scholar] [CrossRef] [Green Version]
- Yao, H.Y.; Jiao, X.D.; Wu, F.Z. Effects of continuous cucumber cropping and alternative rotations under protected cultivation on soil microbial community diversity. Plant Soil 2006, 284, 195–203. [Google Scholar] [CrossRef]
- Chen, M.N.; Li, X.; Yang, Q.L.; Chi, X.Y.; Pan, L.J.; Chen, N. Soil eukaryotic microorganism succession as affected by continuous cropping of peanut-pathogenic and beneficial fungi were selected. PLoS ONE 2012, 7, e40659. [Google Scholar] [CrossRef] [Green Version]
- Xiong, W.; Li, Z.G.; Liu, H.J.; Xue, C.; Zhang, R.F.; Wu, H.S. The Effect of Long-Term Continuous Cropping of Black Pepper on Soil Bacterial Communities as Determined by 454 Pyrosequencing. PLoS ONE 2015, 10, e0136946. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.Y.; Yang, W.X.; Chen, Y.H.; Chen, X.J. Effects of consecutively monocultured Rehmannia glutinosa L. on diversity of fungal community in rhizospheric soil. J. Integr.Agr. 2011, 10, 1374–1384. [Google Scholar] [CrossRef]
- Zhou, X.; Wu, F. Dynamics of the diversity of fungal and Fusarium communities during continuous cropping of cucumber in the greenhouse. FEMS Microbiol. Ecol. 2012, 80, 469–478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, X.J.; Yang, Y.X.; Hu, P.; Zhang, M.; Xia, Y.L. Investigation on the resources of Fritillaria taipaiensis. J. Anhui. Agric. Sci. 2015, 43, 84–85. [Google Scholar] [CrossRef]
- Wu, Z.Z.; Wu, C.S. Effects of different fertilization modes on the growth of Fritillaria taipaiensis. Agric. Eng. 2016, 6, 153–154. [Google Scholar] [CrossRef]
- Hernández, M.; Calabi, M.; Conrad, R.; Dumont, M.G. Analysis of the microbial communities in soils of difffferent ages following volcanic eruptions. Pedosphere 2020, 31, 126–134. [Google Scholar] [CrossRef]
- Lin, X.G. Principles and Methods of Soil Microbial Research, 1st ed.; Higher Education Press: Beijing, China, 2010. [Google Scholar]
- Guan, S.Y. Soil Enzyme and Its Research Method, 1st ed.; Agricultural Press: Beijing, China, 1986. [Google Scholar]
