Abstract
The chickpea chlorotic dwarf virus (CpCDV) (from the genus Mastrevirus and the family Geminiviridae) and tomato leaf curl New Delhi virus (ToLCNDV) (from the genus Begomovirus and the family Geminiviridae) represent an important threat to different crops worldwide, as they are emerging viruses in the Asian continent, were introduced to the Mediterranean region in 2012, and were then reported in Morocco in 2017 and 2018. The present study addresses the dispersion of the CpCDV and the ToLCNDV and evaluates the genetic diversity of the Moroccan isolates of both viruses. A total of 1333 symptomatic leaf plant samples were analyzed by PCR. The study has reported the detection of the ToLCNDV in melon and tomato, as well as the CpCDV in squash crops for the first time in Morocco. Blast analysis of selected representative isolates showed a 97–99% nucleotide identity with the ToLCNDV and the CpCDV infecting different crops in the Mediterranean region. Phylogenetic analysis showed low variability among the Moroccan isolates for the ToLCNDV compared to the Spanish and Italian isolates, whereas the CpCDV strains were variable regarding strains reported in Tunisia and Egypt. Recombination analysis showed the presence of the ToLCNDV recombinant strains with variable parents. The spread of both geminiviruses represents a threat to different crop production, requiring the development of crop protection and management strategies. To prevent viral outbreaks, restrictive phytosanitary measures and the development of resistance strategies are also necessary.
1. Introduction
Geminiviridae is one of the families that have been well-studied in circular single-stranded DNA (ssDNA) plant viruses. Several reviews on the genus Geminivirus have covered different aspects of the biology of these viruses: epidemiology, serologic properties, and molecular biology [1]. However, of the nine genera that define the Geminiviridae family, Begomovirus has the highest number of described species (445), followed by Mastrevirus with 45 species according to the International Committee on Taxonomy of Viruses (ICTV: https://talk.ictvonline.org/taxonomy, accessed on 29 September 2022).
Given that monopartite begomoviruses predominate in the Old World (OW) compared to bipartite begomoviruses, the ToLCNDV’s importance as a bipartite begomovirus with a broad host range in the OW is crucial [2].
After its first description in tomato (Solanum lycopersicum) in India [3], the ToLCNDV was found to be associated with several crops (watermelon (Citrullus lanatus), cucumber (Cucumis sativus), potato (Solanum tuberosum), sponge gourd (Luffa cylindrica), bitter gourd (Momordica charantia), bottle gourd (Lagenaria siceraria), melon (Cucumis melo), and zucchini squash (Cucurbita pepo)) [4,5].
In the beginning, the ToLCNDV was limited to Asian countries (Pakistan, Thailand, Indonesia, and Bangladesh) and the Indian subcontinent. However, recently, the virus has spread to new geographical regions with an expanded known host range and geographic distribution, from the Middle East (Iran) to the Mediterranean Basin (Morocco, Tunisia, Italy, and Spain) [4,6].
Between 2012 and 2017, the ToLCNDV spread in the Mediterranean region. Thus, it was first identified in Spain in 2012, causing leaf curl disease in cucurbits. In 2013, similar symptoms were observed in Almeria [7], then severe damages were observed in 2015 in Tunisia on zucchini, melon, and cucumber. Symptoms consisted of severe yellowing followed by mosaic and curling in young leaves [8]. In Italy in the fall of 2015, the same symptoms were noted [9]. By the fall of 2017, disease symptoms on zucchini crops in the Agadir and Taroudant regions of Morocco were probably similar to those in Italy and Spain [10]. These reports suggest the recent introduction of the virus in North Africa and Southern Europe.
The chickpea chlorotic dwarf virus (CpCDV) is a monopartite virus with a genome ranging from ~2.5–2.7 kb. The virus produces symptoms similar to those of stunts shown to be caused by mastreviruses. Symptoms include yellowing, necrosis, and leaf rolling [1]. The CpCDV was first reported as affecting chickpeas in India [11], and it spread all over the world, including Asia (Pakistan and Iran), Africa (Morocco, Tunisia, Egypt, Burkina Faso, Sudan, South Africa, Nigeria, and Eritiria), the Middle East (Yemen, Turkey, Syria, and Oman) and Australia [12,13,14,15,16]. Besides, five mastreviruses species were recently found in Australia, the chickpea chlorosis virus, chickpea red leaf virus, tobacco yellow dwarf virus, chickpea chlorosis Australia virus, and chickpea yellows virus [1]. The classification of all mastrevirus isolates observed in Africa, Australia, and Asia as infecting chickpeas were reclassified based on a 78% nucleotide identity in the genomic DNA. They were classified into one species, the CpCDV [1,17]. In this study, we report the first detection and molecular characterization of the CpCDV infecting chickpeas, squash, and watermelon, and the ToLCNDV infecting zucchini, melon, and tomato in Morocco.
