Estimation of Vegetative Growth in Strawberry Plants Using Mobile LiDAR Laser Scanner
Abstract
:1. Introduction
2. Materials and Methods
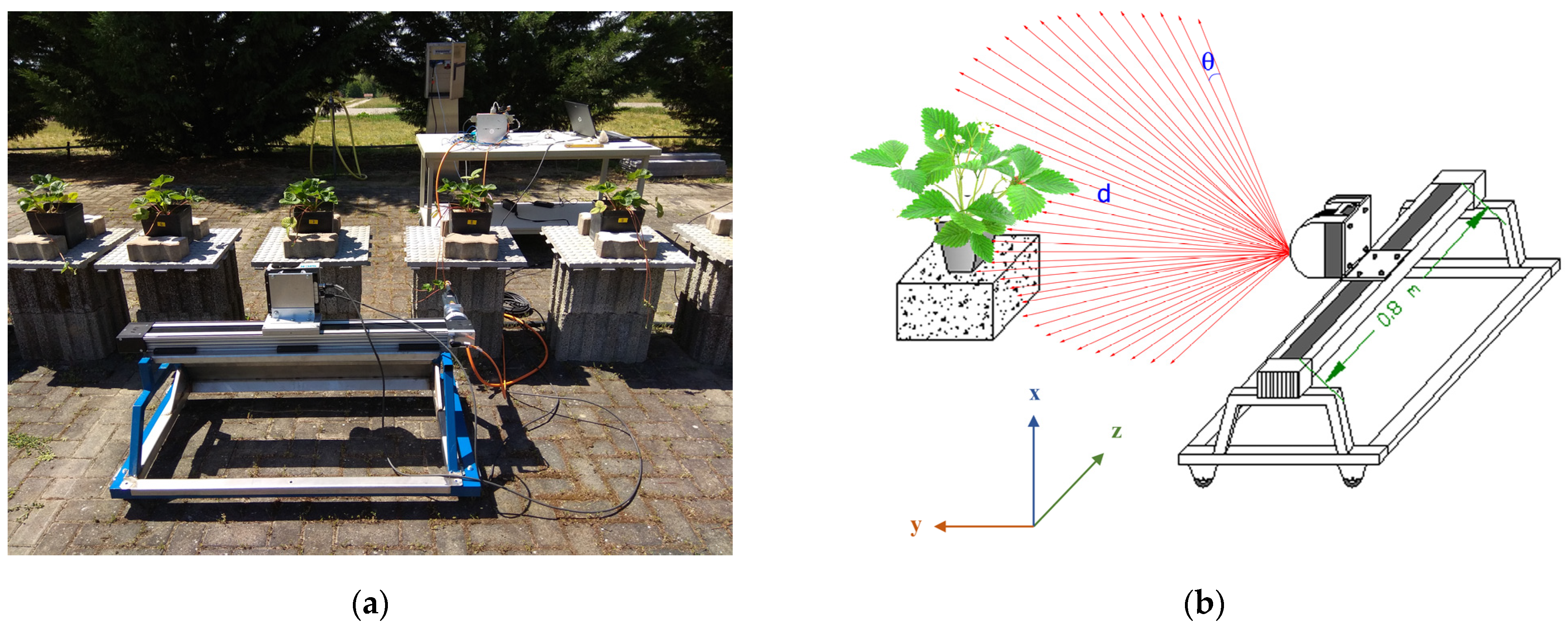
2.1. Experimental Setup
2.2. Plant Reference Data
2.3. LiDAR Data Acquisition
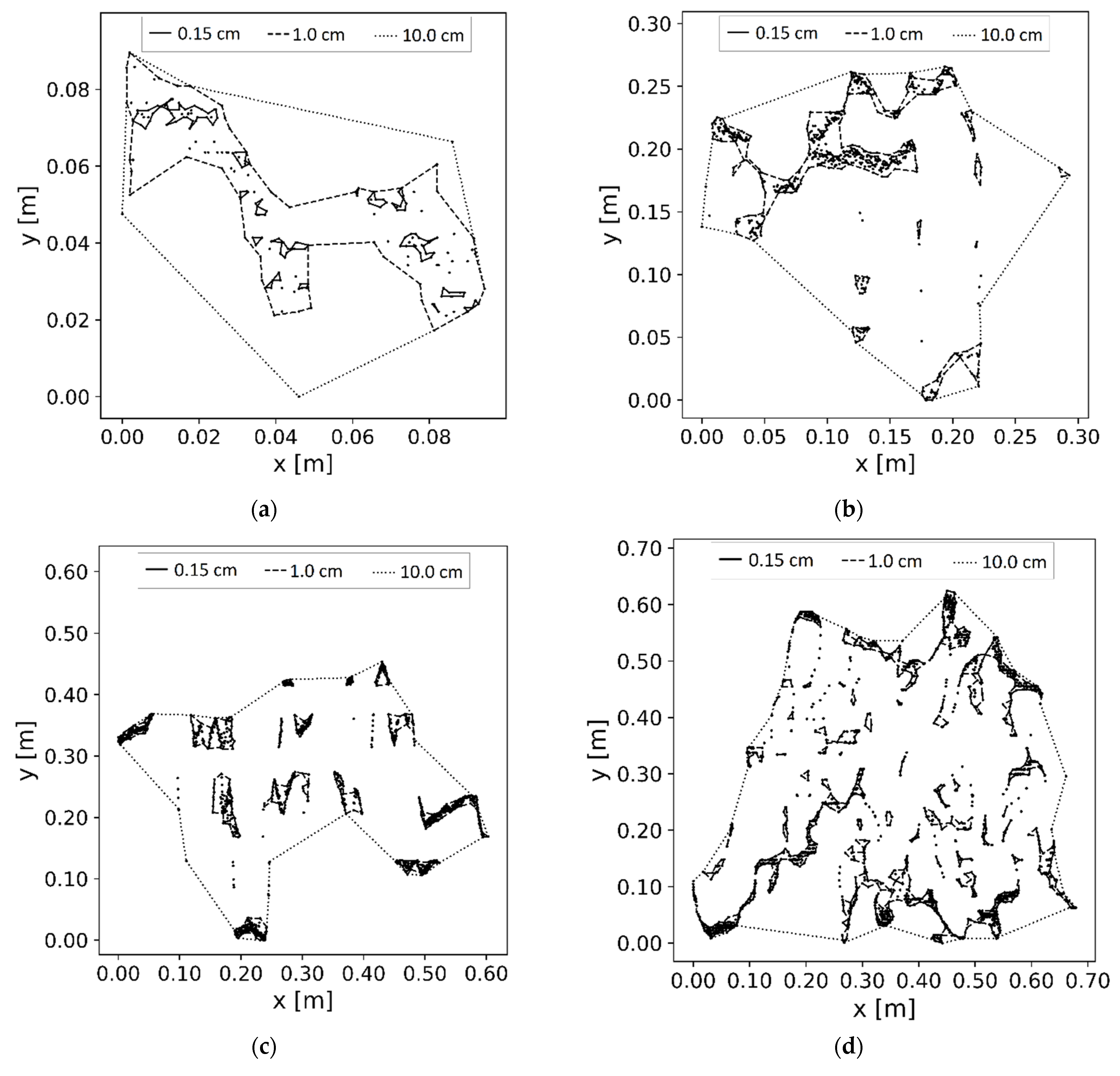
2.4. Reconstruction of 3D Plant Model
2.5. Estimation of Plant Parameters
3. Results
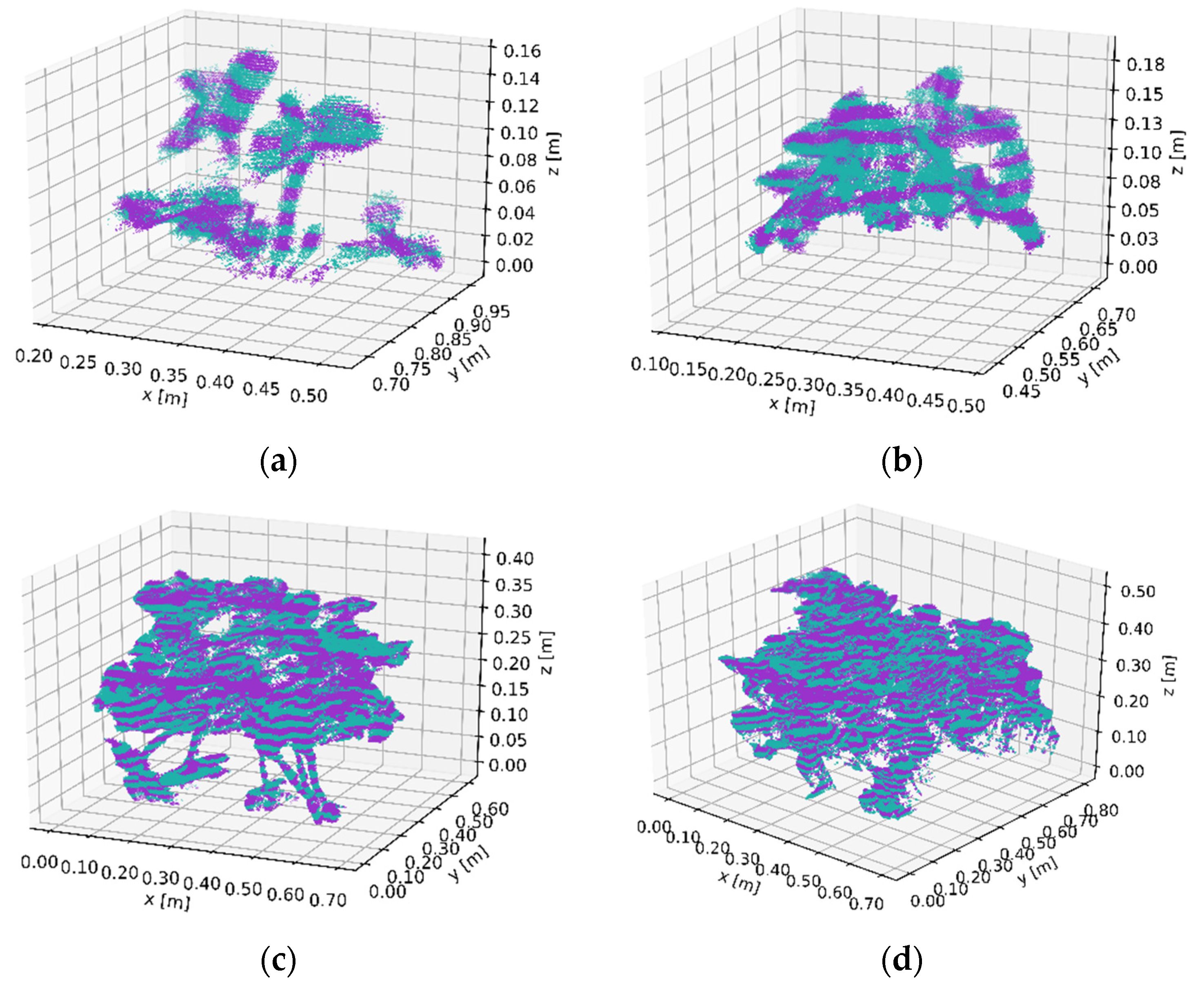
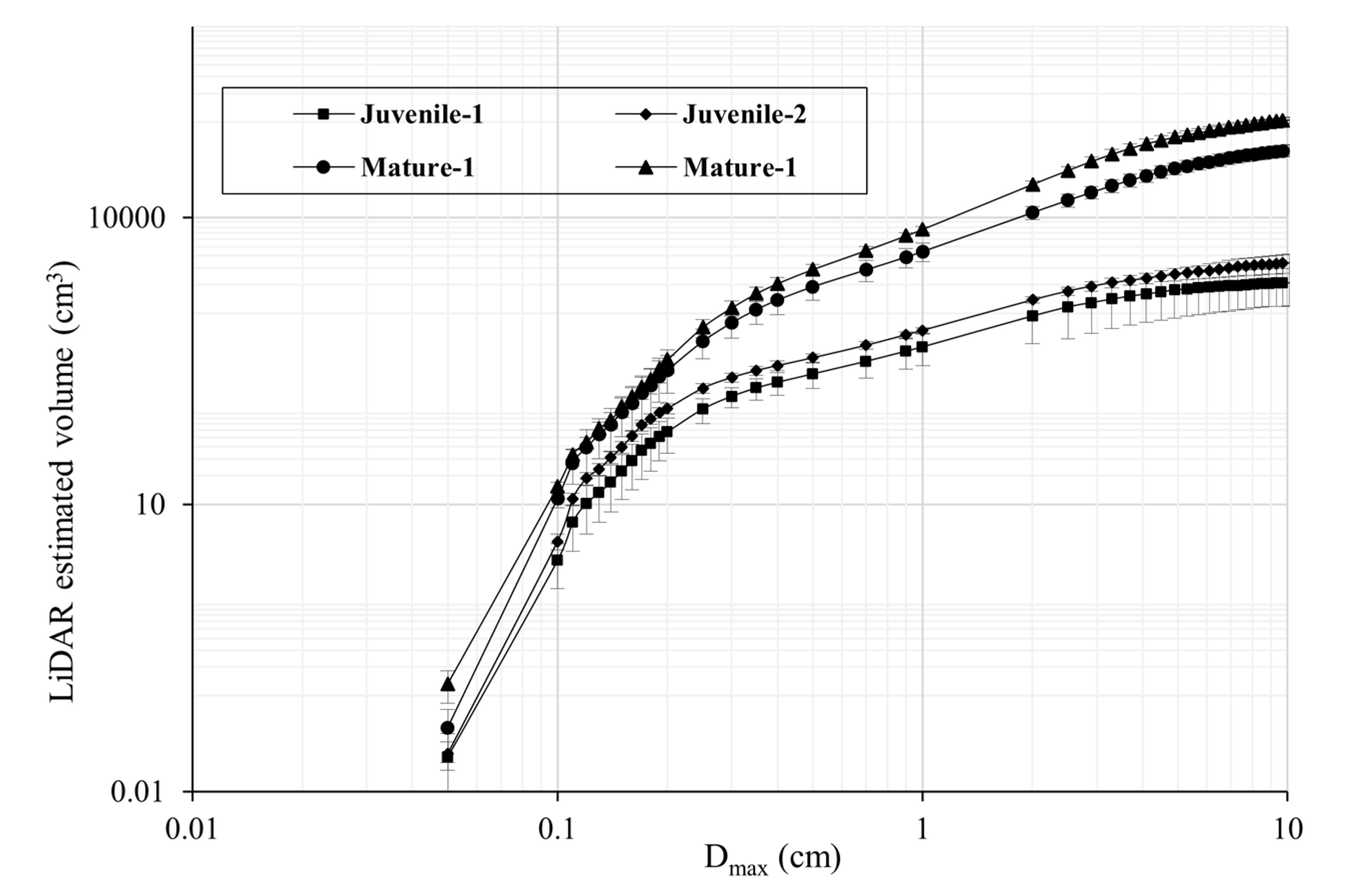
3.1. Canopy Volume Extraction Capturing Four Size Classes
3.2. Comparative Analysis of Different Canopy Volume Estimation Approaches
3.3. Summary Statistics and Correlations of Reference Plant Variables for Four Size Classes
3.4. Temporal Monitoring of Strawberry Canopy
3.4.1. Estimation of Leaf Area with LiDAR-Derived Canopy Variables of Juvenile Plants
3.4.2. Canopy Volume
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- An, X.; Li, Z.; Zude-Sasse, M.; Tchuenbou-Magaia, F.; Yang, Y. Characterization of textural failure mechanics of strawberry fruit. J. Food Eng. 2020, 282, 110016. [Google Scholar] [CrossRef]
- Weng, S.Z.; Yu, S.; Dong, R.L.; Pan, F.F.; Liang, D. Nondestructive detection of storage time of strawberries using visible/near-infrared hyperspectral imaging. Int. J. Food Prop. 2020, 23, 269–281. [Google Scholar] [CrossRef] [Green Version]
- Darnell, R.L.; Cantliffe, D.J.; Kirschbaum, D.S.; Chandler, C.K. The Physiology of Flowering in Strawberry. Hortic. Rev. 2002, 28, 325–349. [Google Scholar] [CrossRef]
- Zude-Sasse, M.; Fountas, S.; Gemtos, T.; Abu-Khalaf, N. Applications of precision agriculture in horticultural crops. Eur. J. Hortic. Sci. 2016, 81, 78–90. [Google Scholar] [CrossRef]
- Johnson, R.S.; Lakso, A.N. Approaches to Modeling Light Interception in Orchards. HortScience 1991, 26, 1002–1004. [Google Scholar] [CrossRef] [Green Version]
- Weiss, M.; Jacob, F.; Duveiller, G. Remote sensing for agricultural applications: A meta-review. Remote Sens. Environ. 2020, 236, 111402. [Google Scholar] [CrossRef]