- Cai, L.T.; Hu, Z.Y.; Luo, Z.Y. Extraction of total DNA of microbes from tobacco diseased-field soil by SDS-CTAB method. Acta Agric. Jiangxi 2011, 44, 641–670. [Google Scholar] [CrossRef] [Green Version]
- Kennedy, A.C.; Smith, K.L. Soil microbial diversity and the sustainability of agricultural soils. Plant Soil 1995, 170, 75–86. [Google Scholar] [CrossRef]
- Song, X.; Pan, Y.; Li, L.; Wu, X.; Wang, Y. Composition and diversity of rhizosphere fungal community in Coptis chinensis Franch. continuous cropping fields. PLoS ONE 2018, 13, e0193811. [Google Scholar] [CrossRef] [Green Version]
- Wen, Y.C.; Li, H.Y.; Lin, Z.A.; Zhao, B.Q.; Sun, Z.B.; Yuan, L. Long-term fertilization alters soil properties and fungal community composition in fuvo-aquic soil of the North China Plain. Sci. Rep. 2020, 10, 7198. [Google Scholar] [CrossRef]
- Wang, S.N.; Cheng, J.K.; Li, T.; Liao, Y.C. Response of soil fungal communities to continuous cropping of fue-cured tobacco. Sci. Rep. 2020, 10, 19911. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.; Dsouza, M.; Gilbert, J.A.; Guo, X.S.; Wang, D.Z.; Guo, Z.B. Fungal community composition in soils subjected to long-term chemical fertilization is most influenced by the type of organic matter. Environ. Microbiol. 2016, 18, 5137–5150. [Google Scholar] [CrossRef] [PubMed]
- Olanrewaju, O.S.; Ayangbenro, A.S.; Glick, B.R.; Babalola, O.O. Plant health: Feedback effect of root exudates-rhizobiome interactions. Appl. Microbiol. Biotechnol. 2019, 103, 1155–1166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haelewaters, D.; Urbina, H.; Brown, S.; Newerth-Henson, S.; Aime, M.C. Isolation and molecular characterization of the Romaine lettuce phylloplane mycobiome. J. Fungi 2021, 7, 227. [Google Scholar] [CrossRef] [PubMed]
- Hunter, P.J.; Pink, D.A.C.; Bending, G.D. Cultivar-level genotype differences inflfluence diversity and composition of lettuce (Lactuca sp.) phyllosphere fungal communities. Fungal Ecol. 2015, 17, 183–187. [Google Scholar] [CrossRef]