2. Materials and Methods
2.1. Survey and Samples Collection
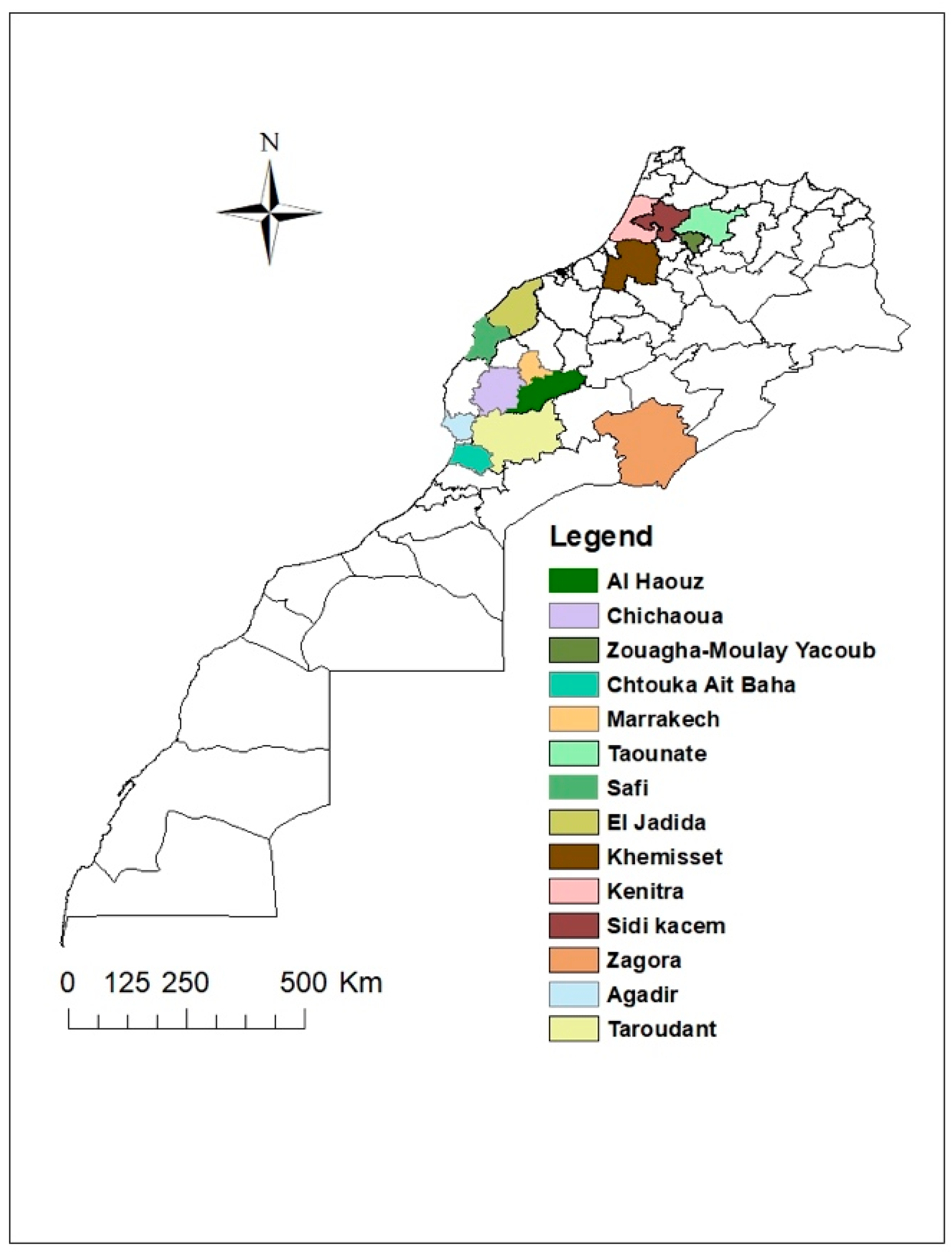
Samples collection was carried out during two growing seasons in the principal cucurbits growing regions of Morocco (Figure 1), in greenhouses or open fields. Leaf samples from plants showing virus-like symptoms of the ToLCNDV and the CpCDV, such as curling, yellowing, and mosaic, were collected.
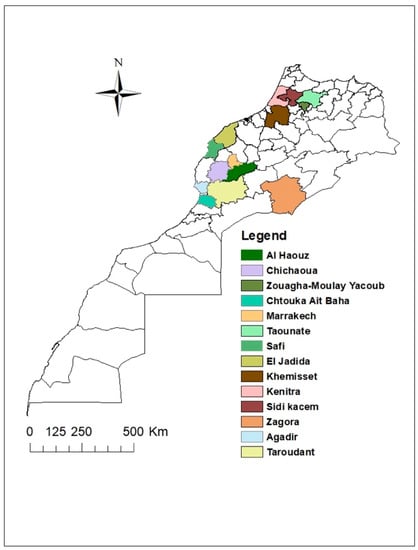
Figure 1.
Map of Morocco showing the sites in which leaf tissues were collected.
During the 2017–2018 and 2018–2019 seasons, 1333 samples were collected from chickpeas, watermelon, melon, zucchini, squash, and tomato. A detailed description of samples collected according to plant species is given in Table 1. All samples were placed in tubes containing calcium chloride and stored at 4 °C.

Table 1.
Description of the 1333 samples collected during two growing seasons from different crop species.
2.2. DNA Isolation and PCR Amplification
The DNA was extracted from 7 mg of infected leaf samples using the cetyltrimethylammonium bromide (CTAB)-based procedure described by Doyle and Doyle (1990) [18]. The samples were crushed in 500 µL of a TE buffer. The quality of the DNA was checked on a 1% agarose gel and stored at −20 °C until further use.
The extracted DNA was subjected to PCR for virus detection, using a set of specific primers for the detection of the ToLCNDV, the ToLCNDV-A1F/R targeting the AC1, AC2, AC3, and AC5 proteins, as well as the coat protein (AV1) [19] and a set of specific capsid proteins (CPs) targeting the primer pair CpCDV-CP-F/R [20] for the detection of the CpCDV. PCR reactions were carried out in 25 μL of volume that contained 2.5 μL of template DNA, 2.5 μL of a 10× PCR buffer, 0.25 μL (10 mM of each) dNTPs, 2,25 μL (50 mM) MgCl2, 1 μL (10 µM) of each primer, and 0.5 μL (5µ/µL) DreamTaq DNA polymerase (Thermo Fisher Scientific, Waltham, MA, USA). The rest of the volume was completed with 15 μL of distilled sterile water. Amplifications were performed in an Eppendorf thermal cycler (Germany). The products were analyzed by 1.5% agarose gel electrophoresis in a TBE buffer of 0.5× for 25 min and stained using Sybersafe. PCR products were then sequenced using the sanger dioxide method (STABVIDA, Portugal).
2.3. Sequence Comparison and Phylogenetic Analysis
Sequences of both viruses were assembled and analyzed using the software BIOEDIT version 7.0 programs [21]. Database searches of similarity were carried out by the NCBI-BLAST program (http://blast.ncbi.nlm.nih.gov, accessed on 23 April 2022).
The phylogenetic tree was constructed using nucleotide sequences of DNA-A of the ToLCNDV and CP sequence of the CpCDV isolates and other selected species reported worldwide. A few other selected begomoviruses and mastreviruses causing yellow mosaic disease in different plants were also taken into consideration to reveal the relationship of the virus isolate under study with its homologs. The phylogenetic tree was constructed using the maximum likelihood method with 1000 bootstrap replications following the Kimura 2-parameter model [22] and viewed with the help of the MEGA X program 10.2.4 (https://www.megasoftware.net/) [23].
2.4. Recombination Analysis
The recombination analysis was performed using 70 complete genome sequences of the ToLCNDV from different geographical regions, with our isolates to detect the presence of recombination sites using the RDP program, GENECONV, BOOTSCAN, MAXIMUM CHISQUARE, CHIMAERA, SISTER SCAN, and 3SEQ non-parametric recombination detection methods, as implemented in the RDP4 software [24]. Only events detectable on more than three different methods were retained for further analysis. The analysis of the hypothetical parental sequences of the recombination sites was assigned using seven non-parametric methods in RDP4 software.
