Phenotypic Diversity in Wild and Cultivated Date Palm (Phoenix, Arecaceae): Quantitative Analysis Using Information Theory
Abstract
:1. Introduction
2. Materials and Methods
- Descriptors of the environment and the locality. Especially those of an ethnobotanical character.
- Characterization descriptors. Essentially, they focus on macromorphological characters easily discernible in the field, although they also include other characters of interest that require the study of samples in the laboratory. With reference to the colors, the Munsell Color Chart for Plant Tissues has been used.
- Evaluation descriptors. These include, among others, resistance to various types of stress.
3. Results
3.1. Phenotypic Heterogeneity
3.1.1. Morphological Heterogeneity and Main Descriptor Categories
3.1.2. Intraindividual/Intrapopulational Variability
3.2. Operative Taxonomic Units’ Heterogeneity and Taxonomy
4. Discussion
4.1. Heterogeneity of Commonly Accepted Phoenix Species
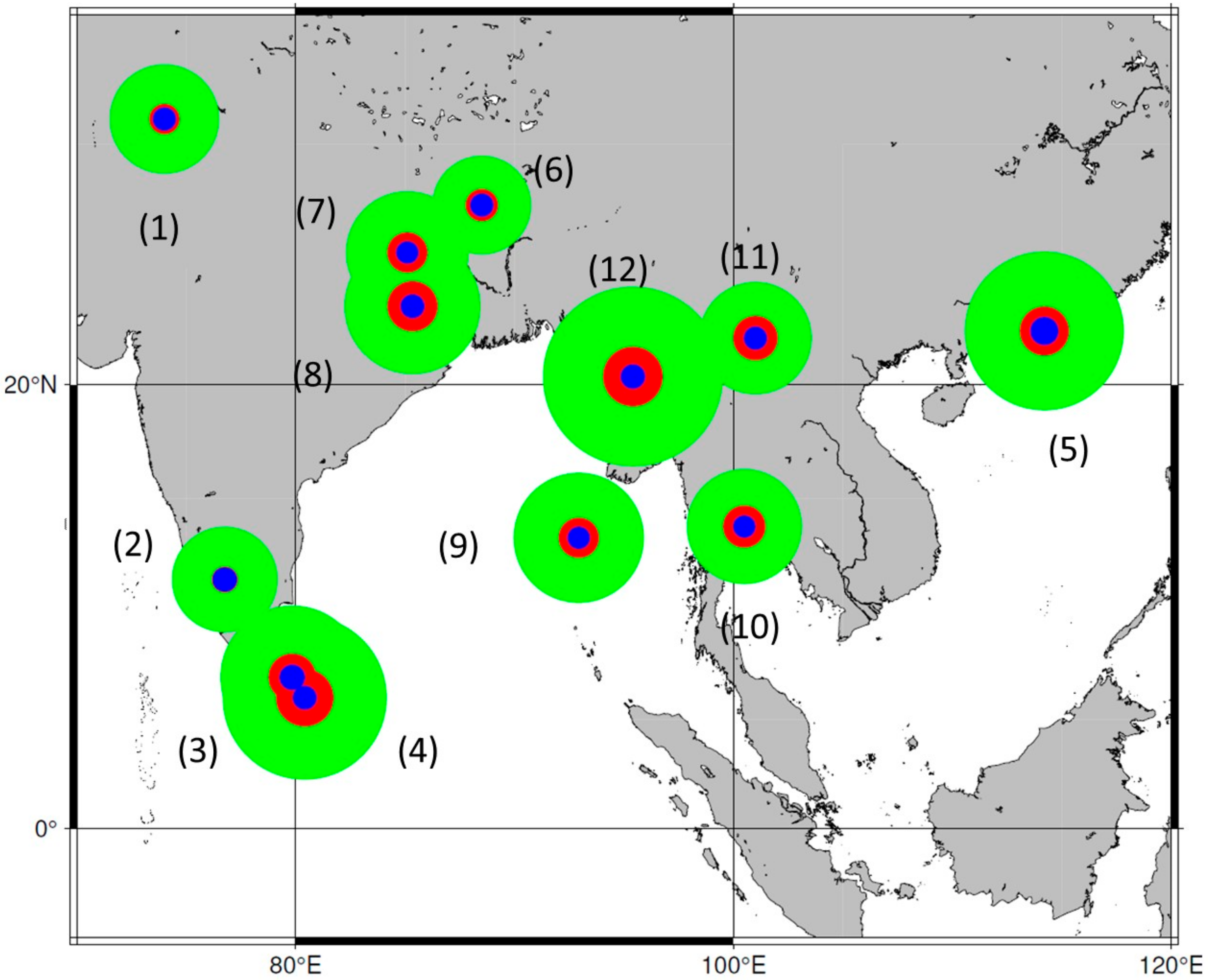
4.2. Heterogeneity of Genus Phoenix Geography and Origins
4.3. Patterns of Date Palm (Phoenix dactylifera) Phenotypic Diversity
4.3.1. Phenotypic Heterogeneity and Heterozygosis in Phoenix dactylifera
4.3.2. Differential Characters for Phoenix dactylifera Western and Eastern Populations
4.4. Role of Hybrids and Hybrid Swarms in Phenotypic Diversity
4.4.1. Large-Scale Hybrid Swarms
4.4.2. Phoenix Interspecific Hybrids
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Roswell, M.; Dushoff, J.; Winfree, R. A conceptual guide to measuring species diversity. Oikos 2021, 130, 321–338. [Google Scholar] [CrossRef]
- Shannon, C.E. A mathematical theory of communication. Bell Syst. Tech. J. 1948, 27, 379–423. [Google Scholar] [CrossRef] [Green Version]
- Shannon, C.E.; Weaver, W. The Mathematical Theory of Communication; The University of Illinois Press: Urbana, IL, USA, 1964. [Google Scholar]
- Xu, S.; Böttcher, L.; Chou, T. Diversity in biology: Definitions, quantification and models. Phys. Biol. 2020, 17, 031001. [Google Scholar] [CrossRef] [PubMed]