- Haynes, K.M.; Preston, M.D.; McLaughlin, J.W.; Webster, K.; Basiliko, N. Dissimilar bacterial and fungal decomposer communities across rich to poor fen peatlands exhibit functional redundancy. Can. J. Soil Sci. 2015, 95, 219–230. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Skinner, M.; Parker, B.L.; Brownbridge, M. Pathogenicity of Beauveria bassiana, Metarhizium anisopliae (Deuteromycotina: Hyphomycetes), and other entomopathogenic fungi against Lygus lineolaris (Hemiptera: Miridae). Brownbridge Source J. Econ. Entomol. 2002, 95, 675–681. [Google Scholar] [CrossRef]
- Ortega-Arbulú, A.S.; Pichler, M.; Vuillemin, A.; Orsi, W.D. Effects of organic matter and low oxygen on the mycobenthos in a coastal lagoon. Environ. Microbiol. 2019, 21, 374–388. [Google Scholar] [CrossRef]
- Eichorst, S.A.; Kuske, C.R. Identification of cellulose-responsive bacterial and fungal communities in geographically and edaphically different soils by using stable isotope probing. Appl. Environ. Microbiol. 2012, 78, 2316–2327. [Google Scholar] [CrossRef] [Green Version]
- Bills, G.F.; Platas, G.; Gams, W. Conspecificity of the cerulenin and helvolic acid producing ‘Cephalosporium caerulens’, and the hypocrealean fungus Sarocladium oryzae. Mycol. Res. 2004, 108, 1291–1300. [Google Scholar] [CrossRef]
- Shoresh, M.; Ilarman, G.E.; Mastouri, F. Induced systemic resistance and plant responses to fungal biocontrol agents. Annu. Rev. Phytopathol. 2010, 48, 21–43. [Google Scholar] [CrossRef] [Green Version]
- Lukešová, T.; Kohout, P.; Větrovský, T.; Vohník, M. The Potential of dark septate endophytes to form root symbioses with ectomycorrhizal and ericoid mycorrhizal middle european forest plants. PLoS ONE 2015, 10, e0124752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Von Arx, J.A. Notes on Monographella and Microdochium. Trans. Br. Mycol. Soc. 1984, 83, 373–374. [Google Scholar] [CrossRef]
- O’Connell, R.J.; Thon, M.R.; Hacquard, S.; Amyotte, S.G.; Kleemann, J.; Torres, M.F. Lifestyle transitions in plant pathogenic Colletotrichum fungi deciphered by genome and transcriptome analyses. Nat. Genet. 2012, 44, 1060–1065. [Google Scholar] [CrossRef] [PubMed]
