Tracing Superior Late-Leafing Genotypes of Persian Walnut for Managing Late-Spring Frost in Walnut Orchards
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
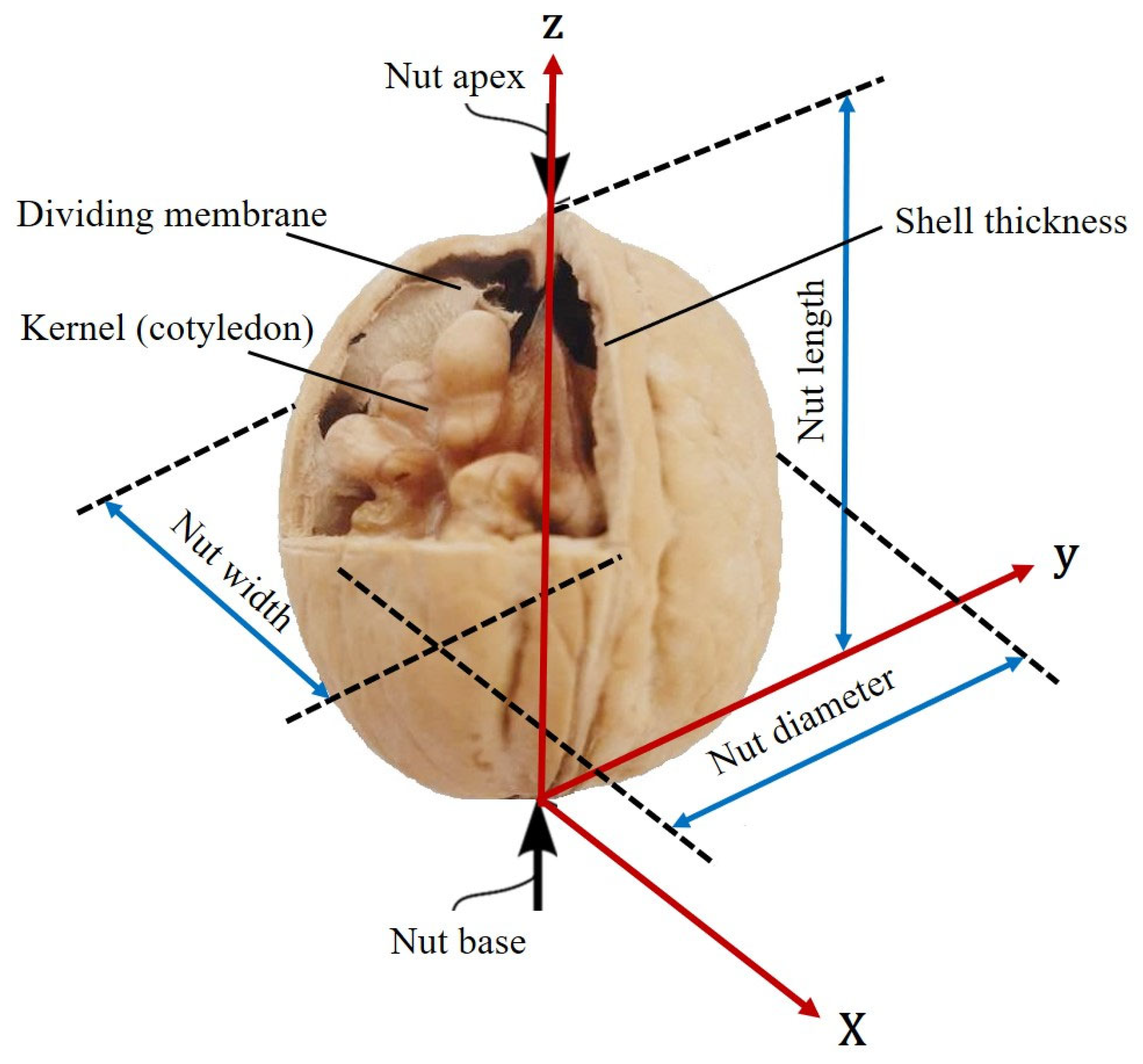
2.2. Pomological and Phenological Traits
2.3. Taste Panel
2.4. Grafting of Superior Genotypes
2.5. Statistical Analysis
3. Results and Discussion
3.1. Description of Genotype Characteristics
3.2. Correlations between Traits
3.3. Multivariate Analysis
3.4. Cluster Analysis (CA)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hoban, S.; Archer, F.I.; Bertola, L.D.; Bragg, J.G.; Breed, M.F.; Bruford, M.W.; Hunter, M.E. Global genetic diversity status and trends: Towards a suite of Essential Biodiversity Variables (EBVs) for genetic composition. Biol. Rev. 2022, 97, 1511–1538. [Google Scholar] [CrossRef] [PubMed]
- Shamlu, F.; Rezaei, M.; Lawson, S.; Ebrahimi, A.; Biabani, A.; Khan-Ahmadi, A. Genetic diversity of superior Persian walnut genotypes in Azadshahr, Iran. Physiol. Mol. Biol. Plants 2018, 24, 939–949. [Google Scholar] [CrossRef]
- Drepper, B.; Bamps, B.; Gobin, A.; Orshoven, J.V. Strategies for managing spring frost risks in orchards: Effectiveness and conditionality—A systematic review protocol. Environ. Evid. 2021, 10, 32. [Google Scholar] [CrossRef]
- Fallah, M.; Vahdati, K.; Hasani, D.; Rasouli, M.; Sarikhani, S. Breeding of Persian walnut: Aiming to introduce late-leafing and early-harvesting varieties by targeted hybridization. Sci. Hortic. 2022, 295, 110885. [Google Scholar] [CrossRef]
- Akca, Y.; Ozongun, S. Selection of late leafing, late flowering, laterally fruitful walnut (Juglans regia) types in Turkey. N. Z. J. Crop Hortic. Sci. 2004, 32, 337–342. [Google Scholar] [CrossRef]
- Khadivi-Khub, A.; Montazeran, A.; Rezaei, M.; Ebrahimi, A. The pomological characterization of walnut (Juglans regia L.) to select the superior genotypes—An opportunity for genetic improvement. Sci. Hortic. 2019, 248, 29–33. [Google Scholar] [CrossRef]
- Sarikhani, S.; Vahdati, K.; Ligterink, W. Biochemical properties of superior Persian walnut genotypes originated from southwest of Iran. Int. J. Hortic. Sci. Technol. 2021, 8, 13–24. [Google Scholar]
- Van Nocker, S.; Gardiner, S.E. Breeding better cultivars, faster: Applications of new technologies for the rapid deployment of superior horticultural tree crops. Hortic. Res. 2014, 1, 14022. [Google Scholar] [CrossRef] [Green Version]
