Genome Wide Identification and Characterization of Apple WD40 Proteins and Expression Analysis in Response to ABA, Drought, and Low Temperature
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification of Apple MdWD40 Genes and Their Chromosomal Location
2.2. Classification and Phylogenetic Analysis of Apple WD40 Proteins
2.3. Exon/Intron Structure of Apple WD40 Genes
2.4. Synteny Analysis of Apple WD40 Genes
2.5. Plant Material and Treatments
2.6. Expression Analysis of WD40 Genes in Apple
3. Results
3.1. Genome-Wide Identification of WD40 Genes in Apple
3.2. Classification and Phylogenetic Analysis of MdWD40 Proteins
3.3. Amplification Pattern of WD40 Genes in Apple
3.4. Synteny Analysis of WD40 Genes in Apple and Arabidopsis
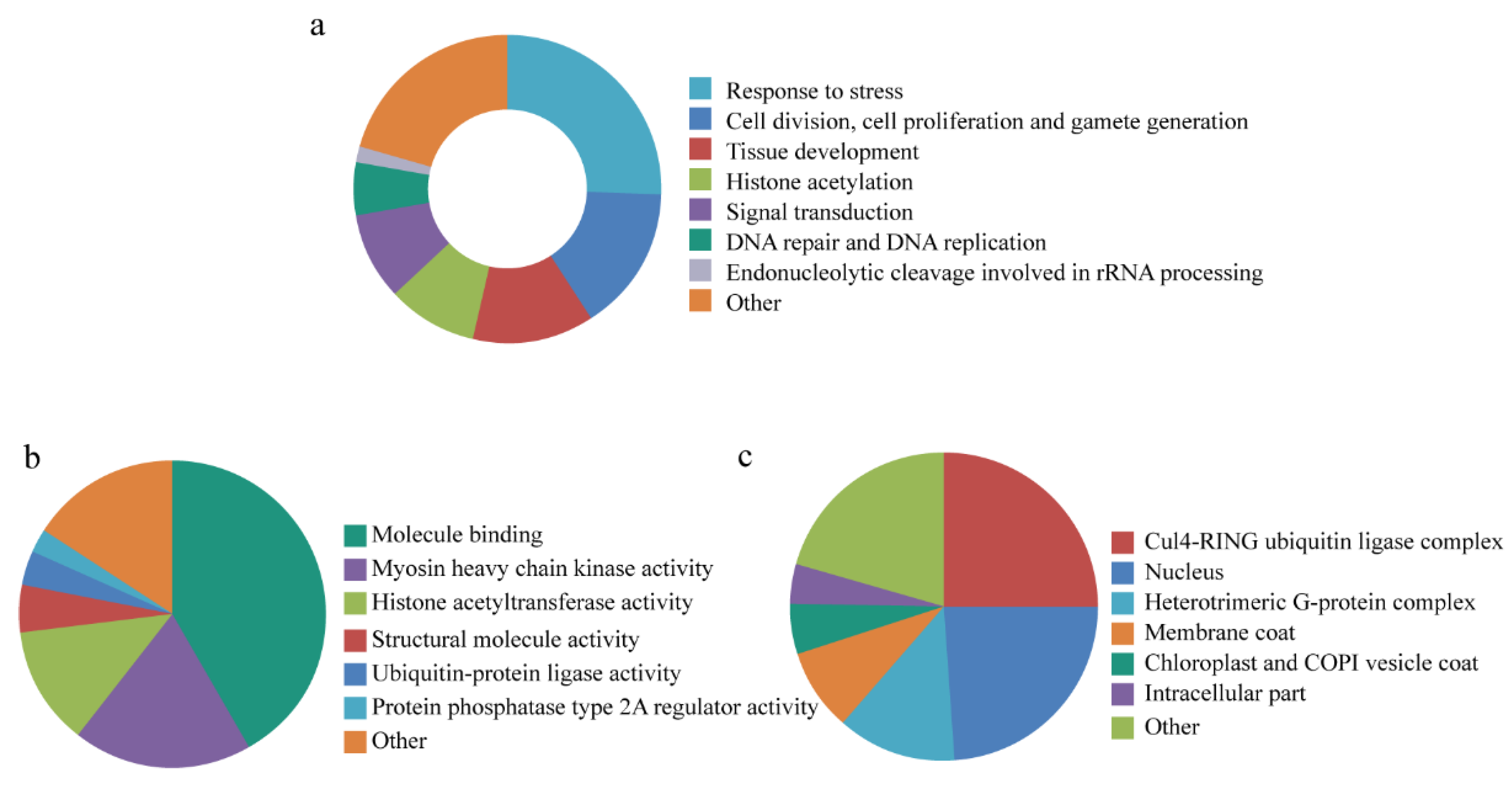
3.5. Gene Ontology Annotation
3.6. Organ Expression Patterns of WD40 Genes in Apple at Different Growth Stages
3.7. Response of WD40 Genes to ABA, Drought, and Low Temperature in Apple
4. Discussion
4.1. Identification, Classification, and Evolutionary Relationships of the WD40 Gene Family in Apple
4.2. Amplification of WD40 Genes in Apple
4.3. Collinearity Analysis of Apple WD40 Genes and Arabidopsis WD40 Genes
4.4. GO Analysis of Apple WD40 Genes
4.5. Apple WD40 Gene-Specific Expression and Response to Exogenous ABA, Drought, and Low Temperature
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Neer, E.; Schmidt, C.; Nambudripad, R.; Smith, T. The ancient regulatory-protein family of WD-repeat proteins. Nature 1994, 371, 297–300. [Google Scholar] [CrossRef] [PubMed]
- Wall, M.A.; Coleman, D.E.; Lee, E.; Iñiguez-Lluhi, J.A.; Posner, B.A.; Gilman, A.G.; Sprang, S.R. The structure of the G protein heterotrimer Giα1β1γ2. Cell 1995, 83, 1047–1058. [Google Scholar] [CrossRef] [Green Version]
- Lambright, D.G.; Sondek, J.; Bohm, A.; Skiba, N.P.; Hamm, H.E.; Sigler, P.B. The 2.0 A crystal structure of a heterotrimeric G protein. Nature 1996, 379, 311–319. [Google Scholar] [CrossRef]
- Xu, C.; Min, J. Structure and function of WD40 domain proteins. Protein Cell 2011, 2, 202–214. [Google Scholar] [CrossRef] [PubMed]
- Chothia, C.; Hubbard, T.; Brenner, S.; Barns, H.; Murzin, A. Protein folds in the all-β and all-α classes. Annu. Rev. Biophys. Biomol. Struct. 1997, 26, 597. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.F.; Gaitatzes, C.; Saxena, K.; Neer, E.J. The WD repeat: A common architecture for diverse functions. Trends Biochem. Sci. 1999, 24, 181. [Google Scholar] [CrossRef]
- Ouyang, Y.; Huang, X.; Lu, Z.; Yao, J. Genomic survey, expression profile and co-expression network analysis of OsWD40 family in rice. BMC Genom. 2012, 13, 100. [Google Scholar] [CrossRef] [Green Version]
- Jain, B.P.; Pandey, S. WD40 repeat proteins: Signalling scaffold with diverse functions. Protein J. 2018, 37, 391–406. [Google Scholar] [CrossRef]
