Analysis of Structure Variations and Expression Characteristics of DMP8 and DMP9 Genes in Brassicaceae
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Identification and Phylogenetic Analysis of DMP9 Homologous Genes
2.3. Transcriptome Sequencing
2.4. Conserved Domain and Gene Structure Analysis
2.5. RNA-Seq Data Analysis
3. Results
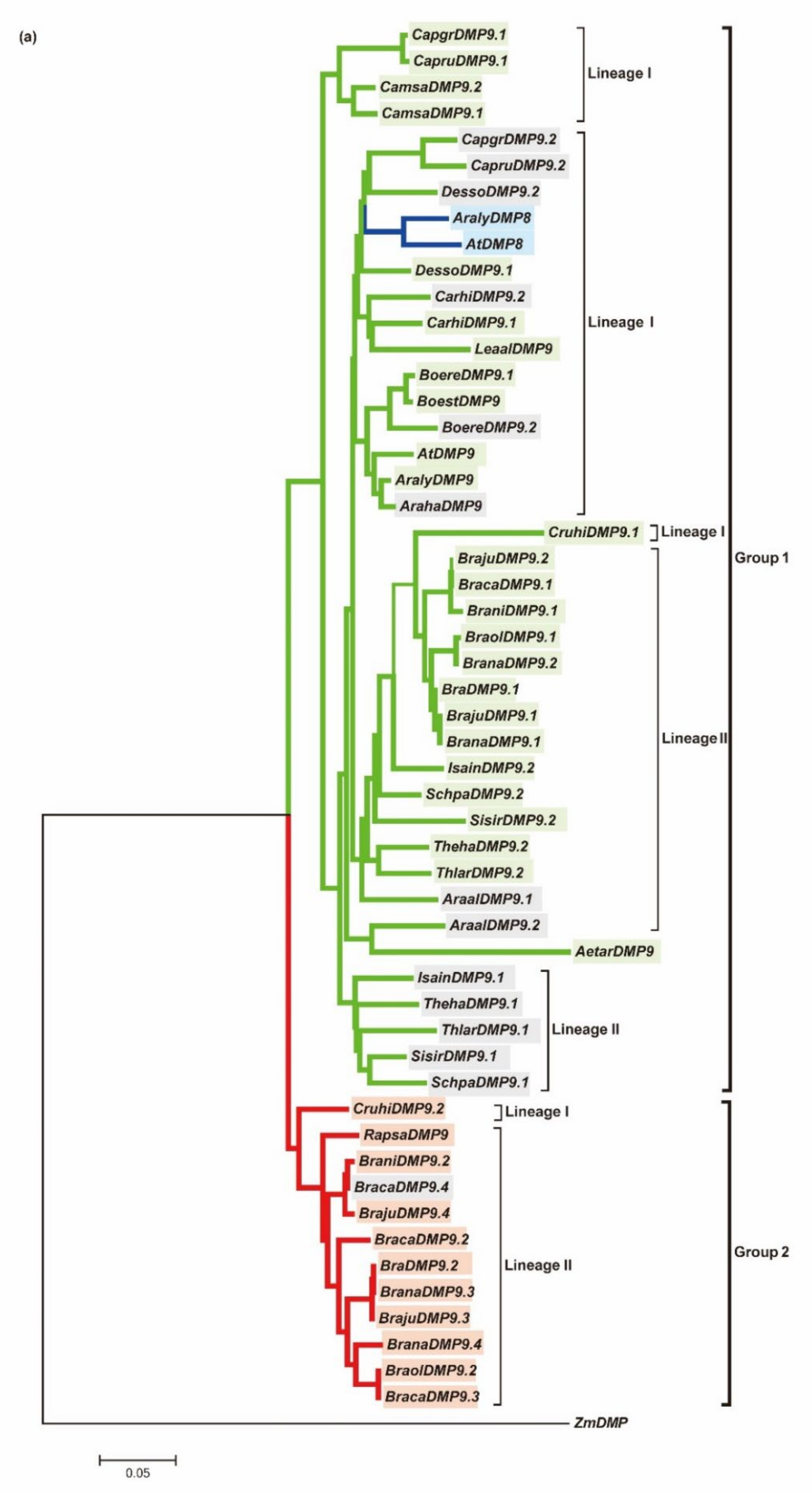
3.1. AtDMP8 and AtDMP9 Homologous Genes in 26 Brassicaceae Genomes
3.2. The Evolution of DMP9 Homologous Genes between Species with an Extra WGT
3.3. DMP9 Homologous Genes in B. rapa
3.4. Transcriptome Analysis of DMP9 Homologous Genes in B. rapa and B. oleracea
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jacquier, N.; Gilles, L.M.; Pyott, D.E.; Martinant, J.P.; Widiez, T. Puzzling out plant reproduction by haploid induction for innovations in plant breeding. Nat. Plants 2020, 6, 610–619. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Wu, J.; Wang, X.W. Genome triplication drove the diversification of Brassica plants. Hortic Res. 2014, 1, 14024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warwick, S.I.; Francis, A.; Al-Shehbaz, I.A. Brassicaceae: Species checklist and database on CD-Rom. Plant Syst. Evol. 2006, 259, 249–258. [Google Scholar] [CrossRef]
- Gilles, L.M.; Khaled, A.; Laffaire, J.B.; Chaignon, S.; Gendrot, G.; Laplaige, J.; Berges, H.; Beydon, G.; Bayle, V.; Barret, P.; et al. Loss of pollen-specific phospholipase NOT LIKE DAD triggers gynogenesis in maize. EMBO J. 2017, 36, 707–717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelliher, T.; Starr, D.; Richbourg, L.; Chintamanani, S.; Delzer, B.; Nuccio, M.L.; Green, J.; Chen, Z.Y.; McCuiston, J.; Wang, W.L.; et al. MATRILINEAL, a sperm-specific phospholipase, triggers maize haploid induction. Nature 2017, 542, 105. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.X.; Li, X.; Meng, D.X.; Zhong, Y.; Chen, C.; Dong, X.; Xu, X.W.; Chen, B.J.; Li, W.; Li, L.; et al. A 4-bp Insertion at ZmPLA1 Encoding a Putative Phospholipase A Generates Haploid Induction in Maize. Mol. Plant 2017, 10, 520–522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, L.; Zhang, Y.; Liu, C.X.; Liu, Y.B.; Wang, Y.L.; Liang, D.W.; Liu, J.T.; Sahoo, G.; Kelliher, T. OsMATL mutation induces haploid seed formation in indica rice. Nat. Plants 2018, 4, 530–533. [Google Scholar] [CrossRef]
- Liu, C.X.; Zhong, Y.; Qi, X.L.; Chen, M.; Liu, Z.K.; Chen, C.; Tian, X.L.; Li, J.L.; Jiao, Y.Y.; Wang, D.; et al. Extension of the in vivo haploid induction system from diploid maize to hexaploid wheat. Plant Biotechnol. J. 2020, 18, 316–318. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.Y.; Wang, K.; Jia, Z.M.; Gong, Q.; Lin, Z.S.; Du, L.P.; Pei, X.W.; Ye, X.G. Efficient induction of haploid plants in wheat by editing of TaMTL using an optimized Agrobacterium-mediated CRISPR system. J. Exp. Bot. 2020, 71, 1337–1349. [Google Scholar] [CrossRef]