3. Results
3.1. Symptoms Description
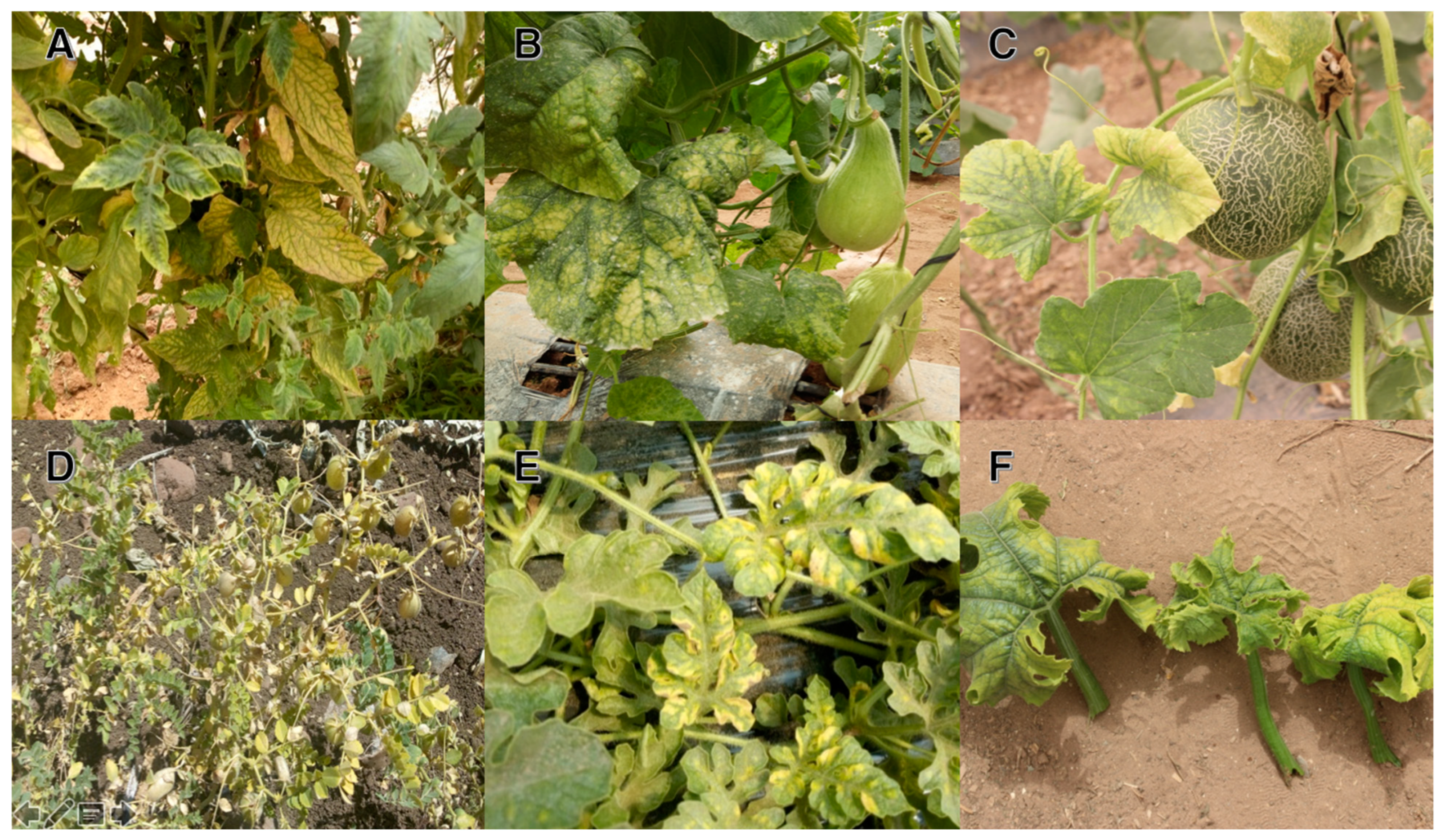
The most common symptoms seen on all of the cucurbit crops ranged from mild to severe mosaics, blistering, shrinkage, leaf curling, yellowing, and a reduction in leaf size and fruit caliber. Due to the multiple infections with several viruses, it was difficult to attribute the symptoms observed in the field to particular viral infections. Watermelon samples taken from Faïja and Ben Dlala in Zagoura showed symptoms of mosaic and yellowing of the leaves (Figure 2E). Symptoms of viral diseases were not too generalized in the plots visited. The symptoms were sporadic, dispersed, and limited in terms of spatial distribution. On melon crops in the region of Chichaoua and Agadir, symptoms of mosaics (Figure 2B,C) were observed on the leaves, with deformations and sometimes with reduced fruit growth. However, the infections were not widespread in the visited plots. The Roumani zone is known for food legumes over large areas. Plots of chickpeas were targeted for visual observations of symptoms and samples for laboratory analysis. During the surveys, symptoms of dwarfism and wilting were observed on the chickpeas in several visited plots (Figure 2E).

Figure 2.
Symptoms of virus infections on solanaceous and cucurbitaceous crops. (A) Tomato plant with the ToLCNDV infection, (B,C) melon plants with the ToLCNDV infection, and (D) chickpea plant with the CpCDV infection. (E) Symptoms of yellowing and curling on watermelon and (F) zucchini leaves collected from a ToLCNDV-infected plant.