- Van Nocker, S.; Ludwig, P. The WD-repeat protein superfamily in Arabidopsis: Conservation and divergence in structure and function. BMC Genom. 2003, 4, 50. [Google Scholar] [CrossRef] [Green Version]
- Li, H.J.; Liu, N.Y.; Shi, D.Q.; Liu, J.; Yang, W.C. YAO is a nucleolar WD40-repeat protein critical for embryogenesis and gametogenesis in Arabidopsis. BMC Plant Biol. 2010, 10, 169. [Google Scholar] [CrossRef] [Green Version]
- Caillaud, M.C.; Paganelli, L.; Lecomte, P.; Deslandes, L.; Quentin, M.; Pecrix, Y.; Bris, M.l.; Marfaing, N.; Abad, P.; Favery, B. Spindle assembly checkpoint protein dynamics reveal conserved and unsuspected roles in plant cell division. PLoS ONE 2009, 4, e6757. [Google Scholar] [CrossRef] [PubMed]
- Saedler, R.; Jakoby, M.; Marin, B.; Galiana-Jaime, E.; Hulskamp, M. The cell morphogenesis gene SPIRRIG in Arabidopsis encodes a WD-BEACH domain protein. Plant J. 2009, 59, 612. [Google Scholar] [CrossRef] [PubMed]
- Kevei, Z.; Baloban, M.; Da, I.O.; Tiricz, H.; Kroll, A.; Regulski, K.; Mergaert, P.; Kondorosi, E. Conserved CDC20 cell cycle functions are carried out by two of the five isoforms in Arabidopsis thaliana. PLoS ONE 2011, 6, e20618. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Gu, X.; He, Y. Establishment of the Winter-Annual Growth Habit via FRIGIDA-Mediated Histone Methylation at FLOWERING LOCUS C in Arabidopsis. Plant Cell 2009, 21, 1733–1746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, D.; Kong, N.; Gu, X.; He, Y. Arabidopsis COMPASS-Like Complexes Mediate Histone H3 Lysine-4 Trimethylation to Control Floral Transition and Plant Development. PLoS Genet. 2011, 7, e1001330. [Google Scholar] [CrossRef] [PubMed]
- Pazhouhandeh, M.; Molinier, J.; Berr, A.; Genschik, P. MSI4/FVE interacts with CUL4–DDB1 and a PRC2-like complex to control epigenetic regulation of flowering time in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 3430–3435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, K.I.; Hoshino, A. A WD40-repeat protein controls proanthocyanidin and phytomelanin pigmentation in the seed coats of the Japanese morning glory. J. Plant Physiol. 2012, 169, 523–528. [Google Scholar] [CrossRef]
- Xiong, Y.; Contento, A.L.; Bassham, D.C. AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant J. 2005, 42, 535–546. [Google Scholar] [CrossRef]
- Chen, Z.-H.; Jenkins, G.I.; Nimmo, H.G. Identification of an F-Box Protein that Negatively Regulates Pi Starvation Responses. Plant Cell Physiol. 2008, 49, 1902–1906. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.; Jeong, J.C.; Zhu, Y.; Sokolchik, I.; Miyazaki, S.; Hasegawa, P.M.; Bohnert, H.J.; Shi, H.; Yun, D.-J.; Bressan, R.A. Involvement of Arabidopsis HOS15 in histone deacetylation and cold tolerance. Proc. Natl. Acad. Sci. USA 2008, 105, 4945–4950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biedermann, S.; Hellmann, H. The DDB1a interacting proteins ATCSA-1 and DDB2 are critical factors for UV-B tolerance and genomic integrity in Arabidopsis thaliana. Plant J. 2010, 62, 404–415. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Brandizzi, F. AtIRE1A/AtIRE1B and AGB1 independently control two essential unfolded protein response pathways in Arabidopsis. Plant J. 2012, 69, 266–277. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Wang, Y.; Shen, J.; Yin, J.; Li, D.; Gao, Y.; Xu, W.; Liang, J. OsRACK1A, encodes a circadian clock-regulated WD40 protein, negatively affect salt tolerance in rice. Rice 2018, 11, 45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, W.; Zheng, S.; Yu, Z.; Gao, X.; Shen, R.; Lu, Y. WD40-REPEAT 5a represses root meristem growth by suppressing auxin synthesis through changes of nitric oxide accumulation in Arabidopsis. Plant J. 2018, 93, 883–893. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Y.; Zhu, W.; Chen, Y.; Ito, S.; Asami, T.; Wang, X. Brassinosteroids control root epidermal cell fate via direct regulation of a MYB-bHLH-WD40 complex by GSK3-like kinases. eLife 2014, 3, e02525. [Google Scholar] [CrossRef]
- Qi, T.; Huang, H.; Wu, D.; Yan, J.; Qi, Y.; Song, S.; Xie, D. Arabidopsis DELLA and JAZ proteins bind the WD-repeat/bHLH/MYB complex to modulate gibberellin and jasmonate signaling synergy. Plant Cell 2014, 26, 1118–1133. [Google Scholar] [CrossRef] [Green Version]
- Maier, A.; Schrader, A.; Kokkelink, L.; Falke, C.; Welter, B.; Iniesto, E.; Rubio, V.; Uhrig, J.; Hülskamp, M.; Hoecker, U. Light and the E3 ubiquitin ligase COP1/SPA control the protein stability of the MYB transcription factors PAP1 and PAP2 involved in anthocyanin accumulation in Arabidopsis. Plant J. 2013, 74, 638–651. [Google Scholar] [CrossRef]
- De Vetten, N.; Quattrocchio, F.; Mol, J.; Koes, R. The an11 locus controlling flower pigmentation in petunia encodes a novel WD-repeat protein conserved in yeast, plants, and animals. Genes Dev. 1997, 11, 1422–1434. [Google Scholar] [CrossRef] [Green Version]
- Walker, A.R.; Davison, P.A.; Bolognesi-Winfield, A.C.; James, C.M.; Srinivasan, N.; Blundell, T.L.; Esch, J.J.; Marks, M.D.; Gray, J.C. The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 1999, 11, 1337–1350. [Google Scholar] [CrossRef] [Green Version]
- Carey, C.C.; Strahle, J.T.; Selinger, D.A.; Chandler, V.L. Mutations in the pale aleurone color1 regulatory gene of the Zea mays anthocyanin pathway have distinct phenotypes relative to the functionally similar TRANSPARENT TESTA GLABRA1 gene in Arabidopsis thaliana. Plant Cell 2004, 16, 450–464. [Google Scholar] [CrossRef] [Green Version]
- Ben-Simhon, Z.; Judeinstein, S.; Nadler-Hassar, T.; Trainin, T.; Bar-Ya’Akov, I.; Borochov-Neori, H.; Holland, D. A pomegranate (Punica granatum L.) WD40-repeat gene is a functional homologue of Arabidopsis TTG1 and is involved in the regulation of anthocyanin biosynthesis during pomegranate fruit development. Planta 2011, 234, 865–881. [Google Scholar] [CrossRef]