- Zhong, Y.; Chen, B.J.; Li, M.R.; Wang, D.; Jiao, Y.Y.; Qi, X.L.; Wang, M.; Liu, Z.K.; Chen, C.; Wang, Y.W.; et al. A DMP-triggered in vivo maternal haploid induction system in the dicotyledonous Arabidopsis. Nat. Plants 2020, 6, 466–472. [Google Scholar] [CrossRef]
- Takahashi, T.; Mori, T.; Ueda, K.; Yamada, P.; Nagahara, S.; Higashiyama, T.; Sawada, H.; Igawa, T. The male gamete membrane protein DMP9/DAU2 is required for double fertilization in flowering plants. Development 2018, 145, dev170076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, N.; Xia, X.Z.; Jiang, T.; Li, L.L.; Zhang, P.C.; Niu, L.F.; Cheng, H.M.; Wang, K.J.; Lin, H. In planta haploid induction by genome editing of DMP in the model legume Medicago truncatula. Plant Biotechnol. J. 2022, 20, 22–24. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Chen, B.J.; Wang, D.; Zhu, X.J.; Li, M.R.; Zhang, J.Z.; Chen, M.; Wang, M.; Riksen, T.; Liu, J.C.; et al. In vivo maternal haploid induction in tomato. Plant Biotechnol. J. 2022, 20, 250–252. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Wang, Y.; Chen, B.; Liu, J.; Wang, D.; Li, M.; Qi, X.; Liu, C.; Boutilier, K.; Chen, S. Establishment of a dmp based maternal haploid induction system for polyploid Brassica napus and Nicotiana tabacum. J. Integr. Plant Biol. 2022, 64, 1281–1294. [Google Scholar] [CrossRef]
- Zhao, X.; Yuan, K.; Liu, Y.; Zhang, N.; Yang, L.; Zhang, Y.; Wang, Y.; Ji, J.; Fang, Z.; Han, F.; et al. In vivo maternal haploid induction based on genome editing of DMP in Brassica oleracea. Plant Biotechnol. J. 2022, 20, 22–24. [Google Scholar] [CrossRef]
- Wu, J.; Liang, J.; Lin, R.; Cai, X.; Zhang, L.; Guo, X.; Wang, T.; Chen, H.; Wang, X. Investigation of Brassica and its relative genomes in the postgenomics era. Hortic. Res. 2022, 9, uhac182. [Google Scholar] [CrossRef]
- Zhu, B.; Liang, Z.; Zang, Y.; Zhu, Z.; Yang, J. Diversity of glucosinolates among common brassicaceae vegetables in China. Hortic. Plant J. 2022. [Google Scholar] [CrossRef]
- Franzke, A.; Lysak, M.A.; Al-Shehbaz, I.A.; Koch, M.A.; Mummenhoff, K. Cabbage family affairs: The evolutionary history of Brassicaceae. Trends Plant Sci. 2011, 16, 108–116. [Google Scholar] [CrossRef]
- Tang, H.B.; Wang, X.Y.; Bowers, J.E.; Ming, R.; Alam, M.; Paterson, A.H. Unraveling ancient hexaploidy through multiply-aligned angiosperm gene maps. Genome Res. 2008, 18, 1944–1954. [Google Scholar] [CrossRef] [Green Version]
- Carroll, S.B. Endless forms: The evolution of gene regulation and morphological diversity. Cell 2000, 101, 577–580. [Google Scholar] [CrossRef]