- Ebrahimi, A.; Khadivi-Khub, A.; Nosrati, Z.; Karimi, R. Identification of superior walnut (Juglans regia) genotypes with late leafing and high kernel quality in Iran. Sci. Hortic. 2015, 193, 195–201. [Google Scholar] [CrossRef]
- Vahdati, K.; Mohseni Pourtaklu, S.; Karimi, R.; Barzehkar, R.; Amiri, R.; Mozaffari, M.; Woeste, K. Genetic diversity and gene flow of some Persian walnut populations in southeast of Iran revealed by SSR markers. Plant Syst. Evol. 2015, 301, 691–699. [Google Scholar] [CrossRef]
- Rigby, J.R.; Porporato, A. Spring frost risk in a changing climate. Geophys. Res. Lett. 2008, 35, 12. [Google Scholar] [CrossRef]
- Liu, J.; Sherif, S.M. Combating spring frost with ethylene. Front. Plant Sci. 2019, 10, 1408. [Google Scholar] [CrossRef] [Green Version]
- Kazemi, N.; Sharifzadeh, M.; Ahmadvand, M. Protecting walnut orchards against frost: A test of extended theory of planned behavior. WCAS 2018, 10, 709–722. [Google Scholar] [CrossRef]
- Vahdati, K.; Arab, M.M.; Sarikhani, S.; Sadat-Hosseini, M.; Leslie, C.A.; Brown, P.J. Advances in Persian walnut (Juglans regia L.) breeding strategies. In Advances in Plant Breeding Strategies: Nut Beverage Crops; Al-Khayri, J., Jain, S., Johnson, D., Eds.; Springer: Cham, Switzerland, 2019; pp. 401–472. [Google Scholar]
- Atefi, J. Evaluation of Walnut Genotypes in Iran. Acta Hortic. 1993, 311, 24–33. [Google Scholar] [CrossRef]
- Hassani, D.; Sarikhani, S.; Dastjerdi, R.; Mahmoudi, R.; Soleimani, A.; Vahdati, K. Situation and recent trends on cultivation and breeding of Persian walnut in Iran. Sci. Hortic. 2020, 270, 109369. [Google Scholar] [CrossRef]
- Hassani, D.; Mozaffari, M.R.; Soleimani, S.; Dastjerdi, R.; Rezaee, R.; Keshavarzi, M.; Vahdati, K.; Fahadan, A.; Atefi, J. Four New Persian Walnut Cultivars of Iran: Persia, Caspian, Chaldoran, and Alvand. Hortic. Sci. 2020, 55, 1162–1163. [Google Scholar] [CrossRef]
- Hajinia, Z.; Sarikhani, S.; Vahdati, K. Exploring low-chill genotypes of Persian walnut (Juglans regia L.) in west of Iran. Genet. Resour. Crop Evol. 2021, 68, 2325–2336. [Google Scholar] [CrossRef]
- Mahmoodi, R.; Dadpour, M.R.; Hassani, D.; Zeinalabedini, M.; Vendramin, E.; Micali, S.; Nahandi, F.Z. Development of a core collection in Iranian walnut (Juglans regia L.) germplasm using the phenotypic diversity. Sci. Hortic. 2019, 249, 439–448. [Google Scholar] [CrossRef]