- Khanal, S.; Kc, K.; Fulton, J.; Shearer, S.; Ozkan, E. Remote Sensing in Agriculture—Accomplishments, Limitations, and Opportunities. Remote Sens. 2020, 12, 3783. [Google Scholar] [CrossRef]
- Fahlgren, N.; A Gehan, M.; Baxter, I. Lights, camera, action: High-throughput plant phenotyping is ready for a close-up. Curr. Opin. Plant Biol. 2015, 24, 93–99. [Google Scholar] [CrossRef] [Green Version]
- Klodt, M.; Herzog, K.; Töpfer, R.; Cremers, D. Field phenotyping of grapevine growth using dense stereo reconstruction. BMC Bioinform. 2015, 16, 143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klose, R.; Penlington, J.; Ruckelshausen, A. Usability study of 3d time-of-flight cameras for automatic plant phenotyping. Bornimer Agrartech. Ber. 2009, 69, 12. [Google Scholar]
- Wang, Z.; Walsh, K.B.; Verma, B. On-Tree Mango Fruit Size Estimation Using RGB-D Images. Sensors 2017, 17, 2738. [Google Scholar] [CrossRef] [Green Version]
- Chéné, Y.; Rousseau, D.; Lucidarme, P.; Bertheloot, J.; Caffier, V.; Morel, P.; Belin, É.; Chapeau-Blondeau, F. On the use of depth camera for 3d phenotyping of entire plants. Comput. Electron. Agric. 2012, 82, 122–127. [Google Scholar] [CrossRef]
- Polo, J.R.R.; Sanz, R.; Llorens, J.; Arnó, J.; Escolà, A.; Ribes-Dasi, M.; Masip, J.; Camp, F.; Gràcia, F.; Solanelles, F.; et al. A tractor-mounted scanning LIDAR for the non-destructive measurement of vegetative volume and surface area of tree-row plantations: A comparison with conventional destructive measurements. Biosyst. Eng. 2009, 102, 128–134. [Google Scholar] [CrossRef] [Green Version]
- Tsoulias, N.; Paraforos, D.S.; Fountas, S.; Zude-Sasse, M. Estimating Canopy Parameters Based on the Stem Position in Apple Trees Using a 2D LiDAR. Agronomy 2019, 9, 740. [Google Scholar] [CrossRef] [Green Version]
- Soudarissanane, S.; Lindenbergh, R.; Menenti, M.; Teunissen, P. Scanning geometry: Influencing factor on the quality of terrestrial laser scanning points. ISPRS J. Photogramm. Remote Sens. 2011, 66, 389–399. [Google Scholar] [CrossRef]