- Gu, Z.L.; Rifkin, S.A.; White, K.P.; Li, W.H. Duplicate genes increase gene expression diversity within and between species. Nat. Genet. 2004, 36, 577–579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lynch, M.; Conery, J.S. The evolutionary fate and consequences of duplicate genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, X.; Chang, L.C.; Zhang, T.T.; Chen, H.X.; Zhang, L.; Lin, R.M.; Liang, J.L.; Wu, J.; Freeling, M.; Wang, X.W. Impacts of allopolyploidization and structural variation on intraspecific diversification in Brassica rapa. Genome Biol. 2021, 22, 166–189. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Cheng, F.; Wu, J.; Fang, L.; Wang, X.W. Syntenic gene analysis between Brassica rapa and other Brassicaceae species. Front. Plant Sci. 2012, 3, 198. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Wang, T.; He, X.; Cai, X.; Lin, R.; Liang, J.; Wu, J.; King, G.; Wang, X. BRAD V3.0: An ungraded Brassicaceae database. Nucleic Acids Res. 2021, 50, D1432–D1441. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An Integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Kovaka, S.; Zimin, A.V.; Pertea, G.M.; Razaghi, R.; Salzberg, S.L.; Pertea, M. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 2019, 20, 1–13. [Google Scholar] [CrossRef]
- Lyons, E.; Pedersen, B.; Kane, J.; Alam, M.; Ming, R.; Tang, H.B.; Wang, X.Y.; Bowers, J.; Paterson, A.; Lisch, D.; et al. Finding and comparing syntenic regions among Arabidopsis and the outgroups papaya, poplar, and grape: CoGe with rosids. Plant Physiol. 2008, 148, 1772–1781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schranz, M.E.; Lysak, M.A.; Mitchell-Olds, T. The ABC’s of comparative genomics in the Brassicaceae: Building blocks of crucifer genomes. Trends Plant Sci. 2006, 11, 535–542. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.W.; Wang, H.Z.; Wang, J.; Sun, R.F.; Wu, J.; Liu, S.Y.; Bai, Y.Q.; Mun, J.H.; Bancroft, I.; Cheng, F.; et al. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 2011, 43, 1035–1157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edger, P.P.; Pires, J.C. Gene and genome duplications: The impact of dosage-sensitivity on the fate of nuclear genes. Chromosome Res. 2009, 17, 699–717. [Google Scholar] [CrossRef] [Green Version]
- Fang, L.; Cheng, F.; Wu, J.; Wang, X. The impact of genome triplication on tandem gene evolution in Brassica rapa. Front. Plant Sci. 2012, 3, 261. [Google Scholar] [CrossRef] [Green Version]
- Engel, W.; Hof, J.O.; Wolf, U. Gene duplication by polyploid evolution: The isoenzyme of the sorbitol dehydrogenase in herring- and salmon-like fishes (Isospondyli). Humangenetik 1970, 9, 157–163. [Google Scholar] [CrossRef]