- Krus, J.; Dietrich, W.; Zimmermann, H.; Klenke, F.; Richter, U.; Richter, H. Ustilago species causing leaf-stripe smut revisited. IMA Fungus 2018, 9, 49–73. [Google Scholar] [CrossRef]
- Quezado-Duval, A.M.; Henz, G.P.; Paz-Lima, M.L.; Medeiros, A.R.; Miranda, B.E.C.; Pfenning, L.H. New hosts of Myrothecium spp. in Brazil and a preliminary In Vitro assay of fungicides. Braz. J. Microbiol. 2010, 41, 246–252. [Google Scholar] [CrossRef] [Green Version]
- Han, J.G.; Shrestha, B.; Hosoya, T.; Lee, K.H.; Sung, G.H.; Shin, H.D. First report of the ash dieback pathogen Hymenoscyphus fraxineus in Korea. Mycobiology 2014, 42, 391–396. [Google Scholar] [CrossRef] [Green Version]
- Nirenberg, H.I.; O’Donnell, K. New Fusarium species and combinations within the Gibberella fujikuroi species complex. Mycologia 1998, 90, 434–458. [Google Scholar] [CrossRef]
- Šafránková, I. Volutella leaf blight and stem canker on Japanese pachysandra in the Czech Republic. Plant Prot. Sci. 2007, 43, 10–12. [Google Scholar] [CrossRef] [Green Version]
- Gomes, T.; Pereira, J.A.; Lino-Neto, T.; Bennett, A.E.; Baptista, P. Bacterial disease induced changes in fungal communities of olive tree twigs depend on host genotype. Sci. Rep. 2019, 9, 5882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Zhang, G.L. Microbial biomass carbon and total organic carbon of soils as affected by rubber cultivation. Pedosphere 2003, 13, 535–537. [Google Scholar] [CrossRef]
- Turner, B.L. Variation in pH optima of hydrolytic enzyme activities in tropical rain forest soils. Appl. Environ. Microbiol. 2010, 76, 6485–6493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mu, M.J.; Zhou, X.Q.; Guo, D.Q.; Wang, Q.; Yang, M.; Zhang, H.; Zhou, N. Effect of growth years to the soil enzyme activities and heavy metal residue of Fritillaria taipaiensis P. Y. Li. Environ. Chem. 2019, 38, 1966–1972. [Google Scholar] [CrossRef]
- Zhang, B.; Liang, C.; He, H.B.; Zhang, X.D. Variations in soil microbial communities and residues along an altitude gradient on the northern slope of changbai mountain, China. PLoS ONE 2013, 8, e66184. [Google Scholar] [CrossRef]