- Herrero-Huerta, M.; Bucksch, A.; Puttonen, E.; Rainey, K.M. Canopy Roughness: A New Phenotypic Trait to Estimate Aboveground Biomass from Unmanned Aerial System. Plant Phenomics 2020, 2020, 1–10. [Google Scholar] [CrossRef]
- Tsoulias, N.; Paraforos, D.; Xanthopoulos, G.; Zude-Sasse, M. Apple Shape Detection Based on Geometric and Radiometric Features Using a LiDAR Laser Scanner. Remote Sens. 2020, 12, 2481. [Google Scholar] [CrossRef]
- Zhou, H.; Zhang, J.; Ge, L.; Yu, X.; Wang, Y.; Zhang, C. Research on volume prediction of single tree canopy based on three-dimensional (3D) LiDAR and clustering segmentation. Int. J. Remote Sens. 2021, 42, 738–755. [Google Scholar] [CrossRef]
- Takahashi, M.; Takayama, S.; Umeda, H.; Yoshida, C.; Koike, O.; Iwasaki, Y.; Sugeno, W. Quantification of Strawberry Plant Growth and Amount of Light Received Using a Depth Sensor. Environ. Control Biol. 2020, 58, 31–36. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, S.; Hayashi, S.; Tsubota, S. Growth Measurement of a Community of Strawberries Using Three-Dimensional Sensor. Environ. Control Biol. 2015, 53, 49–53. [Google Scholar] [CrossRef]
- Guan, Z.; Abd-Elrahman, A.; Fan, Z.; Whitaker, V.M.; Wilkinson, B. Modeling strawberry biomass and leaf area using object-based analysis of high-resolution images. ISPRS J. Photogramm. Remote Sens. 2020, 163, 171–186. [Google Scholar] [CrossRef]
- Han, K.-S.; Kim, S.-C.; Lee, Y.-B.; Kim, S.-C.; Im, D.-H.; Choi, H.-K.; Hwang, H. Strawberry Harvesting Robot for Bench-type Cultivation. J. Biosyst. Eng. 2012, 37, 65–74. [Google Scholar] [CrossRef] [Green Version]
- Ge, Y.; Xiong, Y.; From, P.J. Symmetry-based 3D shape completion for fruit localisation for harvesting robots. Biosyst. Eng. 2020, 197, 188–202. [Google Scholar] [CrossRef]
- Zhou, C.; Hu, J.; Xu, Z.; Yue, J.; Ye, H.; Yang, G. A Novel Greenhouse-Based System for the Detection and Plumpness Assessment of Strawberry Using an Improved Deep Learning Technique. Front. Plant Sci. 2020, 11, 559. [Google Scholar] [CrossRef]
- He, J.Q.; Harrison, R.J.; Li, B. A novel 3D imaging system for strawberry phenotyping. Plant Methods 2017, 13, 1–8. [Google Scholar] [CrossRef]