3.2. Virus Detection and Characterization
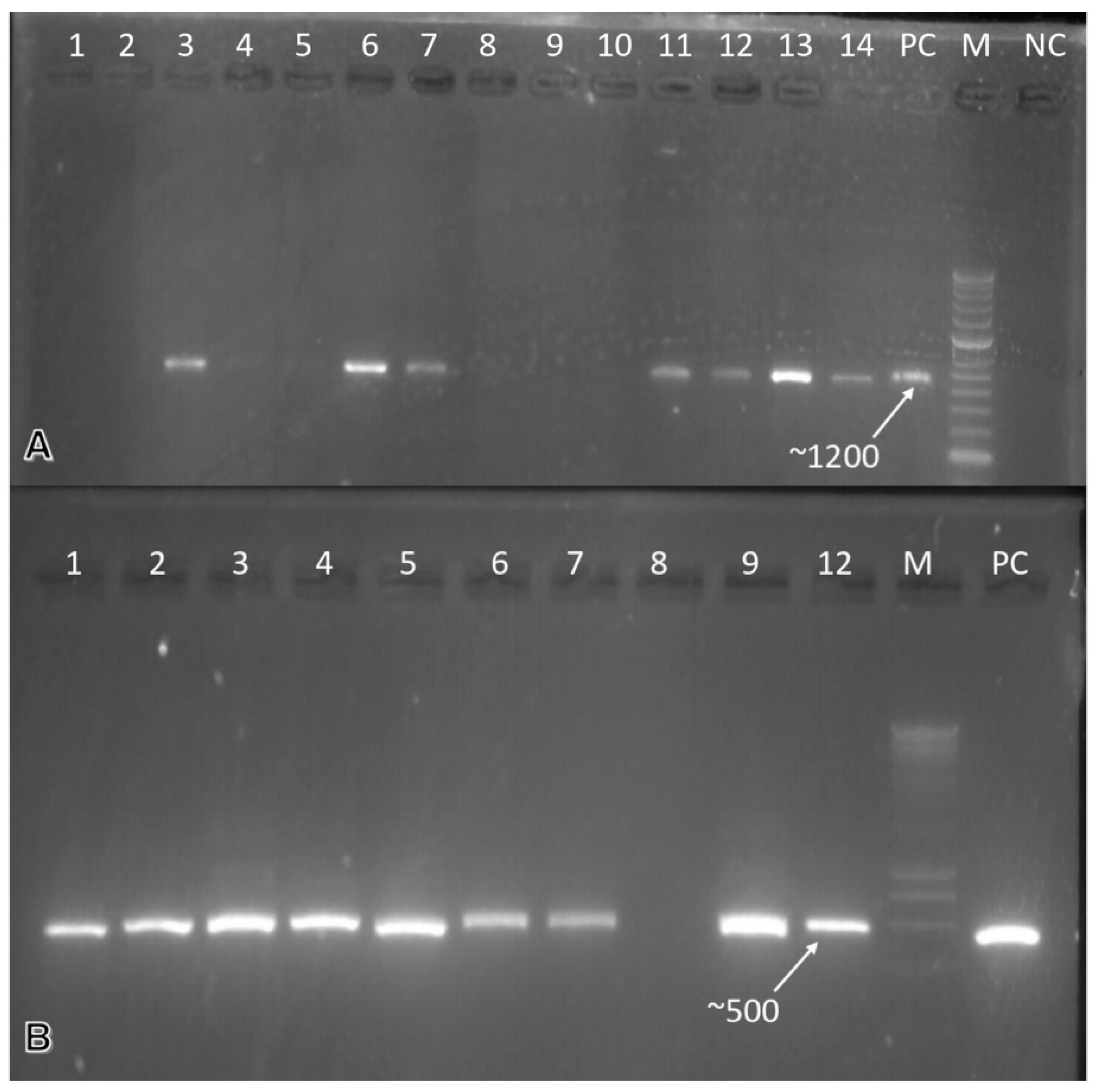
All of the samples that we collected were PCR-tested for the ToLCNDV. Of the 1333 samples analyzed, 45 samples reacted positively to the tested virus. Nineteen out of one-hundred and seventy-seven samples of zucchini were positive for the ToLCNDV while only nine out of four-hundred and forty-seven were for melon, and two were positive for the tomato samples out of fifty-six. A ~1200 bp PCR product was obtained, indicating the virus’ presence in the analyzed samples. It is important to note that the detection of the ToLCNDV was also carried out on asymptomatic samples. The leaf samples of the watermelon, squash, and chickpeas were analyzed by PCR for the detection of the CpCDV. Six out of ninety-four analyzed chickpea samples were positive, while only eight of four-hundred and forty-seven watermelon samples reacted positively to the PCR tests. However, only one sample was positive when analyzing thirty-six samples of squash. A band of ~500 bp was obtained (Figure 3).
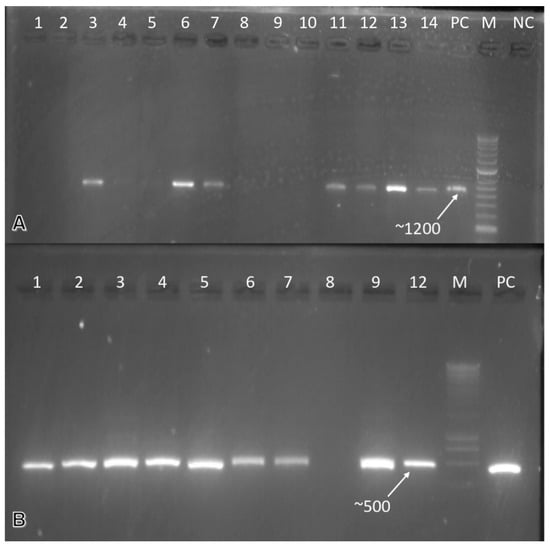
Figure 3.
Gel electrophoresis profile of cucurbit viruses: (A) Molecular detection of the ToLCNDV in two cucurbit crops samples (zucchini and watermelon) using specific primers. Lanes 1–7 are watermelon samples, lanes 8–14 are zucchini samples, lane M is M-1Kb DNA stepladder (Thermo Fisher scientific, Vilnius, Lithuania), lane NC is a negative control, and lane PC is a positive control. (B) Molecular detection of the CpCDV in watermelon and chickpea samples using specific primers. Lanes 1–9 are watermelon samples and 12 chickpea samples, lane M is M-1Kb DNA stepladder, (Thermo Fisher scientific, Vilnius, Lithuania) and lane PC is a positive control.
3.3. Sequence and Phylogenetic Analysis
From the amplified product, 18 representative samples of the DNA-A of the ToLCNDV, and 10 of the CpCDV of the CP gene (Table 2) were sequenced. An analysis of the sequences on the nuclotidic level of the isolated strains from the chickpeas (S4P6 and S3P5) showed an identity of 98–99% of the already published sequences from Morocco affecting watermelons (MH500779-MH500778-MH500777), a 97–98% identity with the CpCDV sequence (KF692356) isolated from squash in Egypt, and 96–97% with the CpCDV isolated from watermelon (KX580024) in Tunisia. However, the CpCDV strains isolated from watermelon (S1Pa1, S3Pa1 CP2, S2Pa2, S1Pa8, S1Pa5, and S1Pa6) were 99% identical to the already published sequence from Morocco affecting watermelon (MH500779-MH500778-MH500777); there was a 98% identity with the sequence (KF692356) from Egypt and 97% with the sequence (KX580024) from Tunisia. Moreover, the last isolated sequence from the squash in Morocco (CgOA1) was 98% identical to the Egyptian strain (KF692356) found to be associated with squash, and 99% identical to our already published sequences (MH500779-MH500778-MH500777).

Table 2.
Sequenced CpCDV and ToLCNDV isolates reported in the present study.