- Cleveland, C.C.; Nemergut, D.R.; Schmidt, S.K.; Townsend, A.R. Increases in soil respiration following labile carbon additions linked to rapid shifs in soil microbial community composition. Biogeochemistry 2007, 82, 229–240. [Google Scholar] [CrossRef]
- Liu, J.J.; Yu, Z.; Wang, X.; Jin, J.; Liu, X.; Wang, G. Soil carbon content drives the biogeographical distribution of fungal communities in the black soil zone of northeast China. Soil Biol. Biochem. 2015, 83, 29–39. [Google Scholar] [CrossRef]
- Li, Y.L.; Tremblay, J.; Bainard, L.D.; Cade-Menun, B.; Hamel, C. Long-term efects of nitrogen and phosphorus fertilization on soil microbial community structure and function under continuous wheat production. Environ. Microbiol. 2019, 22, 1066–1088. [Google Scholar] [CrossRef]
- Deng, S.; Ke, T.; Li, L.; Cai, S.; Zhou, Y.; Liu, Y.; Guo, L.; Chen, L.; Zhang, D. Impacts of environmental factors on the whole microbial communities in the rhizosphere of a metal-tolerant plant: Elsholtzia haichowensis Sun. Environ. Pollut. 2018, 237, 1088–1097. [Google Scholar] [CrossRef]
- Buée, M.; De Boer, W.; Martin, F.; van Overbeek, L.; Jurkevitch, E. The rhizosphere zoo: An overview of plant-associated communities of microorganisms, including phages, bacteria, archaea, and fungi, and of some of their structuring factors. Plant Soil 2009, 321, 189–212. [Google Scholar] [CrossRef]
- Zachow, C.; Berg, C.; Müller, H.; Meincke, R.; Komon-Zelazowska, M.; Druzhinina, I.S.; Kubicek, C.P.; Berg, G. Fungal diversity in the rhizosphere of endemic plant species of Tenerife (Canary Islands): Relationship to vegetation zones and environmental factors. ISME J. 2009, 3, 79–92. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.; Wang, Z.; Lv, X.; Li, Y.; Zhuang, L. High-throughput sequencing reveals the diversity and community structure of rhizosphere fungi of Ferula Sinkiangensis at diferent soil depths. Sci. Rep. 2019, 9, 6558. [Google Scholar] [CrossRef] [PubMed]