- Li, B.; Cockerton, H.M.; Johnson, A.W.; Karlström, A.; Stavridou, E.; Deakin, G.; Harrison, R.J. Defining strawberry shape uniformity using 3d imaging and genetic mapping. Hortic. Res. 2020, 7, 1–13. [Google Scholar] [CrossRef]
- Directive, C. Council directive 91/414/eec of 15 july 1991 concerning the placing of plant protection products on the market. Off. J. Eur. Communities L 1991, 230, 1–32. [Google Scholar]
- Cheein, F.A.A.; Guivant, J.; Sanz, R.; Escolà, A.; Yandún, F.; Torres-Torriti, M.; Rosell-Polo, J.R. Real-time approaches for characterization of fully and partially scanned canopies in groves. Comput. Electron. Agric. 2015, 118, 361–371. [Google Scholar] [CrossRef] [Green Version]
- Colaço, A.F.; Trevisan, R.G.; Molin, J.P.; Rosell-Polo, J.R.; Escolà, A. A Method to Obtain Orange Crop Geometry Information Using a Mobile Terrestrial Laser Scanner and 3D Modeling. Remote Sens. 2017, 9, 763. [Google Scholar] [CrossRef] [Green Version]
- Hosoi, F.; Nakai, Y.; Omasa, K. 3-D voxel-based solid modeling of a broad-leaved tree for accurate volume estimation using portable scanning lidar. ISPRS J. Photogramm. Remote Sens. 2013, 82, 41–48. [Google Scholar] [CrossRef]
- Underwood, J.; Hung, C.; Whelan, B.; Sukkarieh, S. Mapping almond orchard canopy volume, flowers, fruit and yield using lidar and vision sensors. Comput. Electron. Agric. 2016, 130, 83–96. [Google Scholar] [CrossRef]
- Putman, E.B.; Popescu, S.C. Automated Estimation of Standing Dead Tree Volume Using Voxelized Terrestrial Lidar Data. IEEE Trans. Geosci. Remote Sens. 2018, 56, 6484–6503. [Google Scholar] [CrossRef]
- Xu, W.; Su, Z.; Feng, Z.; Xu, H.; Jiao, Y.; Yan, F. Comparison of conventional measurement and LiDAR-based measurement for crown structures. Comput. Electron. Agric. 2013, 98, 242–251. [Google Scholar] [CrossRef]
- Lin, W.; Meng, Y.; Qiu, Z.; Zhang, S.; Wu, J. Measurement and calculation of crown projection area and crown volume of individual trees based on 3D laser-scanned point-cloud data. Int. J. Remote Sens. 2017, 38, 1083–1100. [Google Scholar] [CrossRef]