For the ToLCNDV, according to the nucleotid blast search, the isolates (AM1, AM2, AZ3, AZ4, AZ5, AZ6, AM8, AM12, AZ13, AM14, AZ15, AZ16, and AZ17) have an identity ranging between 98–99% with the sequence published from Morocco affecting zucchini (MG098230). These isolates were also similar to the strains from Spain (MG098230 affecting melon and MH577722 affecting zucchini) and Tunisia (MF967020 affecting squash). Also, the AT10 isolate had a 99% identity with (MH577752) affecting melon and (MH577722) affecting zucchini in Spain and exhibits the same percentage of similarity with a sequence isolated from squash in Tunisia (MF967020). For AZ7, an identity of 98% with (MH577752) affecting melon and (MH577722) affecting zucchini in Spain was reported. Finally, the rest of the isolates (AT9, AZ11, and AZ18) shared 98% of their identity with the MH577752 isolate affecting melon in Spain and the KU145141 isolate affecting zucchini in Italy.
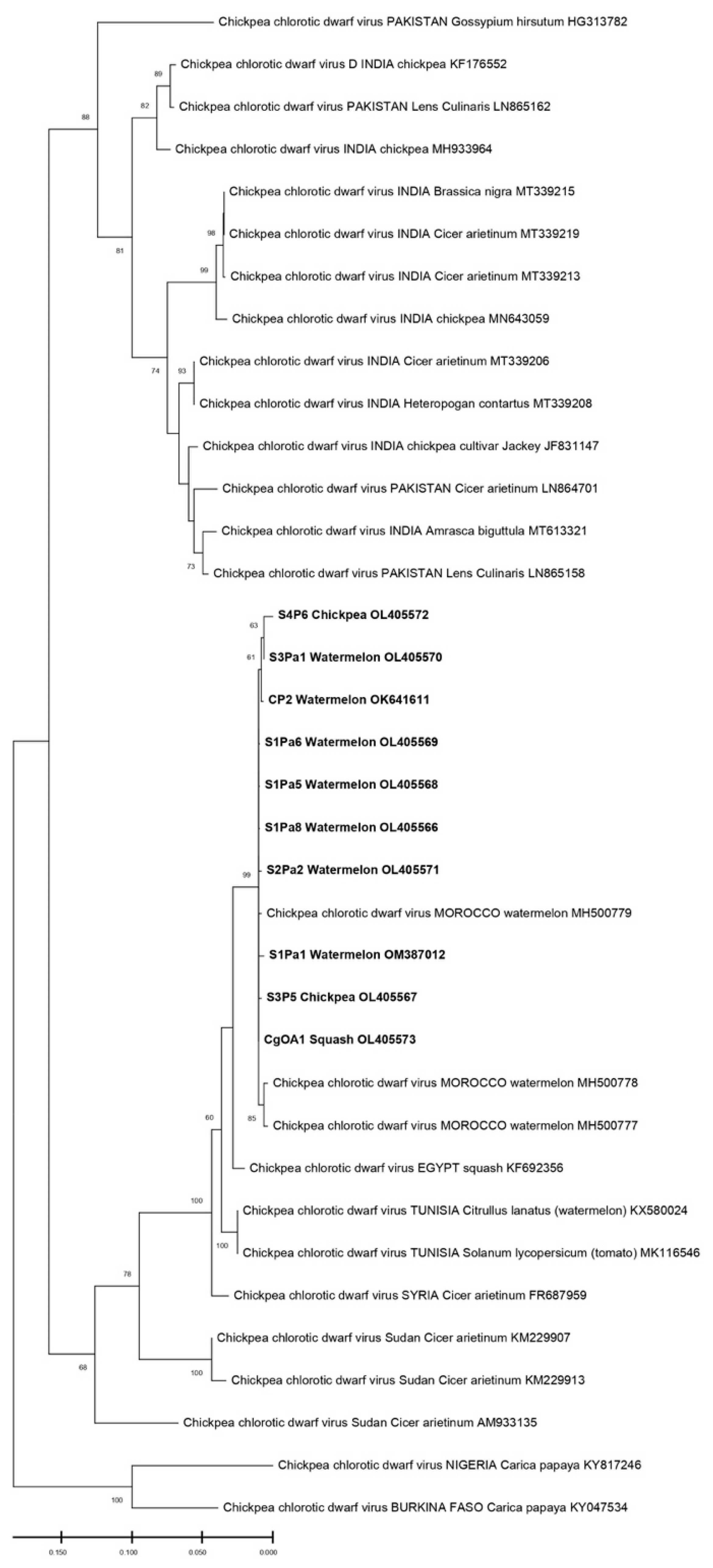
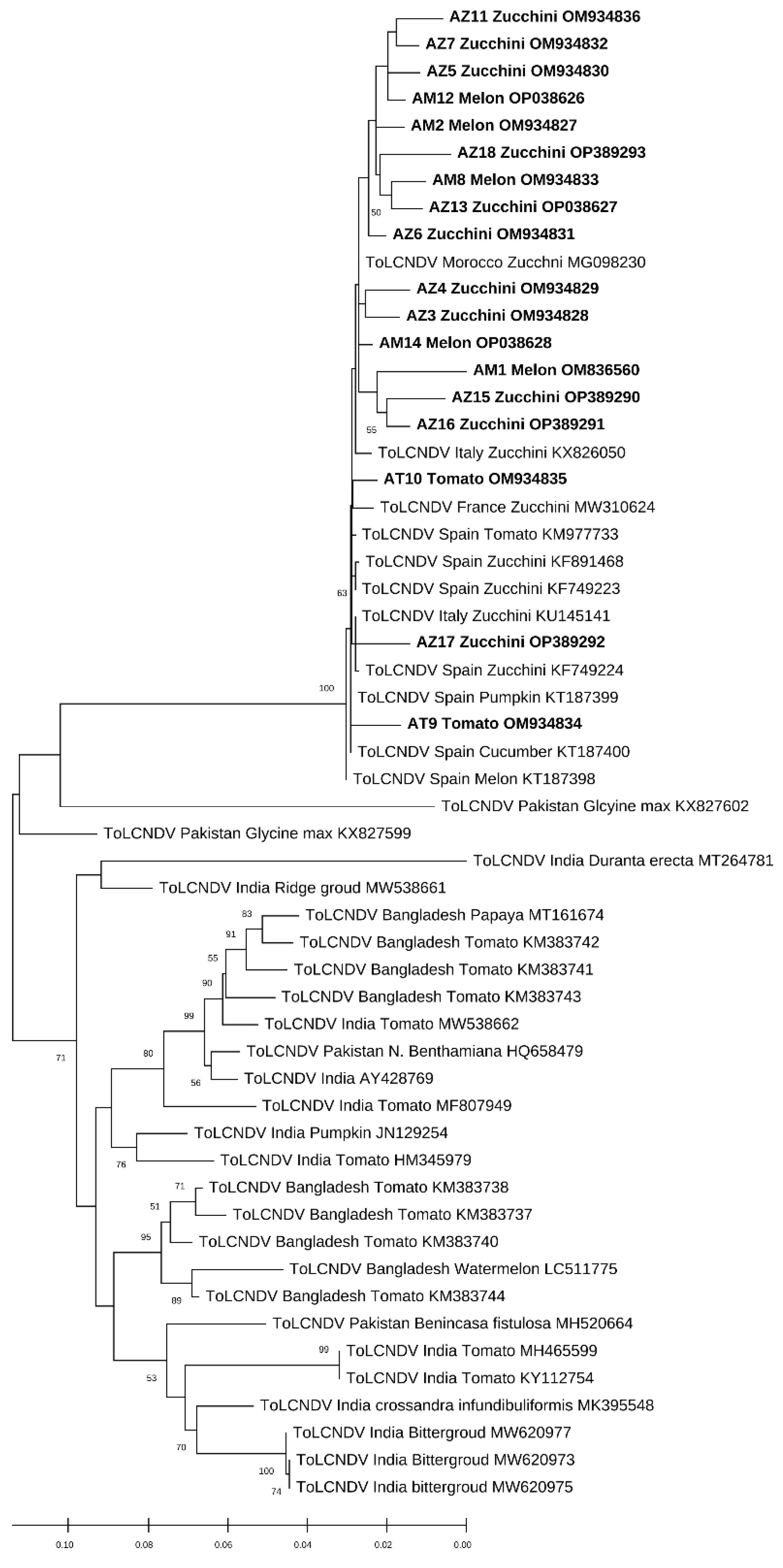
The phylogenetic relationships among the isolates of the present study with those used as references from the world are shown in Figure 4 and Figure 5. During the phylogenetic analysis, the sequences of the ToLCNDV isolates were all clustered with the sequences of the ToLCNDV of Spain, Italy, and Tunisia. The results show that our isolates were highly different from those reported in Asia or even in some countries in Africa, such as Nigeria. However, for the CpCDV, the constructed phylogenic tree based on the CP gene has formed a cluster of a 99% identity with each other; our isolates were close to those of Egypt and Tunisia (Figure 4 and Figure 5).
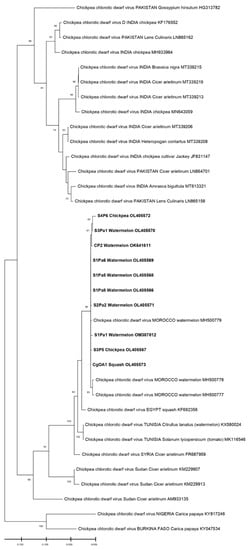
Figure 4.
The evolutionary history was inferred by using the Maximum Likelihood method and the Kimura 2-parameter model for the CpCDV. This analysis involved 36 nucleotide sequences for the CpCDV tree; only the results that have branch support above 50% are shown.
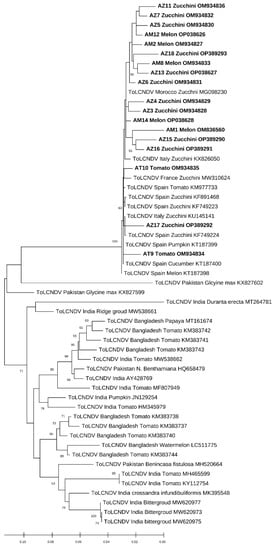
Figure 5.
The evolutionary history was inferred by using the Maximum Likelihood method and the Kimura 2-parameter model for the ToLCNDV. This analysis involved 55 nucleotide sequences for the ToLCNDV; only the results that have branch support above 50% are shown.
3.4. Recombination Analysis
The recombination analysis was carried out using seven different detection methods; they detected the recombination throughout the partial genome of the DNA-A of 18 ToLCNDV sequences compared with 70 sequences from different parts of the world. Six recombination sites were detected in five different isolates from four countries, as shown in Table 3.

Table 3.
Recombinant analysis results.