- Pramanik, M.H.R.; Nagai, M.; Asao, T. Effect of temperature and hotoperiod on the phytotoxic root exudate of cucumber (Cucumis sativus) in hydroponic culture. J. Chem. Ecol. 2000, 28, 1953–1967. [Google Scholar] [CrossRef]
- Zhang, Z.Y.; Lin, W.X. Continuous cropping obstacle and allelopathic autotoxicity of medicinal plants. Chin. J. Eco-Agric. 2009, 1, 189–196. [Google Scholar] [CrossRef]
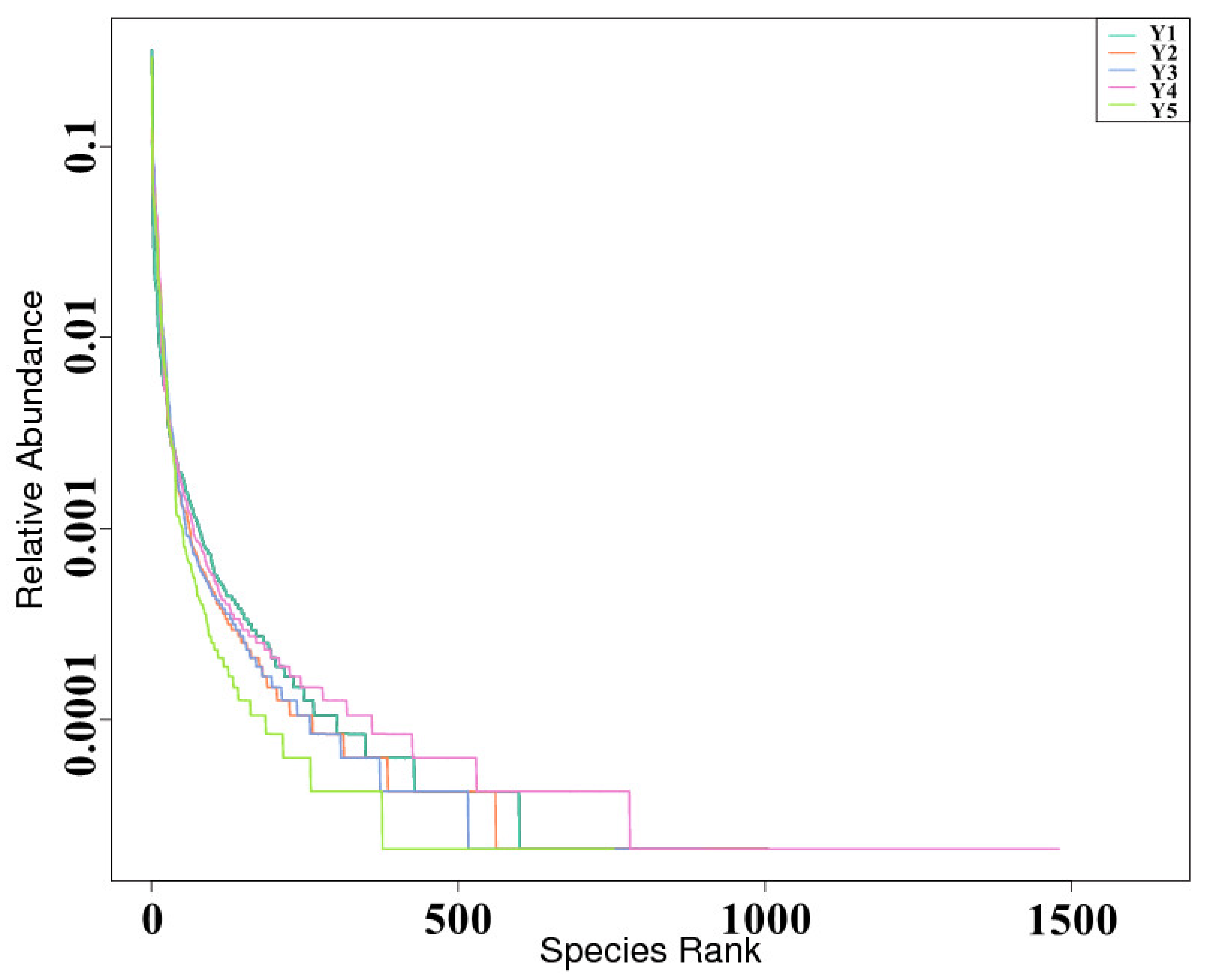
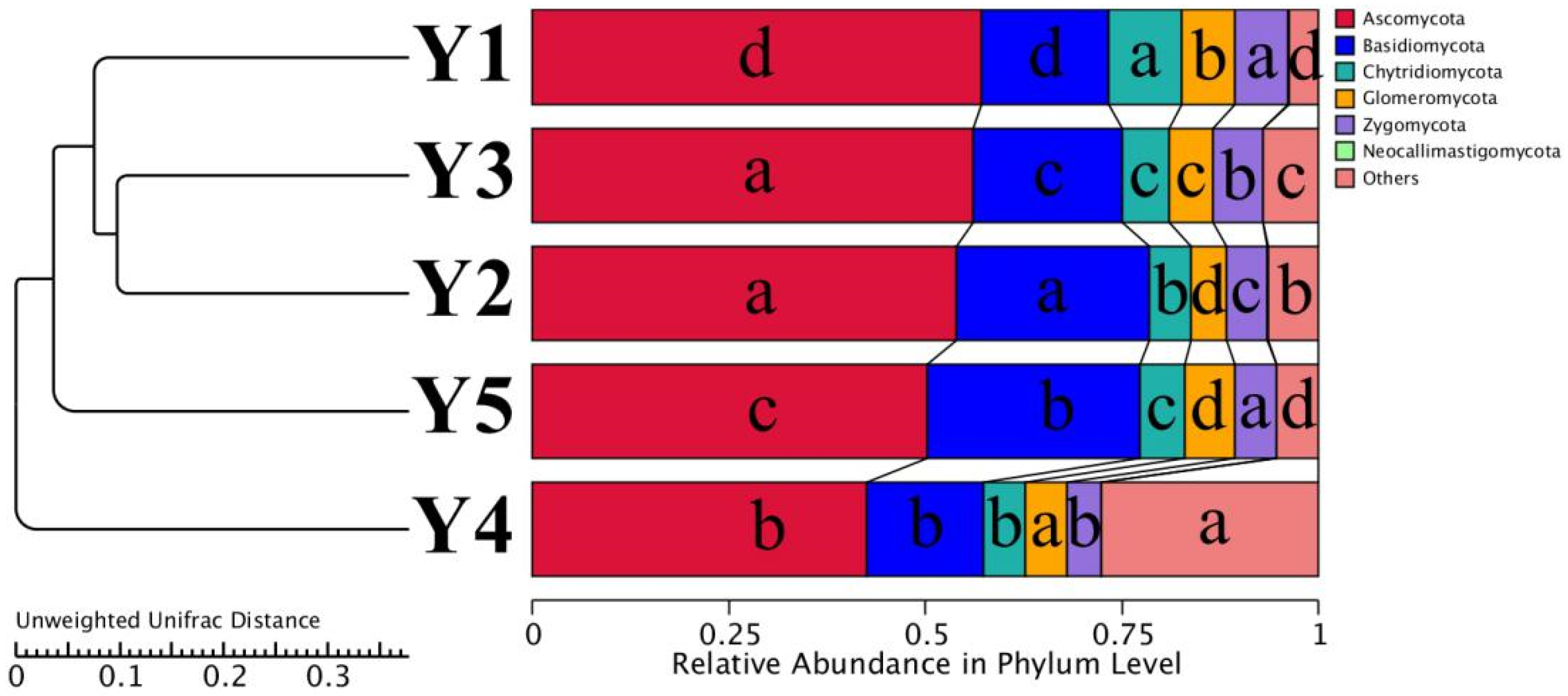
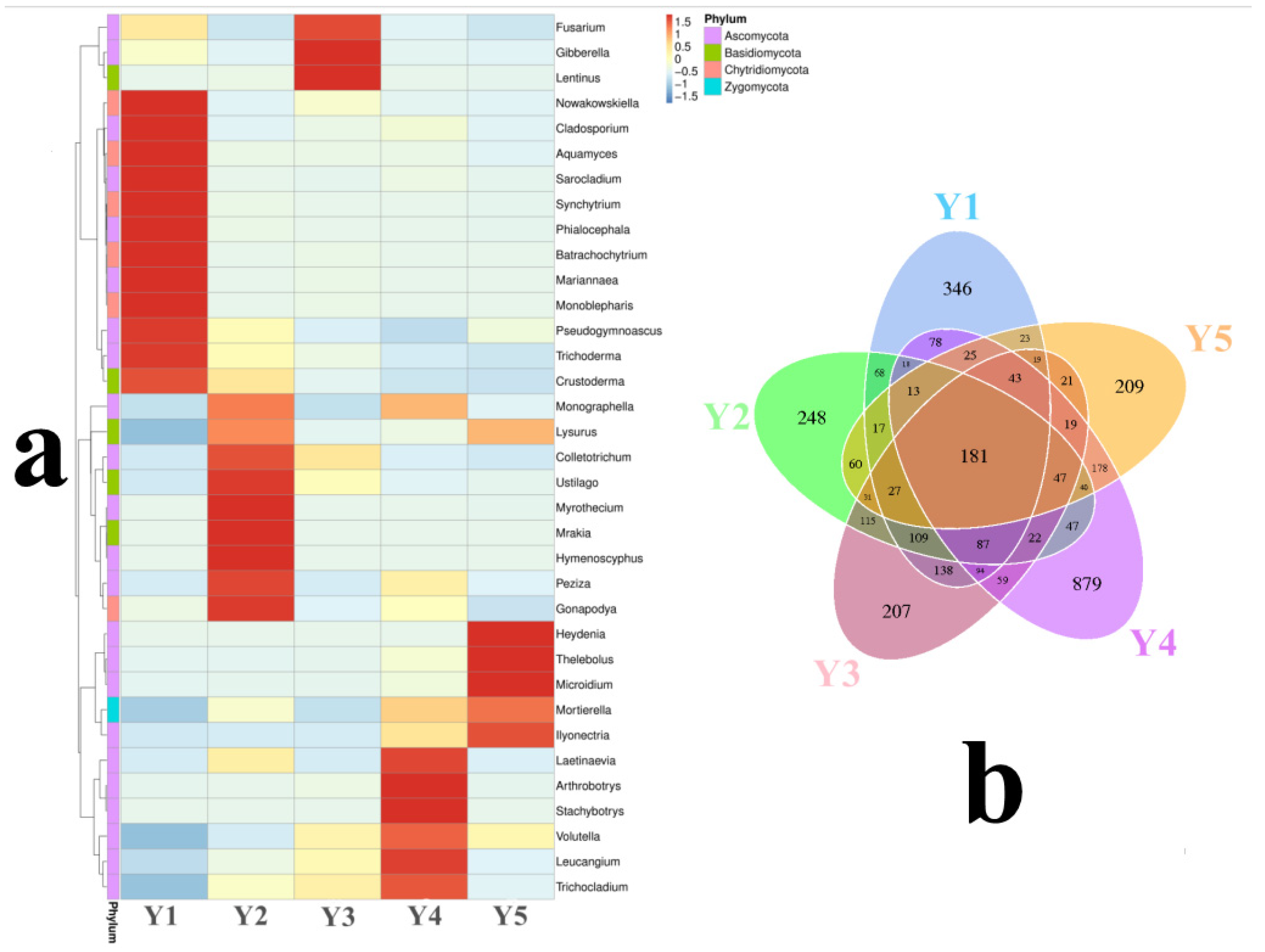
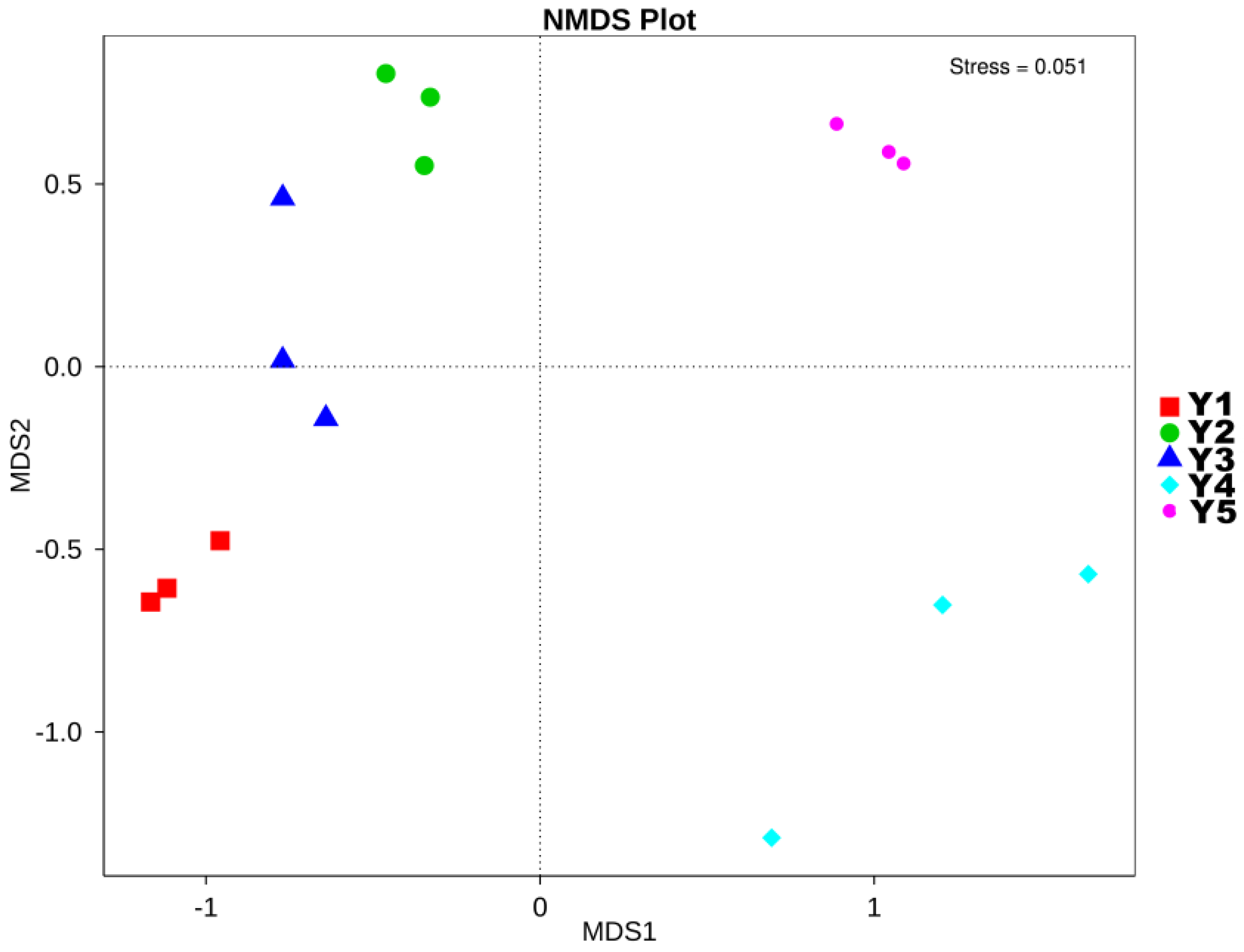

| Sample | Goods-Coverage | Chao1 | ACE | Shannon | Simpson |
|---|---|---|---|---|---|
| Y1 | 0.996 a | 913.377 b | 936.699 b | 4.668 a | 0.855 a |
| Y2 | 0.998 a | 692.037 c | 698.812 c | 5.172 a | 0.937 a |
| Y3 | 0.996 a | 842.382 b | 851.813 b | 4.855 a | 0.911 a |
| Y4 | 0.995 a | 1067.841 a | 1069.508 a | 5.156 a | 0.916 a |
| Y5 | 0.997 a | 612.326 c | 642.342 c | 4.386 a | 0.866 a |
| Phylum | Relative Abundance (%) | ||||
|---|---|---|---|---|---|
| Y1 | Y2 | Y3 | Y4 | Y5 | |
| Ascomycota | 47.20 d | 68.23 a | 65.86 a | 58.23 b | 52.59 c |
| Zygomycota | 36.60 a | 18.46 c | 27.76 b | 28.50 b | 39.30 a |
| Chytridiomycota | 11.12 a | 0.98 b | 0.69 c | 0.96 b | 0.73 c |
| Basidiomycota | 3.30 d | 11.68 a | 4.82 c | 7.28 b | 6.93 b |
| Glomeromycota | 1.61 b | 0.39 d | 0.66 c | 2.71 a | 0.34 d |
| Neocallimastigomycota | 0.03 a | 0.01 b | 0.01 b | 0.01 b | 0.01 b |
| Others | 0.14 d | 0.25 b | 0.20 c | 2.31 a | 0.10 d |
| Genus | Relative Abundance (%) | ||||
|---|---|---|---|---|---|
| Y1 | Y2 | Y3 | Y4 | Y5 | |
| Pseudogymnoascus | 18.26 a | 6.69 b | 1.69 d | 0.23 e | 4.02 c |
| Fusarium | 4.74 b | 0.01 d | 9.63 a | 0.69 c | 0 d |
| Peziza | 0 e | 9.05 a | 0.03 d | 3.73 b | 0.23 c |
| Mortierella | 0.37 e | 3.79 c | 1.21 d | 7.16 b | 9.76 a |
| Arthrobotrys | 0 d | 0.04 c | 0.26 b | 5.15 a | 0 d |
| Colletotrichum | 0.04 d | 4.36 a | 2.23 b | 0.18 c | 0.05 d |
| Myrothecium | 0.04 b | 3.58 a | 0.05 b | 0.02 c | 0 d |
| Laetinaevia | 0.01 e | 2.65 b | 0.06 d | 6.23 a | 0.18 c |
| Gibberella | 0.93 b | 0 d | 5.07 a | 0.12 c | 0 d |
| Microidium | 0.02 d | 0.04 c | 0.06 c | 0.27 b | 3.26 a |