- Yan, Z.; Liu, R.; Cheng, L.; Zhou, X.; Ruan, X.; Xiao, Y. A Concave Hull Methodology for Calculating the Crown Volume of Individual Trees Based on Vehicle-Borne LiDAR Data. Remote Sens. 2019, 11, 623. [Google Scholar] [CrossRef] [Green Version]
- Escolà, A.; Martínez-Casasnovas, J.A.; Rufat, J.; Arnó, J.; Arbonés, A.; Sebé, F.; Pascual, M.; Gregorio, E.; Rosell-Polo, J.R. Mobile terrestrial laser scanner applications in precision fruticulture/horticulture and tools to extract information from canopy point clouds. Precis. Agric. 2017, 18, 111–132. [Google Scholar] [CrossRef] [Green Version]
- Meier, U.; Graf, H.; Hack, H.; Hess, M.; Kennel, W.; Klose, R.; Mappes, D.; Seipp, D.; Stauss, R.; Streif, J. Phenological growth stages of pome fruit (Malus domestica borkh. and Pyrus Communis L.), stone fruit (Prunus species), Currants ribes species and strawberry (Fragaria × ananassa duch.). Nachr. Dtsch. Pflanzenschutzd. 1994, 46, 141–153. [Google Scholar]
- Hobart, M.; Pflanz, M.; Weltzien, C.; Schirrmann, M. Growth Height Determination of Tree Walls for Precise Monitoring in Apple Fruit Production Using UAV Photogrammetry. Remote Sens. 2020, 12, 1656. [Google Scholar] [CrossRef]
- Lawlor, D.W.; Boyle, F.A.; Keys, A.J.; Kendall, A.C.; Young, A.T. Nitrate Nutrition and Temperature Effects on Wheat: A Synthesis of Plant Growth and Nitrogen Uptake in Relation to Metabolic and Physiological Processes. J. Exp. Bot. 1988, 39, 329–343. [Google Scholar] [CrossRef]
- Harrington, J.T.; Mexal, J.G.; Fisher, J.T. Volume displacement provides a quick and accurate way to quantify new root production. Seedling 1994, 121, 124. [Google Scholar]
- Rusu, R.B.; Marton, Z.C.; Blodow, N.; Dolha, M.; Beetz, M. Towards 3D Point cloud based object maps for household environments. Robot. Auton. Syst. 2008, 56, 927–941. [Google Scholar] [CrossRef]
- Girardeau-Montaut, D. Cloudcompare, v. 2.10. 2019. Available online: https://cloudcompare.org (accessed on 2 May 2020).