The first detected ToLCNDV isolate was AZ15 with the Moroccan isolate MG098230 as a major parent and the Indian isolate MK395548 as a minor parent. The second isolates were AZ18 and AM2 with the French isolate MW310624 as a major parent for both of them, while the minor was unknown. For AM12 and AZ16, the major parents detected were the French isolate MW310624 and the Moroccan isolate MG098230, respectively, with an unknown minor parent for both strains.
4. Discussion
The emergence of viruses belonging to the family of Geminiviridae affecting several crops has occurred recently, leading to global outbreaks and severe economic losses in the Mediterranean [25,26]. In the present study, we have revealed the occurrence of two Geminiviridae viruses, the CpCDV and the ToLCNDV, in association with several cucurbits, solanaceous, and fabaceous crops in Morocco. During a survey of two years, the symptoms of leaf curling, mosaic and yellowing, vein clearing, and growth reduction of plants were observed and found to be common in several viral occurrences in the Mediterranean region; these symptoms were generally associated with Begomovirus and Mastrevirus infection [6,15].
The CpCDV, which is a leafhopper-transmitted virus, was formerly reported in Morocco as causing severe losses in watermelon production in the Zagoura region [16]. Thereafter, a survey was conducted to study the emergence of the disease on a large scale and different crops produced in the country, especially cucurbits, to explain whether it constituted an emerging strain since cucurbit crops are not a preferred host of the CpCDV, which brings into question the degree of evolution needed for the virus to enhance its adaptative response to affect such hosts [27].
Sequence analysis of the CpCDV revealed a genetic relatedness with different strains isolated from various crops (squash, watermelon, and chickpea). These isolates showed significant relatedness also with the CpCDV strains reported in neighboring countries, such as Tunisia and Egypt [20,27]. However, our strains did not cluster with the strains reported in both countries in the phylogenetic tree, but they formed a distinct clade among the diver’s population of the CpCDV used as a reference in the study. This could be explained by the fact that the Moroccan isolates have evolved from an already existing population, affecting primarily chickpeas to watermelon and squash to form a new population of the virus. The only report of the CpCDV in squash was in Egypt in 2015 [27] in which it was found that the CpCDV-Eg was infecting squash in a mixed infection with SqLCV; the resulting symptoms were mainly thought to be associated with Betasatellites and also recombination events that were found to be the major contributor to these symptoms. In the present study, the squash sample reacted positively and was cultivated close to the watermelon, which was also positive for the CpCDV. These may be explained by the high pressure of the infection, which led to the host expansion.