| Synchytrium | 4.99 a | 0.14 b | 0.08 c | 0.06 c | 0.01 d |
| Lysurus | 0.14 d | 4.21 a | 1.24 c | 1.49 c | 3.67 b |
| Trichocladium | 0.28 d | 1.19 b | 1.52 b | 2.61 a | 0.79 c |
| Volutella | 0.13 d | 0.81 c | 1.90 b | 3.53 a | 1.86 b |
| Monoblepharis | 2.03 a | 0.01 b | 0.04 b | 0.02 b | 0.01 b |
| Lentinus | 0.01 b | 0.02 b | 0.99 a | 0 c | 0 c |
| Heydenia | 0 b | 0 b | 0 b | 0 b | 2.07 a |
| Aquamyces | 1.67 a | 0.14 b | 0.13 b | 0.15 b | 0.04 c |
| Trichoderma | 0.92 a | 0.33 b | 0.15 c | 0.05 d | 0.02 d |
| Ilyonectria | 0 d | 0.01 c | 0.02 c | 0.37 b | 0.71 a |
| Vs Groups | Df | Sums of Sqs | Mean Sqs | F. Model | R2 | p |
|---|---|---|---|---|---|---|
| Y1/Y2 | 1 | 1.026 | 1.026 | 22.382 | 0.848 | 0.100 |
| Y1/Y3 | 1 | 0.723 | 0.723 | 7.076 | 0.639 | 0.001 |
| Y1/Y4 | 1 | 1.162 | 1.162 | 12.104 | 0.752 | 0.100 |
| Y1/Y5 | 1 | 1.131 | 1.131 | 19.166 | 0.827 | 0.100 |
| Y2/Y3 | 1 | 0.581 | 0.581 | 5.050 | 0.558 | 0.001 |
| Y2/Y4 | 1 | 0.566 | 0.566 | 5.194 | 0.565 | 0.100 |
| Y2/Y5 | 1 | 0.495 | 0.495 | 6.871 | 0.632 | 0.100 |
| Y3/Y4 | 1 | 0.728 | 0.728 | 4.407 | 0.524 | 0.100 |
| Y3/Y5 | 1 | 0.738 | 0.738 | 5.754 | 0.590 | 0.001 |
| Y4/Y5 | 1 | 0.510 | 0.510 | 4.175 | 0.511 | 0.001 |
| Enzyme Activity/Content/pH | Samples | ||||
|---|---|---|---|---|---|
| Y1 | Y2 | Y3 | Y4 | Y5 | |
| pH | 6.43 c | 6.50 c | 6.52 c | 7.47 a | 6.85 b |
| Enzyme activity/content | g/kg | ||||
| Acid phosphatase | 0.728 a | 0.577 ab | 0.473 b | 0.441 b | 0.683 a |
| Alkaline phosphatase | 0.128 a | 0.101 b | 0.093 bc | 0.078 c | 0.055 d |
| Catalase | 0.674 a | 0.539 b | 0.528 b | 0.677 a | 0.669 a |
| Invertase | 12.815 c | 9.558 d | 9.239 d | 21.772 b | 28.948 a |
| Organic matter | 36.038 a | 28.825 b | 26.124 b | 21.423 c | 21.323 c |
| Protease | 0.280 c | 0.270 c | 0.277 c | 0.350 b | 0.469 a |
| Urease | 5.407 ab | 5.662 a | 5.235 ab | 4.389 c | 4.733 bc |
| mg/kg | |||||
| Available nitrogen | 112.000 b | 134.303 a | 113.044 b | 80.312 c | 78.212 c |
| Available phosphorus | 149.466 a | 95.478 b | 96.076 b | 43.716 c | 38.413 d |