- Besl, P.J.; McKay, N.D. Method for registration of 3-D shapes. Proceedings of Sensor Fusion IV: Control Paradigms and Data Structures, Boston, MA, USA, 14–15 November 1991; pp. 586–606. [Google Scholar]
- Moreira, A.; Santos, M.Y. Concave Hull: A K-Nearest Neighbors Approach for the Computation of the Region Occupied by A Set of Points. In Proceedings of the GRAPP 2007 International Conference on Computer Graphics Theory and Applications, Barcelona, Spain, 8–11 March 1991; Volume 2, pp. 61–68. [Google Scholar]
- Awrangjeb, M. Using point cloud data to identify, trace, and regularize the outlines of buildings. Int. J. Remote Sens. 2016, 37, 551–579. [Google Scholar] [CrossRef]
- Saha, K.; Tsoulias, N.; Zude-Sasse, M. Estimation of leaf area of sweet cherry trees trained as spindle using ground based 2D mobile LiDAR system. Acta Hortic. 2021, 8, 429–436. [Google Scholar] [CrossRef]
- Hunt, R. Plant Growth Curves. The Functional Approach to Plant Growth Analysis; Edward Arnold Ltd.: London, UK, 1982. [Google Scholar]
- Lecigne, B.; Delagrange, S.; Messier, C. Exploring trees in three dimensions: VoxR, a novel voxel-based R package dedicated to analysing the complex arrangement of tree crowns. Ann. Bot. 2017, 121, 589–601. [Google Scholar] [CrossRef] [PubMed]
- Walter, J.D.C.; Edwards, J.; McDonald, G.; Kuchel, H. Estimating Biomass and Canopy Height with LiDAR for Field Crop Breeding. Front. Plant Sci. 2019, 10, 1145. [Google Scholar] [CrossRef] [PubMed]