- Khorami, S.S.; Arzani, K.; Karimzadeh, G.; Shojaeiyan, A.; Ligterink, W. Genome size: A novel predictor of nut weight and nut size of walnut trees. HortScience 2018, 53, 275–282. [Google Scholar] [CrossRef] [Green Version]
- Sutyemez, M.; Bükücü, Ş.B.; Özcan, A. ‘Helete Güneşi’, a New Walnut Cultivar with Late Leafing, Early Harvest Date, and Superior Nut Traits. Agriculture 2021, 11, 991. [Google Scholar] [CrossRef]
- Akca, Y.; Yuldaşulu, Y.B.; Murad, E.; Vahdati, K. Exploring of walnut genetic resources in Kazakhstan and evaluation of promising selections. Int. J. Hortic. Sci. Technol. 2020, 7, 93–102. [Google Scholar]
- IPGRI. Descriptors for Walnut (Juglans spp.); International Plant Genetic Resources Institute: Rome, Italy, 1994. [Google Scholar]
- Ingels, C.A.; McGranahan, G.H.; Noble, A.C. Sensory Evaluation of Selected Walnut Cultivars. HortScience 1990, 25, 1446–1447. [Google Scholar] [CrossRef]
- Zokaee-Khosroshahi, M.R.; Esna-Ashari, M.; Ershadi, A. Effect of exogenous putrescine on post-harvest life of strawberry (Fragaria ananassa Duch.) fruit, cultivar Selva. Sci. Hortic. 2007, 114, 27–32. [Google Scholar] [CrossRef]
- Kaiser, H.F. The varimax criterion for analytic rotation in factor analysis. Psychometrika 1958, 23, 187–200. [Google Scholar] [CrossRef]
- Rasouli, M.; Fattahi Moghadam, M.; Zamani, Z.; Imani, A.; Ebadi, A. A Study of the Phenotypic Diversity of some Almond Cultivars and Genotypes, using Morphological Traits. Int. J. Hortic. Sci. 2012, 43, 357–370. [Google Scholar]
- Janick, J.; Moore, J.N. (Eds.) Fruit Breeding, Tree and Tropical Fruits; John Wiley and Sons: Hoboken, NJ, USA, 1996; Volume 1. [Google Scholar]
- Aslantas, R. Identification of superior walnut (Juglans regia) genotypes in north-eastern Anatolia, Turkey. N. Z. J. Crop Hortic. Sci. 2006, 34, 231–237. [Google Scholar] [CrossRef]
- Arzani, K.; Mansouri-Ardakan, H.; Vezvaei, A.; Roozban, M.R. Morphological variation among Persian walnut (Juglans regia) genotypes from central Iran. N. Z. J. Crop Hortic. Sci. 2008, 36, 159–168. [Google Scholar] [CrossRef] [Green Version]
- Khadivi-Khub, A.; Ebrahimi, A.; Sheibani, F.; Esmaeili, A. Phenological and pomological characterization of Persian walnut to select promising trees. Euphytica 2015, 205, 557–567. [Google Scholar] [CrossRef]
- Bayazit, S.; Sumbul, A. Determination of fruit quality and fatty acid composition of Turkish walnut (Juglans regia) cultivars and genotypes grown in subtropical climate of eastern Mediterranean region. Int. J. Agric. Biol. 2012, 14, 419–424. [Google Scholar]