- Ferris, S.D.; Whitt, G.S. Evolution of the differential regulation of duplicate genes after polyploidization. J. Mol. Evol. 1979, 12, 267–317. [Google Scholar] [CrossRef]
- Li, Y.; Li, D.; Xiao, Q.; Wang, H.; Wen, J.; Tu, J.; Shen, J.; Fu, T.; Yi, B. An in planta haploid induction system in Brassica napus. J. Integr. Plant Biol. 2022, 64, 1140–1144. [Google Scholar] [CrossRef]
- Kane, J.; Freeling, M.; Lyons, E. The Evolution of a High Copy Gene Array in Arabidopsis. J. Mol. Evol. 2010, 70, 531–544. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Xiong, H.; Zhao, P.; Sun, M. DMP8 and 9 regulate HAP2/GCS1 trafficking for the timely acquisition of sperm fusion competence. Proc. Natl. Acad. Sci. USA 2022, 45, 119. [Google Scholar] [CrossRef]
- Philipp, C.; Maria, L.; Stefanie, S. Gamete fusion is facilitated by two sperm cell-expressed DUF679 membrane proteins. Nat. Plant 2019, 5, 253–257. [Google Scholar] [CrossRef]
- Song, Y.; Guo, X.; Wu, Y.; Liang, J.; Lin, R.; Yan, Z.; Wang, X. An optimized protocol for detecting guard cell specific gene expression by in situ RT-PCR in Brassica rapa. Hortic. Plant J. 2022, 8, 311–318. [Google Scholar] [CrossRef]
- Chen, R.; Chang, L.; Cai, X.; Wu, J.; Lin, R.; Song, Y.; Wang, X. Development of InDel markers for Brassica rapa based on a Highp-resolution melting curve. Hortic. Plant J. 2021, 7, 31–37. [Google Scholar] [CrossRef]
- Kaul, S.; Koo, H.L.; Jenkins, J.; Rizzo, M.; Rooney, T.; Tallon, L.J.; Feldblyum, T.; Nierman, W.; Benito, M.I.; Lin, X.Y.; et al. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 2000, 408, 796–815. [Google Scholar] [CrossRef] [Green Version]
- Hu, T.T.; Pattyn, P.; Bakker, E.G.; Cao, J.; Cheng, J.F.; Clark, R.M.; Fahlgren, N.; Fawcett, J.A.; Grimwood, J.; Gundlach, H.; et al. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat. Genet. 2011, 43, 476–481. [Google Scholar] [CrossRef]
- Kliver, S.; Rayko, M.; Komissarov, A.; Bakin, E.; Zhernakova, D.; Prasad, K.; Rushworth, C.; Baskar, R.; Smetanin, D.; Schmutz, J.; et al. Assembly of the Boechera retrofracta genome and evolutionary analysis of apomixis-associated genes. Genes 2018, 9, 185. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Bi, C.; Tu, J.; Lu, Z. The complete mitochondrial genome sequence of Boechera stricta. Mitochondrial DNA B Resour. 2018, 3, 896–897. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.; Qiao, Q.; Novikova, P.Y.; Wang, Q.; Yue, J.; Guan, Y.; Ming, S.; Liu, T.; De, J.; Liu, Y.; et al. Genome of Crucihimalaya himalaica, a close relative of Arabidopsis, shows ecological adaptation to high altitude. Proc. Natl. Acad. Sci. USA 2019, 116, 7137–7146. [Google Scholar] [CrossRef] [Green Version]
- Slotte, T.; Hazzouri, K.M.; Agren, J.A.; Koenig, D.; Maumus, F.; Guo, Y.L.; Steige, K.; Platts, A.E.; Escobar, J.S.; Newman, L.K.; et al. The Capsella rubella genome and the genomic consequences of rapid mating system evolution. Nat. Genet. 2013, 45, 831–835. [Google Scholar] [CrossRef] [Green Version]