| Available potassium | 404.853 a | 173.790 d | 209.447 c | 168.293 d | 301.979 b |
| pH | Acid Phosphatase | Alkaline Phosphatase | Catalase | Invertase | Organic Matter | Protease | Urease | Available Nitrogen | Available Phosphorus | |
|---|---|---|---|---|---|---|---|---|---|---|
| Available potassium | −0.429 | 0.870 ** | 0.372 | 0.522 | 0.135 | 0.592 | 0.134 | 0.184 | −0.087 | 0.528 |
| Available phosphorus | −0.766 * | 0.405 | 0.959 ** | −0.217 | −0.758 * | 0.980 ** | −0.763 | 0.784 * | 0.716 * | |
| Available nitrogen | −0.770 * | 0.063 | 0.715 * | −0.745 * | −0.911 ** | 0.681 * | −0.835 ** | 0.950 ** | ||
| Urease | −0.917 * | 0.348 | 0.709 * | −0.631 | −0.805 ** | 0.771 * | −0.695 | |||
| Protease | 0.489 | 0.235 | −0.859 ** | 0.557 | 0.967 ** | −0.686 * | ||||
| Organic matter | −0.733 | 0.527 | 0.949 ** | −0.095 | −0.664 * | |||||
| Invertase | 0.646 * | 0.185 | −0.789 * | 0.734 * | ||||||
| Catalase | 0.529 | 0.394 | −0.180 | |||||||
| Alkaline phosphatase | −0.593 | 0.244 | ||||||||
| Acid phosphatase | −0.514 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, N.; Mu, M.; Xie, H.; Wu, Y.; Zhou, Y.; Li, W. Rhizospheric Fungal Diversities and Soil Biochemical Factors of Fritillaria taipaiensis over Five Cultivation Years. Horticulturae 2021, 7, 560. https://doi.org/10.3390/horticulturae7120560
Zhou N, Mu M, Xie H, Wu Y, Zhou Y, Li W. Rhizospheric Fungal Diversities and Soil Biochemical Factors of Fritillaria taipaiensis over Five Cultivation Years. Horticulturae. 2021; 7(12):560. https://doi.org/10.3390/horticulturae7120560
Chicago/Turabian StyleZhou, Nong, Maojun Mu, Hui Xie, Yu Wu, You Zhou, and Weidong Li. 2021. "Rhizospheric Fungal Diversities and Soil Biochemical Factors of Fritillaria taipaiensis over Five Cultivation Years" Horticulturae 7, no. 12: 560. https://doi.org/10.3390/horticulturae7120560
APA StyleZhou, N., Mu, M., Xie, H., Wu, Y., Zhou, Y., & Li, W. (2021). Rhizospheric Fungal Diversities and Soil Biochemical Factors of Fritillaria taipaiensis over Five Cultivation Years. Horticulturae, 7(12), 560. https://doi.org/10.3390/horticulturae7120560