- Gonzalez, A.; Zhao, M.; Leavitt, J.M.; Lloyd, A.M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008, 53, 814–827. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Dubos, C.; Lepiniec, L. Transcriptional control of flavonoid biosynthesis by MYB–bHLH–WDR complexes. Trends Plant Sci. 2015, 20, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Xian-Hui, W.; De-Lin, Z.; Wang, Y.; Xiong, Y.; Wu, X.-H.; Zhang, D.-L.; Ye, Z.-Q.; Wu, Y.-D. Prokaryotic and Highly-Repetitive WD40 Proteins: A Systematic Study. Sci. Rep. 2017, 7, 10585. [Google Scholar] [CrossRef] [Green Version]
- He, S.; Tong, X.; Han, M.; Hu, H.; Dai, F. Genome-Wide Identification and Characterization of WD40 Protein Genes in the Silkworm, Bombyx mori. Int. J. Mol. Sci. 2018, 19, 527. [Google Scholar] [CrossRef] [Green Version]
- Zou, X.D.; Hu, X.J.; Ma, J.; Li, T.; Ye, Z.Q.; Wu, Y.D. Genome-wide Analysis of WD40 Protein Family in Human. Sci. Rep. 2016, 6, 39262. [Google Scholar] [CrossRef] [Green Version]
- Mishra, A.K.; Muthamilarasan, M.; Khan, Y.; Parida, S.K.; Prasad, M. Genome-wide investigation and expression analyses of WD40 protein family in the model plant foxtail millet (Setaria italica L.). PLoS ONE 2014, 9, e86852. [Google Scholar]
- Li, Q.; Zhao, P.; Li, J.; Zhang, C.; Wang, L.; Ren, Z. Genome-wide analysis of the WD-repeat protein family in cucumber and Arabidopsis. Mol. Genet. Genom. 2014, 289, 103–124. [Google Scholar] [CrossRef]
- Feng, R.; Zhang, C.; Ma, R.; Cai, Z.; Lin, Y.; Yu, M. Identification and characterization of WD40 superfamily genes in peach. Gene 2019, 710, 291–306. [Google Scholar] [CrossRef]
- Salih, H.; Gong, W.; Mkulama, M.; Du, X. Genome-wide characterization, identification, and expression analysis of the WD40 protein family in cotton. Genome 2018, 61, 539–547. [Google Scholar] [CrossRef]
- Chen, Z.; Tan, J.L.H.; Ingouff, M.; Sundaresan, V.; Berger, F. Chromatin assembly factor 1 regulates the cell cycle but not cell fate during male gametogenesis in Arabidopsis thaliana. Development 2007, 135, 65–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, I.; Ambaru, B.; Thakkar, P.; Marcotte, E.; Rhee, S. Rational association of genes with traits using a genome-scale gene network for Arabidopsis thaliana. Nat. Biotechnol. 2010, 28, 149–156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daccord, N.; Celton, J.-M.; Linsmith, G.; Becker, C.; Choisne, N.; Schijlen, E.; Van De Geest, H.; Bianco, L.; Micheletti, D.; Velasco, R.; et al. High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat. Genet. 2017, 49, 1099–1106. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.; Lee, T.; Cheng, C.H.; Buble, K.; Zheng, P.; Yu, J.; Humann, J.; Ficklin, S.P.; Gasic, K.; Scott, K.; et al. 15 years of GDR: New data and functionality in the Genome Database for Rosaceae. Nucleic Acids Res. 2019, 47, D1137–D1145. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Gao, M.; Huang, L.; Wang, Y.; van Nocker, S.; Wan, R.; Guo, C.; Wang, X.; Gao, H. Identification and expression analysis of the apple (Malus × domestica) basic helix-loop-helix transcription factor family. Sci. Rep. 2017, 7, 28. [Google Scholar] [CrossRef] [Green Version]