- Greaves, H.E.; Vierling, L.A.; Eitel, J.U.H.; Boelman, N.T.; Magney, T.S.; Prager, C.M.; Griffin, K.L. Estimating aboveground biomass and leaf area of low-stature Arctic shrubs with terrestrial LiDAR. Remote Sens. Environ. 2015, 164, 26–35. [Google Scholar] [CrossRef]
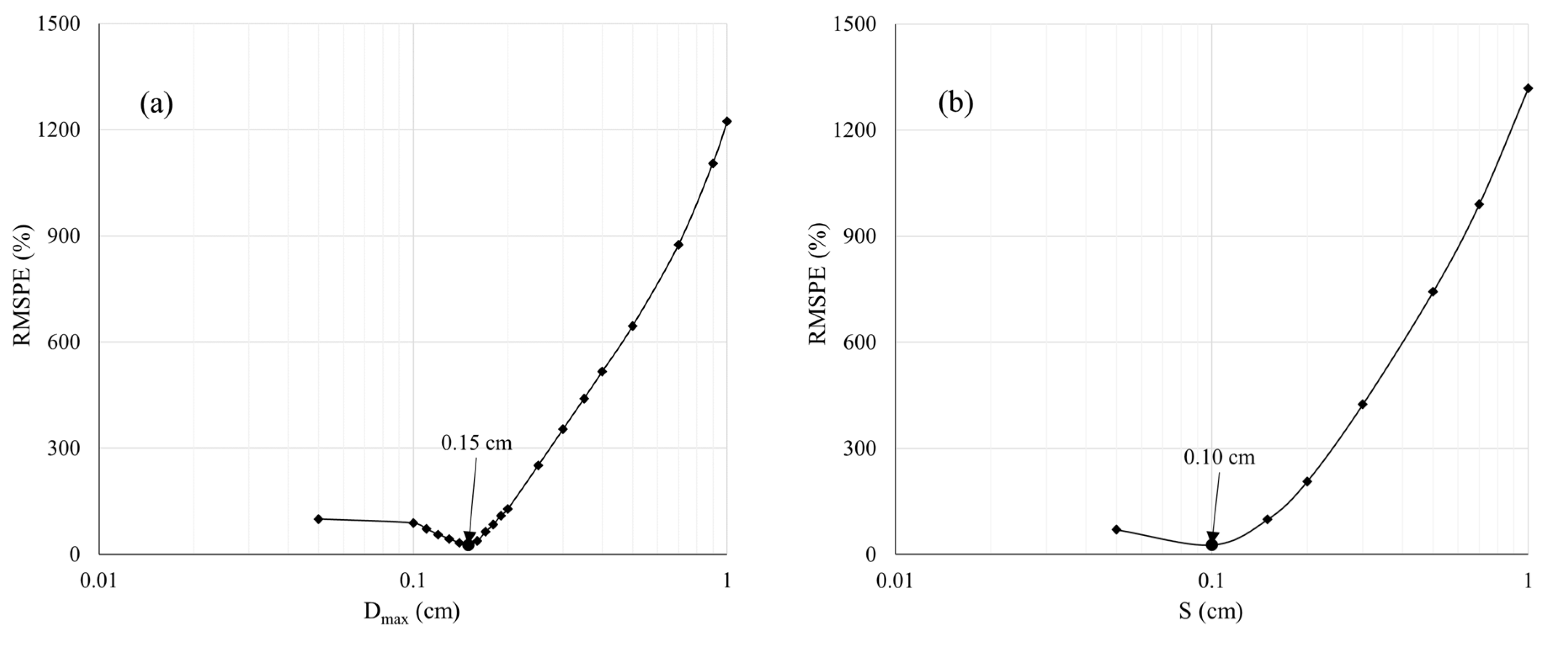

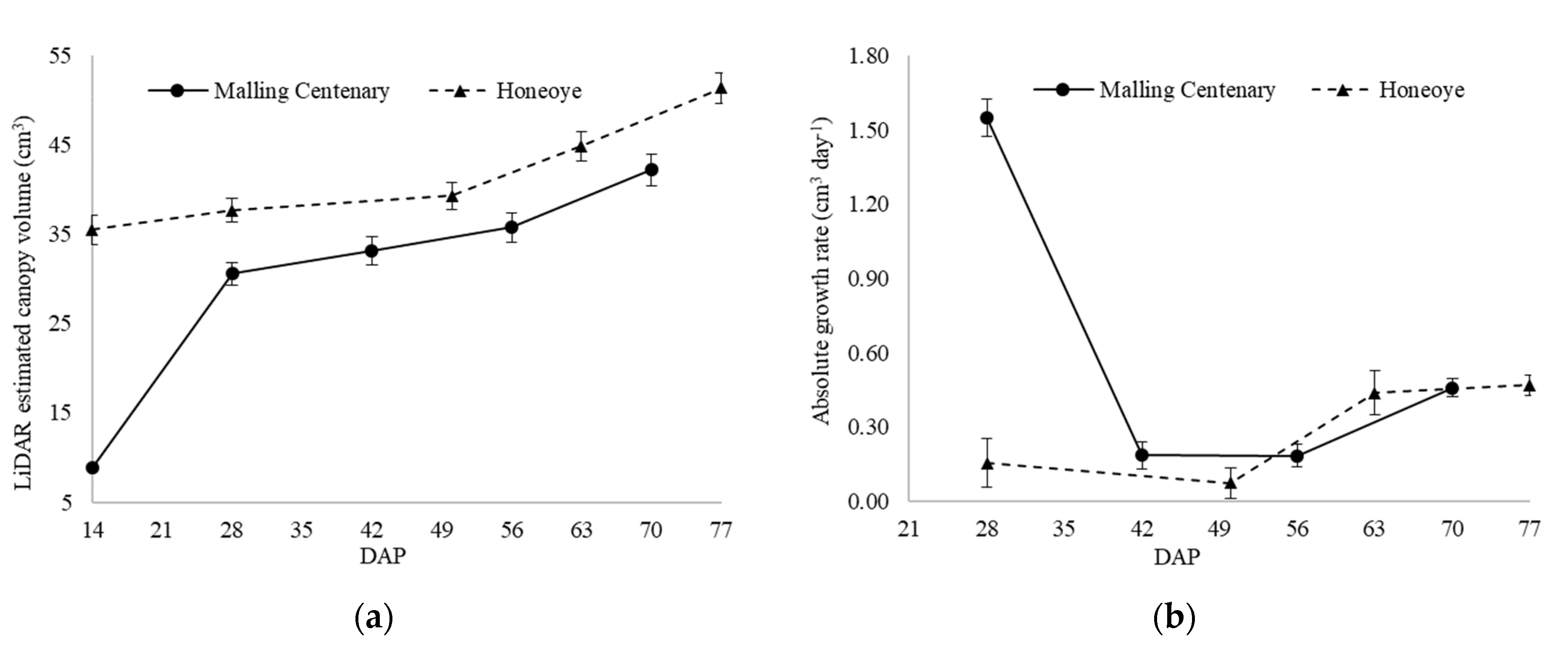
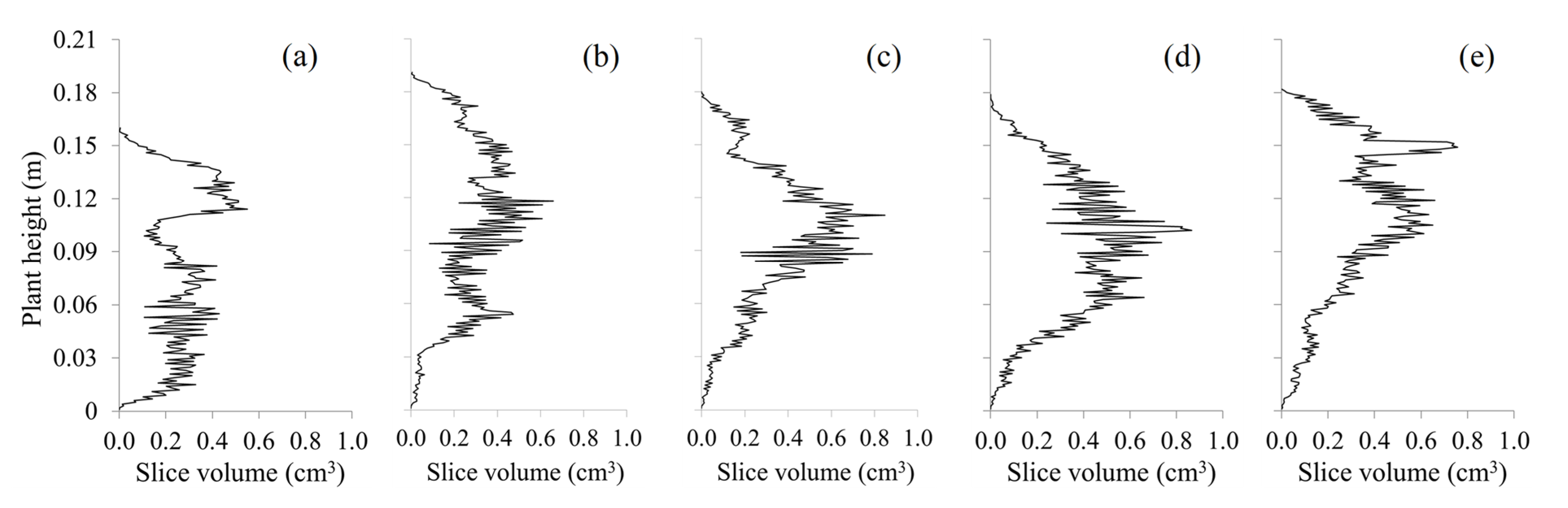
| Approach | Min (cm3) | Max (cm3) | SD (cm3) | Mean (cm3) | MBE (cm3) | RMSE (cm3) | RMSPE (%) | R2 | Computational Time (s/plant) |
|---|---|---|---|---|---|---|---|---|---|
| Slicing and summing slices | 43.9 | 97.2 | 13.7 | 71.0 | −10.1 | 18.8 | 20.4 | 0.79 | 35.70 |
| Voxel-grid | 1692.0 | 2451.0 | 212.7 | 2134.3 | 2053.3 | 2062.3 | 2929.7 | 0.62 | 2534.00 |
| 3D convex hull | 6064.2 | 17,341.9 | 2889.7 | 10,563.2 | 10,482.1 | 10,868.6 | 1,086,864 | 0.41 | 0.85 |
| Descriptive Statistics | LiDAR Estimated Variables | Manually Measured Variables | ||||
|---|---|---|---|---|---|---|
| PPP | h (cm) | Canopy Area (cm2) | FM (g) | DM (g) | LA (cm2) | |
| Min | 34,459 | 8.73 | 311.59 | 11.15 | 3.86 | 409.92 |
| Max | 658,840 | 52.06 | 4555.31 | 382.09 | 131.92 | 19,336.00 |
| Mean | 243,200 | 27.38 | 1905.97 | 127.84 | 44.53 | 6522.55 |
| Median | 84,333 | 17.24 | 768.62 | 37.79 | 11.97 | 1369.32 |
| Standard deviation | 226,807 | 15.54 | 1659.92 | 130.66 | 45.40 | 7223.58 |
| Skewness | 0.80 | 0.39 | 0.48 | 0.91 | 0.80 | 0.87 |
| Kurtosis | −0.82 | −1.64 | −1.54 | −0.55 | −0.78 | −0.72 |
| Model | LiDAR-Estimated Variables | MBE | RMSE | RMSPE (%) | R2 |
|---|---|---|---|---|---|
| Fresh mass (g) | No. of points per plant | −0.0006 | 3.37 | 5.44 | 0.99 |
| Volume (cm3) | −0.0174 | 10.56 | 37.38 | 0.91 | |