- Zeneli, G.; Kola, H.; Maxhum, D. Phenotypic variation in native walnut populations of Northern Albania. Sci. Hortic. 2005, 105, 91–100. [Google Scholar] [CrossRef]
- Korac, M.; Cerovic, S.; Golosin, B.; Miletic, R. Collecting, evaluation and utilization of walnut (Juglans regia L.) in Yugoslavia. Bull. Des Ressour. Phytogenet. (IPGRI/FAO) Not. De Recur. Fitogeneticos (IPGRI/FAO) 1997, 111, 72–74. [Google Scholar]
- Germain, E. Genetic improvement of the Persian walnut (Juglans regia L.). Acta Hortic. 1997, 442, 21–31. [Google Scholar] [CrossRef]
- Cosmulescu, S.; Botu, M. Walnut biodiversity in southwestern Romania resource for perspective cultivars. Pak. J. Bot. 2012, 44, 307–311. [Google Scholar]
- Bukucu, S.B.; Ozcan, A.; Sutyemez, M.; Yildirim, E. Determination in the phenological difference levels of seedlings of some walnut genotypes (Juglans regia L.). Appl. Ecol. Environ. Res. 2020, 8, 4807–4815. [Google Scholar] [CrossRef]
- Hansche, P.E.; Beres, V.; Forde, H.I. Estimates of quantitative genetic properties of walnut and their implications for cultivar improvement. Hortic. Sci. 1972, 97, 279–285. [Google Scholar]
- Ramos, D.E. Walnut Production Manual; UCANR Publications: Oakland, CA, USA, 1997; Volume 3373, pp. 19–23. [Google Scholar]
- Sharma, O.C.; Sharma, S.D. Correlation between nut and kernel characters of Persian walnut seedlings trees of Garsa valley in Kullu district of Himachal Pradesh. Acta Hortic. 2001, 544, 129–132. [Google Scholar] [CrossRef]
- Amiri, R.; Vahdati, K.; Mohsenipoor, S.; Mozaffari, M.R.; Leslie, C. Correlations between some horticultural traits in walnut. HortScience 2010, 45, 1690–1694. [Google Scholar] [CrossRef]
- Sarikhani Khorami, S.; Arzani, K.; Roozban, M.R. Correlations of certain high-heritability horticultural traits in Persian walnut (Juglans regia L.). Acta Hortic. 2014, 1050, 61–68. [Google Scholar] [CrossRef]
- Poggetti, L.; Ermacora, P.; Cipriani, G.; Pavan, F.; Testolin, R. Morphological and carpological variability of walnut germplasm (Juglans regia L.) collected in North Eastern Italy and selection of superior genotypes. Hortic. Sci. 2017, 225, 615–619. [Google Scholar] [CrossRef]
- Pop, F.I.; Cristina-Vicol, A.; Botu, M.; Andrei Raica, P.; Vahdati, K.; Pamfila, D. Relationships of walnut cultivars in a germplasm collection: Comparative analysis of phenotypic and molecular data. Sci. Hortic. 2013, 153, 124–135. [Google Scholar] [CrossRef]