- Fan, S.; Zhang, D.; Gao, C.; Zhao, M.; Wu, H.; Li, Y.; Shen, Y.; Han, M. Identification, Classification, and Expression Analysis of GRAS Gene Family in Malus domestica. Front. Physiol. 2017, 8, 253. [Google Scholar] [CrossRef] [Green Version]
- Liu, R.-H.; Meng, J.-L. MapDraw: A microsoft excel macro for drawing genetic linkage maps based on given genetic linkage data. Yi Chuan Hered. 2003, 25, 317–321. [Google Scholar]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2020, 49, D412–D419. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [Green Version]
- Yupeng, W.; Haibao, T.; Debarry, J.D.; Xu, T.; Jingping, L.; Xiyin, W.; Tae-Ho, L.; Huizhe, J.; Barry, M.; Hui, G. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar]
- Zhao, J.; Guo, R.; Guo, C.; Hou, H.; Wang, X.; Gao, H. Evolutionary and Expression Analyses of the Apple Basic Leucine Zipper Transcription Factor Family. Front. Plant Sci. 2016, 7, 376. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Guo, R.; Li, J.; Singer, S.; Zhang, Y.; Yin, X.; Zheng, Y.; Fan, C.; Wang, X. Genome-wide identification and analysis of the aldehyde dehydrogenase (ALDH) gene superfamily in apple (Malus × domestica Borkh.). Plant Physiol. Biochem. 2013, 71, 268–282. [Google Scholar] [CrossRef] [PubMed]
- An, J.P.; Wang, X.F.; Zhang, X.W.; Xu, H.F.; Bi, S.Q.; You, C.X.; Hao, Y.J. An apple MYB transcription factor regulates cold tolerance and anthocyanin accumulation and undergoes MIEL1-mediated degradation. Plant Biotechnol. J. 2020, 18, 337–353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, N.; Gong, X.-q.; Ma, F.-w. Genome-wide identification of the radiation sensitivity protein-23 (RAD23) family members in apple (Malus×domestica Borkh.) and expression analysis of their stress responsiveness. J. Integr. Agric. 2017, 16, 820–827. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Hou, H.; Li, X.; Xiang, J.; Yin, X.; Gao, H.; Zheng, Y.; Bassett, C.L.; Wang, X. Genome-wide identification and analysis of the SBP-box family genes in apple (Malus × domestica Borkh.). Plant Physiol. Biochem. 2013, 70, 100–114. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCTMethod. Methods A Companion Methods Enzymol. 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Holub, E.B. The arms race is ancient history in Arabidopsis, the wildflower. Nat. Rev. Genet. 2001, 2, 516. [Google Scholar] [CrossRef]