- Kagale, S.; Koh, C.; Nixon, J.; Bollina, V.; Clarke, W.E.; Tuteja, R.; Spillane, C.; Robinson, S.J.; Links, M.G.; Clarke, C.; et al. The emerging biofuel crop Camelina sativa retains a highly undifferentiated hexaploid genome structure. Nat. Commun. 2014, 5, 3706. [Google Scholar] [CrossRef]
- Akama, S.; Shimizu-Inatsugi, R.; Shimizu, K.K.; Sese, J. Genome-wide quantification of homeolog expression ratio revealed nonstochastic gene regulation in synthetic allopolyploid Arabidopsis. Nucleic Acids Res. 2014, 42, e46. [Google Scholar] [CrossRef] [PubMed]
- Gan, X.; Hay, A.; Kwantes, M.; Haberer, G.; Hallab, A.; Ioio, R.D.; Hofhuis, H.; Pieper, B.; Cartolano, M.; Neumann, U.; et al. The Cardamine hirsuta genome offers insight into the evolution of morphological diversity. Nat. Plants 2016, 2, 16167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haudry, A.; Platts, A.E.; Vello, E.; Hoen, D.R.; Leclercq, M.; Williamson, R.J.; Forczek, E.; Joly-Lopez, Z.; Steffen, J.G.; Hazzouri, K.M.; et al. An atlas of over 90,000 conserved noncoding sequences provides insight into crucifer regulatory regions. Nat. Genet. 2013, 45, 891–898. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Cai, X.; Wu, J.; Liu, M.; Grob, S.; Cheng, F.; Liang, J.; Cai, C.; Liu, Z.; Liu, B.; et al. Erratum: Author Correction: Improved Brassica rapa reference genome by single-molecule sequencing and chromosome conformation capture technologies. Hortic. Res. 2019, 6, 124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, S.; Liu, Y.; Yang, X.; Tong, C.; Edwards, D.; Parkin, I.A.; Zhao, M.; Ma, J.; Yu, J.; Huang, S.; et al. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 2014, 5, 3930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paritosh, K.; Pradhan, A.K.; Pental, D. A highly contiguous genome assembly of Brassica nigra (BB) and revised nomenclature for the pseudochromosomes. BMC Genom. 2020, 21, 887. [Google Scholar] [CrossRef]
- Yang, J.; Liu, D.; Wang, X.; Ji, C.; Cheng, F.; Liu, B.; Hu, Z.; Chen, S.; Pental, D.; Ju, Y.; et al. The genome sequence of allopolyploid Brassica juncea and analysis of differential homoeolog gene expression influencing selection. Nat. Genet. 2016, 48, 1225–1232. [Google Scholar] [CrossRef]
- Rousseau-Gueutin, M.; Belser, C.; Da Silva, C.; Richard, G.; Istace, B.; Cruaud, C.; Falentin, C.; Boideau, F.; Boutte, J.; Delourme, R.; et al. Long-read assembly of the Brassica napus reference genome Darmor-bzh. Gigascience 2020, 9, giaa137. [Google Scholar] [CrossRef]