- Rasouli, M.; Ershady, G.B. Study on genetic diversity of 33 walnut genotypes (Juglans regia) using morphological and pomological markers for introducing of superior genotypes. Res. Pomol. 2019, 3, 27–39. [Google Scholar]
- Khadivi-Khub, A.; Anjam, K. Morphological characterization of Prunus scoparia using multivariate analysis. Plant Syst. Evol. 2014, 300, 1361–1372. [Google Scholar] [CrossRef]
- Eskandari, S.; Hassani, D.; Abdi, A. Investigation on genetic diversity of Persian walnut and evaluation of promising genotypes. Acta Hortic. 2005, 705, 159–166. [Google Scholar] [CrossRef]






| Origin (City) | Latitude | Longitude | Altitude (m) | Annual Rainfall (mm) | High Temp Average (°C) | Low Temp Average (°C) | Annual Temp Average (°C) |
|---|---|---|---|---|---|---|---|
| Tuyserkan | 34°54′98″ | 48°45′37″ | 1780 | 355 | 17.75 | 7 | 12.35 |
| Razan | 35°39′14″ | 49°03′37″ | 1850 | 338 | 16.33 | 5.58 | 10.95 |
| Korzan | 34°54′48″ | 48°35′08″ | 1790 | 361 | 18.08 | 7 | 12.54 |
| Genotype | Origin (City) | Genotype | Origin (City) | Genotype | Origin (City) | Genotype | Origin (City) |
|---|---|---|---|---|---|---|---|
| TAL1 | Tuyserkan | TAL19 | Tuyserkan | TB11 | Tuyserkan | KG5 | Korzan |
| TAL2 | Tuyserkan | TAL20 | Tuyserkan | TAL12 | Tuyserkan | KG6 | Korzan |
| TAL3 | Tuyserkan | TAL21 | Tuyserkan | TAL13 | Tuyserkan | KG10 | Korzan |
| TAL4 | Tuyserkan | TAL22 | Tuyserkan | TB2 | Tuyserkan | KG16 | Korzan |
| TAL5 | Tuyserkan | TAL23 | Tuyserkan | TB3 | Tuyserkan | KG18 | Korzan |
| TAL6 | Tuyserkan | TAL24 | Tuyserkan | TB4 | Tuyserkan | KG19 | Korzan |
| TAL8 | Tuyserkan | RD1 | Razan | TB5 | Tuyserkan | KG26 | Korzan |
| TAL9 | Tuyserkan | RD2 | Razan | TB6 | Tuyserkan | KG27 | Korzan |
| TAL10 | Tuyserkan | RD3 | Razan | KG1 | Korzan | KG70 | Korzan |
| TAL11 | Tuyserkan | RD4 | Razan | KG2 | Korzan | KG111 | Korzan |
| TAL14 | Tuyserkan | RDGH5 | Razan | KG3 | Korzan | KG373 | Korzan |
| TAL15 | Tuyserkan | TB1 | Tuyserkan | KG4 | Korzan |
| Trait | Abbr. | Unit | Min. | Max. | Mean | Std. Deviation | Variance | CV% |
|---|---|---|---|---|---|---|---|---|
| Nut weight | NWT | g | 7.15 | 21.05 | 13.01 | 2.54 | 6.48 | 11.05 |
| Nut length | NL | mm | 32.59 | 51.13 | 38.58 | 3.74 | 14.03 | 15.35 |
| Nut width | NWI | mm | 24.79 | 41.20 | 33.98 | 2.48 | 6.17 | 13.66 |
| Nut diameter | ND | mm | 25.00 | 41.73 | 34.51 | 2.98 | 8.93 | 14.27 |
| Kernel weight | KW | g | 3.40 | 10.84 | 6.44 | 1.33 | 1.77 | 12.64 |