- Lee, J.H.; Yoon, H.J.; Terzaghi, W.; Martinez, C.; Dai, M.; Li, J.; Byun, M.O.; Deng, X.W. DWA1 and DWA2, two Arabidopsis DWD protein components of CUL4-based E3 ligases, act together as negative regulators in ABA signal transduction. Plant Cell 2010, 22, 1716–1732. [Google Scholar] [CrossRef] [Green Version]
- Alexandre, C.; Möller-Steinbach, Y.; Schönrock, N.; Gruissem, W.; Hennig, L. Arabidopsis MSI1 Is Required for Negative Regulation of the Response to Drought Stress. Mol. Plant 2009, 2, 675–687. [Google Scholar] [CrossRef]
- Mehdi, S.; Derkacheva, M.; Ramstrom, M.; Kralemann, L.; Bergquist, J.; Hennig, L. The WD40 Domain Protein MSI1 Functions in a Histone Deacetylase Complex to Fine-Tune Abscisic Acid Signaling. Plant Cell 2016, 28, 42–54. [Google Scholar] [CrossRef] [Green Version]
- Jiang, S.; Kumar, S.; Eu, Y.J.; Jami, S.K.; Stasolla, C.; Hill, R.D. The Arabidopsis mutant, fy-1, has an ABA-insensitive germination phenotype. J. Exp. Bot. 2012, 63, 2693–2703. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Terzaghi, W.; Deng, X.W. DWA3, an Arabidopsis DWD protein, acts as a negative regulator in ABA signal transduction. Plant Sci. 2011, 180, 352–357. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Wang, S.; Valerius, O.; Hall, H.; Zeng, Q.; Li, J.F.; Weston, D.J.; Ellis, B.E.; Chen, J.G. Involvement of Arabidopsis RACK1 in protein translation and its regulation by abscisic acid. Plant Physiol. 2011, 155, 370–383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stirnimann, C.U.; Petsalaki, E.; Russell, R.B.; Müller, C.W. WD40 proteins propel cellular networks. Trends Biochem. Sci. 2010, 35, 565–574. [Google Scholar] [CrossRef] [PubMed]
- Fanfan, Y.; Yougen, C.; Bingjing, L.; Defang, G.; Hua, W.; Dongliang, W. Identification of WD40 gene family in Prunus mume and its expression level under light illumination. PeerJ Prepr. 2018, 6, e27106v1. [Google Scholar]
- Strygina, K.; Khlestkina, E. Structural and Functional Organization and Evolution of the WD40 Genes Involved in the Regulation of Flavonoid Biosynthesis in the Triticeae Tribe. Russ. J. Genet. 2019, 55, 1398–1405. [Google Scholar] [CrossRef]
- Blanc, G.; Hokamp, K.; Wolfe, K. A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res. 2003, 13, 137–144. [Google Scholar] [CrossRef] [Green Version]
- Ling, J.; Jiang, W.; Zhang, Y.; Yu, H.; Mao, Z.; Gu, X.; Huang, S.; Xie, B. Genome-wide analysis of WRKY gene family in Cucumis sativus. BMC Genom. 2011, 12, 471. [Google Scholar] [CrossRef] [Green Version]
- Velasco, R.; Zharkikh, A.; Affourtit, J.; Dhingra, A.; Cestaro, A.; Kalyanaraman, A.; Fontana, P.; Bhatnagar, S.; Troggio, M.; Pruss, D.; et al. The genome of the domesticated apple (Malus × domestica Borkh.). Nat. Genet. 2010, 42, 833–839. [Google Scholar] [CrossRef]
- Lyons, E.; Pedersen, B.; Kane, J.; Alam, M.; Ming, R.; Tang, H.; Wang, X.; Bowers, J.; Paterson, A.; Lisch, D.; et al. Finding and Comparing Syntenic Regions among Arabidopsis and the Outgroups Papaya, Poplar, and Grape: CoGe with Rosids. Plant Physiol. 2008, 148, 1772–1781. [Google Scholar] [CrossRef] [Green Version]