- Song, X.; Wei, Y.; Xiao, D.; Gong, K.; Sun, P.; Ren, Y.; Yuan, J.; Wu, T.; Yang, Q.; Li, X.; et al. Brassica carinata genome characterization clarifies U’s triangle model of evolution and polyploidy in Brassica. Plant Physiol. 2021, 186, 388–406. [Google Scholar] [CrossRef]
- Kitashiba, H.; Li, F.; Hirakawa, H.; Kawanabe, T.; Zou, Z.; Hasegawa, Y.; Tonosaki, K.; Shirasawa, S.; Fukushima, A.; Yokoi, S.; et al. Draft sequences of the radish (Raphanus sativus L.) genome. DNA Res. 2014, 21, 481–490. [Google Scholar] [CrossRef]
- Kang, M.; Wu, H.; Yang, Q.; Huang, L.; Hu, Q.; Ma, T.; Li, Z.; Liu, J. A chromosome-scale genome assembly of Isatis indigotica, an important medicinal plant used in traditional Chinese medicine: An Isatis genome. Hortic. Res. 2020, 7, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Willing, E.M.; Rawat, V.; Mandakova, T.; Maumus, F.; James, G.V.; Nordstrom, K.J.; Becker, C.; Warthmann, N.; Chica, C.; Szarzynska, B.; et al. Genome expansion of Arabis alpina linked with retrotransposition and reduced symmetric DNA methylation. Nat. Plants 2015, 1, 14023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dorn, K.M.; Fankhauser, J.D.; Wyse, D.L.; Marks, M.D. A draft genome of field pennycress (Thlaspi arvense) provides tools for the domestication of a new winter biofuel crop. DNA Res. 2015, 22, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Jarvis, D.E.; Chen, H.; Beilstein, M.A.; Grimwood, J.; Jenkins, J.; Shu, S.; Prochnik, S.; Xin, M.; Ma, C.; et al. The Reference genome of the halophytic plant Eutrema salsugineum. Front. Plant Sci. 2013, 4, 46. [Google Scholar] [CrossRef] [PubMed]



| Sample | Name | Description | E-Value | Identity | Genomic Block | Accession |
|---|---|---|---|---|---|---|
| CCA | ssp. pekinensis | Chinese cabbage | 0 | 80.30% | S | CCADMP9.1 |
| 0 | 68.30% | S | CCADMP9.2 | |||
| 0 | 85.8% | R | CCADMP9.3 | |||
| CCB | ssp. pekinensis | Chinese cabbage | 0 | 91.00% | S | CCBDMP9.1 |
| 0 | 85.60% | R | CCBDMP9.2 | |||
| Chiifu # | ssp. pekinensis | Chinese cabbage | 0 | 91.00% | S | BraDMP9.1 |
| 0 | 85.80% | R | BraDMP9.2 | |||
| CXA | ssp. parachinensis | Caixin | 0 | 90.30% | S | CXADMP9.1 |
| 0 | 85.60% | R | CXADMP9.2 | |||
| CXB | ssp. parachinensis | Caixin | 0 | 90.30% | S | CXBDMP9.1 |
| 0 | 85.80% | R | CXBDMP9.2 | |||
| OIA | ssp. oleifera | Oil seeds | 0 | 91.00% | S | OIADMP9.1 |
| 0 | 85.80% | R | OIADMP9.2 | |||
| OIB | ssp. oleifera | Rapid cycling | 0 | 90.30% | S | OIBDMP9.1 |
| 0 | 90.30% | S | OIBDMP9.2 | |||
| 0 | 85.60% | R | OIBDMP9.3 | |||
| OIC | ssp. oleifera | Rapid cycling | 0 | 90.30% | S | OICDMP9.1 |
| 0 | 85.60% | R | OICDMP9.2 | |||
| Z1 | ssp. oleifera | Sarson type | 0 | 90.30% | S | Z1DMP9.1 |
| 0 | 85.60% | R | Z1DMP9.2 | |||
| TUA | ssp. rapa | Turnip | 0 | 90.70% | S | TUADMP9.1 |
| 0 | 85.80% | R | TUADMP9.2 | |||