| Kernel percentage | KP | % | 30.94 | 69.75 | 49.69 | 5.23 | 27.40 | 7.30 |
| Shell thickness | STH | mm | 0.80 | 2.20 | 1.29 | 0.26 | 0.07 | 12.97 |
| Packing tissue thickness | PTT | mm | 0.04 | 0.80 | 0.19 | 0.12 | 0.01 | 49.93 |
| Shape of nut base | SNB | Code | 3.00 | 9.00 | 7.56 | 1.58 | 2.50 | 11.17 |
| Ease in removal of kernel halves | ERKH | Code | 1.00 | 6.00 | 2.66 | 1.54 | 2.39 | 17.51 |
| Lateral Bearing | LB | % | 50 | 100 | 82.18 | 12.32 | 151.9 | 4.63 |
| Leafing date | LD | Day | 0 | 40 | 25.44 | 9.12 | 83.20 | 3.22 |
| Kernel Brittleness | KB | Code | 4 | 10 | 8.81 | 1.36 | 1.87 | 5.69 |
| Kernel Taste | KT | Code | 4 | 10 | 8.42 | 1.60 | 2.56 | 3.34 |
| Genotype | NWT (g) | KW (g) | KP (%) | NL (mm) | NWI (mm) | ND (mm) | STH (mm) | PTT (mm) |
|---|---|---|---|---|---|---|---|---|
| TAL8 | 12.76 ± 0.82 | 6.53 ± 0.41 | 51.18 ± 1.44 | 41.42 ± 0.79 | 34.17 ± 0.58 | 33.87 ± 0.46 | 1.08 ± 0.21 | 0.21 ± 0.07 |
| TAL9 | 14.47 ± 2.34 | 7.79 ± 0.97 | 53.83 ± 2.29 | 43.90 ± 0.69 | 35.06 ± 1.32 | 37.94 ± 1.01 | 1.20 ± 0.05 | 0.12 ± 0.02 |
| TAL10 | 14.85 ± 1.33 | 7.84 ± 0.87 | 52.80 ± 1.89 | 38.58 ± 1.09 | 34.71 ± 1.45 | 35.20 ± 1.35 | 1.10 ± 0.9 | 0.24 ± 0.19 |
| TAL14 | 16.82 ± 1.29 | 8.15 ± 1.40 | 48.45 ± 5.36 | 37.58 ± 2.92 | 38.77 ± 1.40 | 40.19 ± 1.09 | 1.13 ± 0.24 | 0.18 ± 0.09 |
| TAL19 | 15.72 ± 1.38 | 8.07 ± 0.74 | 51.37 ± 1.62 | 37.32 ± 1.23 | 34.76 ± 0.75 | 36.44 ± 1.08 | 1.23 ± 0.03 | 0.14 ± 0.02 |
| TAL22 | 15.84 ± 0.56 | 7.98 ± 0.51 | 50.39 ± 1.45 | 38.02 ± 0.46 | 36.60 ± 0.38 | 38.36 ± 0.95 | 1.19 ± 0.08 | 0.11 ± 0.03 |
| TB2 | 12.77 ± 1.00 | 6.87 ± 0.44 | 53.83 ± 1.89 | 40.28 ± 3.18 | 34.23 ± 0.87 | 33.38 ± 0.67 | 1.06 ± 0.12 | 0.13 ± 0.02 |
| TB4 | 13.70 ± 1.52 | 7.56 ± 1.09 | 55.20 ± 2.82 | 46.74 ± 1.88 | 33.48 ± 1.17 | 32.80 ± 1.44 | 1.13 ± 0.06 | 0.14 ± 0.05 |
| TB6 | 12.52 ± 1.30 | 6.79 ± 0.70 | 54.24 ± 1.27 | 37.71 ± 2.12 | 35.06 ± 1.01 | 33.49 ± 1.35 | 1.14 ± 0.21 | 0.09 ± 0.01 |
| RDGH5 | 12.91 ± 0.30 | 6.55 ± 0.69 | 50.71 ± 4.86 | 39.91 ± 1.72 | 31.82 ± 0.96 | 32.63 ± 0.71 | 1.25 ± 0.13 | 0.10 ± 0.03 |
| Genotype | SNB | ERKH | LB | LD | KB | KT | SS | KC |
| TAL8 | 9 | 2 | 100 | 27 (11–April) | 10 | 10 | Triangular | Extra light |
| TAL9 | 7 | 3 | 80 | 28 (12–April) | 9 | 8 | Broad elliptic | Extra light |
| TAL10 | 9 | 1 | 100 | 35 (19–April) | 10 | 10 | Round | Extra light |
| TAL14 | 7 | 3 | 80 | 25 (9–April) | 8 | 9 | Short trapezoid | Light |