- Guo, J.; Wang, J.; Xi, L.; Huang, W.D.; Liang, J.; Chen, J.G. RACK1 is a negative regulator of ABA responses in Arabidopsis. J. Exp. Bot. 2009, 60, 3819–3833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Jia, W.; Yang, J.; Ismail, A.M. Role of ABA in integrating plant responses to drought and salt stresses. Field Crops Res. 2006, 97, 111–119. [Google Scholar] [CrossRef]
- Vishwakarma, K.; Upadhyay, N.; Kumar, N.; Yadav, G.; Singh, J.; Mishra, R.K.; Kumar, V.; Verma, R.; Upadhyay, R.; Pandey, M.; et al. Abscisic acid signaling and abiotic stress tolerance in plants: A review on current knowledge and future prospects. Front. Plant Sci. 2017, 8, 161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El-Maarouf-Bouteau, H.; Bailly, C. Oxidative signaling in seed germination and dormancy. Plant Signal. Behav. 2008, 3, 175–182. [Google Scholar] [CrossRef] [Green Version]
- Van den Berg, L.; Zeng, Y. Response of South African indigenous grass species to drought stress induced by polyethylene glycol (PEG) 6000. South Afr. J. Bot. 2006, 72, 284–286. [Google Scholar] [CrossRef] [Green Version]
- Seki, M.; Umezawa, T.; Urano, K.; Shinozaki, K. Regulatory metabolic networks in drought stress responses. Curr. Opin. Plant Biol. 2007, 10, 296–302. [Google Scholar] [CrossRef] [PubMed]
- Nordin, K.; Heino, P.; Palva, E.T. Separate signal pathways regulate the expression of a low-temperature-induced gene in Arabidopsis thaliana (L.) Heynh. Plant Mol. Biol. 1991, 16, 1061–1071. [Google Scholar] [CrossRef] [PubMed]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, B.; Qu, D.; Yang, H.; Long, X.; Zhu, Z.; Yang, Y.; Zhao, Z. Genome Wide Identification and Characterization of Apple WD40 Proteins and Expression Analysis in Response to ABA, Drought, and Low Temperature. Horticulturae 2022, 8, 141. https://doi.org/10.3390/horticulturae8020141
Zhang B, Qu D, Yang H, Long X, Zhu Z, Yang Y, Zhao Z. Genome Wide Identification and Characterization of Apple WD40 Proteins and Expression Analysis in Response to ABA, Drought, and Low Temperature. Horticulturae. 2022; 8(2):141. https://doi.org/10.3390/horticulturae8020141
Chicago/Turabian StyleZhang, Bo, Dong Qu, Huijuan Yang, Xiaogang Long, Zhenzhen Zhu, Yazhou Yang, and Zhengyang Zhao. 2022. "Genome Wide Identification and Characterization of Apple WD40 Proteins and Expression Analysis in Response to ABA, Drought, and Low Temperature" Horticulturae 8, no. 2: 141. https://doi.org/10.3390/horticulturae8020141
APA StyleZhang, B., Qu, D., Yang, H., Long, X., Zhu, Z., Yang, Y., & Zhao, Z. (2022). Genome Wide Identification and Characterization of Apple WD40 Proteins and Expression Analysis in Response to ABA, Drought, and Low Temperature. Horticulturae, 8(2), 141. https://doi.org/10.3390/horticulturae8020141