| TUE | ssp. rapa | Turnip | 0 | 91.00% | S | TUEDMP9.1 |
| 0 | 85.60% | R | TUEDMP9.2 | |||
| PCA | ssp. chinensis | Pak choi | 0 | 90.30% | S | PCADMP9.1 |
| 0 | 85.80% | R | PCADMP9.2 | |||
| PCB | ssp. chinensis | Pak choi | 0 | 90.30% | S | PCBDMP9.1 |
| 0 | 85.70% | R | PCBDMP9.2 | |||
| WTC | ssp. narinosa | Wutacai | 0 | 91.00% | S | WTCDMP9.1 |
| 0 | 85.60% | R | WTCDMP9.2 | |||
| TCA | ssp. chinensis var.tai-tsai | Taicai | 0 | 91.00% | S | TCADMP9.1 |
| 0 | 85.80% | R | TCADMP9.2 | |||
| BRO | Broccolieto | Broccolleto | 0 | 68.30% | S | BroDMP9.1 |
| 0 | 90.70% | S | BroDMP9.2 | |||
| 0 | 85.80% | R | BroDMP9.3 | |||
| MIZ | ssp. nipposinica | Mizuna | 0 | 91.00% | S | MIZDMP9.1 |
| 0 | 85.70% | R | MIZDMP9.2 | |||
| TBA (WLD) | unknown | From Tibet, China | 0 | 90.70% | S | TBADMP9.1 |
| 0 | 85.80% | R | TBADMP9.2 | |||
| 0 | 85.60% | unknown | TBADMP9.3 * | |||
| 0 | 85.60% | unknown | TBADMP9.4 * |
| Gene Name | Chr | Gene ID | Start | End | Gene Block | Species | Varieties/ Subspecies |
|---|---|---|---|---|---|---|---|
| AtDMP9 | A05 | At5g39650 | 15875199 | 15876116 | S | A. thaliana | A. thaliana |
| BraDMP9.1 | A04 | BraA04g012030.3C | 9524183 | 9524908 | S | B. rapa | Chiifu |
| BraDMP9.2 | A03 | BraA03g003970.3C | 1691698 | 1692426 | R | B. rapa | Chiifu |
| CCADMP9.1 * | A04 | A04p12560.1_BraCCA | 8999489 | 9000040 | S | B. rapa | B9008 |
| CCADMP9.2 * | A04 | A04p12570.1_BraCCA | 9000509 | 9001147 | S | B. rapa | B9008 |
| CCADMP9.3 | A03 | A03p04040.1_BraCCA | 1746905 | 1747628 | R | B. rapa | B9008 |
| BolDMP9.1 | C04 | BolC04g044930.2J | 46361838 | 46362575 | S | B. oleracea | JZS |
| BolDMP9.2 | C03 | BolC03g004320.2J | 2118435 | 2119158 | R | B. oleracea | JZS |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, T.; Liang, J.; Cai, X.; Zhang, L.; Wu, J.; Wang, X. Analysis of Structure Variations and Expression Characteristics of DMP8 and DMP9 Genes in Brassicaceae. Horticulturae 2022, 8, 1095. https://doi.org/10.3390/horticulturae8111095
Zhang T, Liang J, Cai X, Zhang L, Wu J, Wang X. Analysis of Structure Variations and Expression Characteristics of DMP8 and DMP9 Genes in Brassicaceae. Horticulturae. 2022; 8(11):1095. https://doi.org/10.3390/horticulturae8111095
Chicago/Turabian StyleZhang, Tingting, Jianli Liang, Xu Cai, Lei Zhang, Jian Wu, and Xiaowu Wang. 2022. "Analysis of Structure Variations and Expression Characteristics of DMP8 and DMP9 Genes in Brassicaceae" Horticulturae 8, no. 11: 1095. https://doi.org/10.3390/horticulturae8111095
APA StyleZhang, T., Liang, J., Cai, X., Zhang, L., Wu, J., & Wang, X. (2022). Analysis of Structure Variations and Expression Characteristics of DMP8 and DMP9 Genes in Brassicaceae. Horticulturae, 8(11), 1095. https://doi.org/10.3390/horticulturae8111095