| TAL19 | 9 | 1 | 80 | 25 (9–April) | 10 | 10 | Round | Extra light |
| TAL22 | 8 | 2 | 80 | 35 (19–April) | 10 | 10 | Round | Light |
| TB2 | 9 | 1 | 75 | 34 (18–April) | 10 | 10 | Elliptic | Extra light |
| TB4 | 9 | 1 | 90 | 40 (24–April) | 10 | 9 | Elliptic | Light |
| TB6 | 8 | 1 | 85 | 36 (20–April) | 10 | 10 | Short trapezoid | Extra light |
| RDGH5 | 9 | 1 | 80 | 30 (14–April) | 9 | 10 | Broad elliptic | Light |
| Trait | Factor | ||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | |
| Nut weight (NWT) | 0.842 | 0.398 | −0.043 | 0.021 | 0.034 |
| Nut length (NL) | 0.53 | 0.245 | 0.308 | −0.198 | −0.164 |
| Nut width (NWI) | 0.828 | 0.2 | −0.206 | −0.080 | 0.054 |
| Nut diameter (ND) | 0.792 | 0.281 | −0.231 | −0.118 | 0.117 |
| Kernel weight (KW) | 0.894 | −0.019 | −0.120 | −0.153 | −0.021 |
| Kernel percentage (KP) | 0.152 | −0.813 | −0.166 | −0.334 | −0.103 |
| Shell thickness (STT) | −0.178 | 0.733 | 0.196 | 0.4 | −0.056 |
| Packing tissue thickness (PTT) | −0.242 | 0.488 | 0.421 | −0.085 | 0.221 |
| Shape of nut base (SNB) | 0.318 | −0.029 | 0.102 | 0.315 | −0.768 |
| Ease of removal of kernel halves (ERKH) | −0.149 | 0.481 | −0.526 | −0.282 | 0.18 |
| Lateral Bearing (LB) | 0.043 | −0.126 | 0.722 | −0.395 | 0.232 |
| Leafing date (LD) | 0.583 | −0.128 | 0.485 | −0.028 | −0.059 |
| Kernel brittleness (KB) | 0.595 | −0.313 | 0.136 | 0.302 | 0.27 |
| Kernel Taste (KT) | 0.302 | −0.385 | −0.033 | 0.653 | 0.443 |
| Variability (%) | 32.1 | 17.07 | 10.81 | 8.68 | 7.54 |
| Cumulative (%) | 32.1 | 49.8 | 60.61 | 69.29 | 76.83 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fallah, M.; Rasouli, M.; Hassani, D.; Lawson, S.S.; Sarikhani, S.; Vahdati, K. Tracing Superior Late-Leafing Genotypes of Persian Walnut for Managing Late-Spring Frost in Walnut Orchards. Horticulturae 2022, 8, 1003. https://doi.org/10.3390/horticulturae8111003
Fallah M, Rasouli M, Hassani D, Lawson SS, Sarikhani S, Vahdati K. Tracing Superior Late-Leafing Genotypes of Persian Walnut for Managing Late-Spring Frost in Walnut Orchards. Horticulturae. 2022; 8(11):1003. https://doi.org/10.3390/horticulturae8111003
Chicago/Turabian StyleFallah, Mehdi, Mousa Rasouli, Darab Hassani, Shaneka S. Lawson, Saadat Sarikhani, and Kourosh Vahdati. 2022. "Tracing Superior Late-Leafing Genotypes of Persian Walnut for Managing Late-Spring Frost in Walnut Orchards" Horticulturae 8, no. 11: 1003. https://doi.org/10.3390/horticulturae8111003