The CpCDV has a wide host range and has expanded its geographical distribution, making the virus a serious threat to several crops in the Mediterranean countries and the world. The vector of the virus Orosius orientalis was reported as present in both Tunisia and Morocco. The virus may also be present in southern Europe, which threatens the cucurbits crops’ production and makes the presence of the virus a real problem for farmers [20,27,28].
Symptoms of yellowing and curling were observed in tomato crops growing in a greenhouse in the region of Fes. The observed symptoms of yellowing and apical leaf curling of zucchini plants were both related to the ToLCNDV. This is the first time that the virus has been found in tomato plants in the country; it had previously only been reported in tomatoes in Spain and Tunisia in the Mediterranean region [29,30]. Observations in the field in Morocco, over the last five years since the declaration of the ToLCNDV in Morocco, suggest that zucchini and melon crops were more severely affected than other cucurbit crops [10]. Similar observations were reported in Spain [5]. The partial genome sequencing of the Moroccan isolates shared nucleotide identities of 98–99% with isolates reported from Tunisia, Spain, and Italy. Further, the phylogenic analysis showed the population of the Moroccan isolates belong to those reported in the Mediterranean, as they clustered in the same clade by 100%. The group of the Mediterranean region is highly different from the population of the virus in the Asia continent. These differences are mainly derived from recombination events that seem to be a major force driving the evolution of the ToLCNDV, which led to the emergence of these isolates and cause epidemics [25].
Several recombination events were identified within our isolates, and several events were already reported, mainly in Spain and Italy [31,32,33]. This exchange of DNA might occur between species of the same genus, which determines the aggressiveness of the virus and the expansion of its host range [25]. Begomoviruses have an important potential of increasing their genetic variability through several processes, such as mutations and recombination. The presence of a high intra-population diversity allows the rapid accumulation of virus variants during the infection, resulting in the diversification and rapid evolution of the virus in response to new conditions [33].
The ToLCNDV has been spread widely in the region of Agadir in Morocco, as this region is widely known for cucurbits production in the country. To avoid the spread of the virus to other regions or even the introduction of other pests and pathogens, it is very important to carry out a correct management strategy, surveillance, and the identification of pathogens through molecular and serological tools, as well as develop genetic resistance varieties against not only geminiviruses (Begomovirus and Mastrevirus) but also other pathogens that may cause important economic losses to the country’s crops.
5. Conclusions
The CpCDV, which is a new pathogen to cucurbit crops, is now becoming a real threat to the region. The genetic characterization of both viruses is an essential move to know the information needed and required for taking the right measures for the control of different pathogens and their potential vectors, as well as the development of breeding programs so that farmers can befit from the introduction of a resistance cultivar.
Author Contributions
Conceptualization, N.R. and A.T.; methodology, N.R.; software, N.R. and S.E.; validation, A.T, Z.B., R.L. and M.B.; writing—original draft preparation, N.R. and M.D.; writing—review and editing, R.L., Z.B. and E.A.B.; supervision, A.T. and M.B.; funding acquisition, A.T. and R.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data used for the analyses in this study are available within the article.
Acknowledgments
This research was financially supported by the Phytopathology Unit of the Department of Plant Protection (ENA-Meknès). The first author wishes to acknowledge the technical staff of “Société Prograines” who assisted us during the sampling for this study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Marwal, A.; Sahu, A.K.; Gaur, R.K. Transmission and Host Interaction of Geminivirus in Weeds; Academic Press: Cambridge, MA, USA, 2014; pp. 143–161. ISBN 9780124115842. [Google Scholar]
- Briddon, R.W.; Akbar, F.; Iqbal, Z.; Amrao, L.; Amin, I.; Saeed, M.; Mansoor, S. Effects of genetic changes to the begomovirus/betasatellite complex causing cotton leaf curl disease in South Asia post-resistance breaking. Virus Res. 2014, 186, 114–119. [Google Scholar] [CrossRef] [PubMed]
- Padidam, R.; Beachy, R.N.; Fauquet, C.M.; Padidam, M.; Beachy, R.N.; Fauquet, C.M.; Jolla, L. Tomato leaf curl geminivirus from India has a bipartite genome and coat protein is not essential for infectivity. J. Gen. Virol. 1995, 76, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Moriones, E.; Praveen, S.; Chakraborty, S. Tomato Leaf Curl New Delhi Virus: An Emerging Virus Complex Threatening Vegetable and Fiber Crops. Viruses 2017, 9, 264. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, L.; Simon, A.; Velasco, L.; Janssen, D. Biological characterization of Tomato leaf curl New Delhi virus from Spain. Plant Pathol. 2016, 66, 376–382. [Google Scholar] [CrossRef]
- Radouane, N.; Ezrari, S.; Belabess, Z.; Tahiri, A.; Tahzima, R.; Massart, S.; Jijakli, H.; Benjelloun, M.; Lahlali, R. Viruses of cucurbit crops: Current status in the Mediterranean Region. Phytopathol. Mediterr. 2021, 60, 493–519. [Google Scholar] [CrossRef]
- López, C.; Ferriol, M.; Picó, M.B. Mechanical transmission of Tomato leaf curl New Delhi virus to cucurbit germplasm: Selection of tolerance sources in Cucumis melo. Euphytica 2015, 204, 679–691. [Google Scholar] [CrossRef]