| Height (cm) | 0.0073 | 8.67 | 30.72 | 0.93 | |
| Projected canopy area (cm2) | 0.0020 | 7.21 | 11.97 | 0.95 | |
| Dry mass (g) | No. of points per plant | −0.0003 | 1.00 | 4.45 | 0.99 |
| Volume (cm3) | −0.0010 | 3.36 | 37.09 | 0.92 | |
| Height (cm) | 0.0011 | 2.50 | 29.91 | 0.95 | |
| Projected canopy area (cm2) | 0.0002 | 2.09 | 10.87 | 0.97 | |
| Leaf area (cm²) | No. of points per plant | 0.0010 | 258.03 | 12.84 | 0.98 |
| Volume (cm3) | 35.1019 | 604.49 | 64.78 | 0.90 | |
| Height (cm) | −1.3308 | 509.56 | 50.61 | 0.93 | |
| Projected canopy area (cm2) | 0.4246 | 370.43 | 22.66 | 0.96 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saha, K.K.; Tsoulias, N.; Weltzien, C.; Zude-Sasse, M. Estimation of Vegetative Growth in Strawberry Plants Using Mobile LiDAR Laser Scanner. Horticulturae 2022, 8, 90. https://doi.org/10.3390/horticulturae8020090
Saha KK, Tsoulias N, Weltzien C, Zude-Sasse M. Estimation of Vegetative Growth in Strawberry Plants Using Mobile LiDAR Laser Scanner. Horticulturae. 2022; 8(2):90. https://doi.org/10.3390/horticulturae8020090
Chicago/Turabian StyleSaha, Kowshik Kumar, Nikos Tsoulias, Cornelia Weltzien, and Manuela Zude-Sasse. 2022. "Estimation of Vegetative Growth in Strawberry Plants Using Mobile LiDAR Laser Scanner" Horticulturae 8, no. 2: 90. https://doi.org/10.3390/horticulturae8020090
APA StyleSaha, K. K., Tsoulias, N., Weltzien, C., & Zude-Sasse, M. (2022). Estimation of Vegetative Growth in Strawberry Plants Using Mobile LiDAR Laser Scanner. Horticulturae, 8(2), 90. https://doi.org/10.3390/horticulturae8020090