- Mnari-Hattab, M.; Zammouri, S.; Belkadhi, M.S.; Bellon Doña, D.; ben Nahia, E.; Hajlaoui, M.R. First report of Tomato leaf curl New Delhi virus infecting cucurbits in Tunisia. New Dis. Rep. 2015, 31, 21. [Google Scholar] [CrossRef]
- Panno, S.; Iacono, G.; Davino, M.; Marchione, S.; Zappardo, V.; Bella, P.; Tomassoli, L.; Accotto, G.P.; Davino, S. First report of Tomato leaf curl New Delhi virus affecting zucchini squash in an important horticultural area of southern Italy. New Dis. Rep. 2016, 33, 6. [Google Scholar] [CrossRef]
- Radouane, N.; Tahiri, A.; El Ghadraoui, L.; Al Figuigui, J.; Lahlali, R. First report of Tomato Leaf Curl New Delhi virus in Morocco. New Dis. Rep. 2018, 37, 2. [Google Scholar] [CrossRef]
- Horn, N.M.; Reddy, S.V.; Roberts, I.M.; Reddy, D.V.R. Chickpea chlorotic dwarf virus, a new leafhopper-transmitted geminivirus of chickpea in India. Ann. Appl. Biol. 1993, 122, 467–479. [Google Scholar] [CrossRef]
- Kraberger, S.; Harkins, G.W.; Kumari, S.G.; Thomas, J.E.; Schwinghamer, M.W.; Sharman, M.; Collings, D.A.; Briddon, R.W.; Martin, D.P.; Varsani, A. Evidence that dicot-infecting mastreviruses are particularly prone to inter-species recombination and have likely been circulating in Australia for longer than in Africa and the Middle East. Virology 2013, 444, 282–291. [Google Scholar] [CrossRef] [PubMed]
- Kraberger, S.; Mumtaz, H.; Claverie, S.; Martin, D.P.; Briddon, R.W.; Varsani, A. Identification of an Australian-like dicot-infecting mastrevirus in Pakistan. Arch. Virol. 2015, 160, 825–830. [Google Scholar] [CrossRef] [PubMed]
- Zaagueri, T.; Mnari-Hattab, M.; Zammouri, S.; Hajlaoui, M.R.; Accotto, G.P.; Vaira, A.M. First report of chickpea chlorotic dwarf virus in watermelon (Citrullus lanatus) in Tunisia. Plant Dis. 2017, 101, 392. [Google Scholar] [CrossRef]
- Kanakala, S.; Kuria, P. Chickpea chlorotic dwarf virus: An emerging monopartite dicot infecting mastrevirus. Viruses 2019, 11, 5. [Google Scholar] [CrossRef] [PubMed]
- Radouane, N.; Ezrari, S.; Accotto, G.P.; Benjelloun, M.; Lahlali, R.; Tahiri, A.; Vaira, A.M. First report of Chickpea chlorotic dwarf virus in watermelon (Citrullus lanatus) in Morocco. New Dis. Rep. 2019, 39, 4404. [Google Scholar] [CrossRef]
- Muhire, B.; Martin, D.P.; Brown, J.K.; Navas-Castillo, J.; Moriones, E.; Zerbini, F.M.; Rivera-Bustamante, R.; Malathi, V.G.; Briddon, R.W.; Varsani, A. A genome-wide pairwise-identity-based proposal for the classification of viruses in the genus Mastrevirus (family Geminiviridae). Arch. Virol. 2013, 158, 1411–1424. [Google Scholar] [CrossRef]
- Doyle, J.; Doyle, J. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Radouane, N.; Ermadi, S.; Ezrari, S.; Al Figuigui, J.; Benjelloune, M.; Tahiri, A.; Lahlali, R. Occurrence and distribution of viruses infecting Zucchini and Watermelon in Morocco. Arch. Phytopathol. Plant Prot. 2020, 54, 375–387. [Google Scholar] [CrossRef]
- Zaagueri, T.; Miozzi, L.; Mnari-Hattab, M.; Noris, E.; Accotto, G.P.; Vaira, A.M. Deep sequencing data and infectivity assays indicate that chickpea chlorotic dwarf virus is the etiological agent of the “hard fruit syndrome” of watermelon. Viruses 2017, 9, 311. [Google Scholar] [CrossRef]
- Hall, T. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Stecher, G.; Tamura, K.; Kumar, S. Molecular Evolutionary Genetics Analysis (MEGA) for macOS. Mol. Biol. Evol. 2020, 37, 1237–1239. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.P.; Murrell, B.; Golden, M.; Khoosal, A.; Muhire, B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015, 1, vev003. [Google Scholar] [CrossRef] [PubMed]
- Fortes, I.M.; Sánchez-Campos, S.; Fiallo-Olivé, E.; Díaz-Pendón, J.A.; Navas-Castillo, J.; Moriones, E. A novel strain of tomato leaf curl New Delhi virus has spread to the Mediterranean basin. Viruses 2016, 8, 307. [Google Scholar] [CrossRef]
- Gilbertson, R.L.; Batuman, O.; Webster, C.G.; Adkins, S. Role of the Insect Supervectors Bemisia tabaci and Frankliniella occidentalis in the Emergence and Global Spread of Plant Viruses. Annu. Rev. Virol. 2015, 2, 67–93. [Google Scholar] [CrossRef]
- Fahmy, I.F.; Taha, O.; El-Ashry, A.N. First genome analysis and molecular characterization of Chickpea chlorotic dwarf virus Egyptian isolate infecting squash. VirusDisease 2015, 26, 33–41. [Google Scholar] [CrossRef]
- Horn, N.M.; Makkouk, K.M.; Kumari, S.G.; Van den Heuvel, J.F.J.M.; Reddy, D.V.R. Survey of chickpea (Cicer arientinum L.) for chickpea stunt disease and associated viruses in Syria, Turkey and Lebanon. Phytopathol. Mediterr. 1995, 34, 192–198. [Google Scholar]
- Ruiz, M.L.; Simón, A.; Velasco, L.; García, M.C.; Janssen, D. First Report of Tomato leaf curl New Delhi virus Infecting Tomato in Spain. Plant Dis. 2015, 99, 894. [Google Scholar] [CrossRef]
- Zammouri, S.; Zaagueri, T.; Eddouzi, J.; Belkhadhi, M.S.; Hajlaoui, M.R.; Mnari-Hattab, M. First report of tomato leaf curl New Delhi virus on tomato crop in Tunisia. J. Plant Pathol. 2017, 99, 813. [Google Scholar] [CrossRef]
- Kil, E.J.; Thi, T.; Vo, B.; Fadhila, C.; Ho, P.T.; Lal, A.; Bich Vo, T.T.; Fadhila, C.; Thi Ho, P.; Lal, A.; et al. Seed Transmission of Tomato Leaf Curl New Delhi Virus from Zucchini Squash in Italy. Plants 2020, 9, 563. [Google Scholar] [CrossRef]
- Panno, S.; Caruso, A.G.; Troiano, E.; Luigi, M.; Manglli, A.; Vatrano, T.; Iacono, G.; Marchione, S.; Bertin, S.; Tomassoli, L.; et al. Emergence of tomato leaf curl New Delhi virus in Italy: Estimation of incidence and genetic diversity. Plant Pathol. 2019, 68, 601–608. [Google Scholar] [CrossRef]
- Juárez, M.; Rabádan, M.P.; Martínez, L.D.; Tayahi, M.; Grande-Pérez, A.; Gómez, P. Natural hosts and genetic diversity of the emerging tomato leaf curl New Delhi virus in Spain. Front. Microbiol. 2019, 10, 140. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).