- Mitchell-Olds, T.; Willis, J.H.; Goldstein, D.B. Which evolutionary processes influence natural genetic variation for phenotypic traits? Nat. Rev. Genet. 2007, 8, 845–856. [Google Scholar] [CrossRef]
- Hansson, B.; Westerberg, L. On the correlation between heterozygosity and fitness in natural populations. Mol. Ecol. 2002, 11, 2467–2474. [Google Scholar] [CrossRef]
- FAOSTAT. Available online: https://www.fao.org/faostat/en/#data/QCL (accessed on 7 December 2021).
- Rivera, D.; Obón, C.; Alcaraz, F.; Laguna, E.; Johnson, D. Date-palm (Phoenix, Arecaceae) iconography in coins from the Mediterranean and West Asia (485 BC–1189 AD). J. Cult. Herit. 2019, 37, 199–214. [Google Scholar] [CrossRef]
- Rivera, D.; Abellán, J.; Palazón, J.A.; Obón, C.; Alcaraz, F.; Carreño, E.; Laguna, E.; Ruiz, A.; Johnson, D. Modelling ancient areas for date palms (Phoenix species: Arecaceae): Bayesian analysis of biological and cultural evidence. Bot. J. Linn. Soc. 2020, 193, 228–262. [Google Scholar] [CrossRef]
- UNESCO. Palmeral of Elche. Available online: https://whc.unesco.org/en/list/930/ (accessed on 7 December 2021).
- IPGRI; INRAA; INRAM; INRAT; FEM; PNUD. Descripteurs du Palmier dattier (Phoenix dactylifera L.); IPGRI: Rome, Italy, 2005. [Google Scholar]
- Stone, J.V. Information Theory a Tutorial Introduction; Sebtel Press: Milton Keynes, UK, 2015. [Google Scholar]
- MacKay, D. Information Theory, Inference, and Learning Algorithms; Cambridge University Press: Cambridge, UK, 2005. [Google Scholar]
- Partridge, D. Information Theory and Redundancy. Philos. Sci. 1981, 48, 308–316. [Google Scholar] [CrossRef]
- Hiller, L.; Bean, C. Information Theory Analyses of Four Sonata Expositions. J. Music Theory 1966, 10, 96–137. [Google Scholar] [CrossRef]
- Cohen, J.E. Information Theory and Music. Behav. Sci. 1962, 7, 137–163. [Google Scholar] [CrossRef]
- Perrier, X.; Flori, A.; Bonnot, F. Data analysis methods. In Genetic Diversity of Cultivated Tropical Plants; Hamon, P., Seguin, M., Perrier, X., Glaszmann, J., Eds.; Enfield, Science Publishers: Montpellier, France, 2003; pp. 43–76. [Google Scholar]
- Perrier, X.; Jacquemoud-Collet, J. DARwin Software. Available online: http://darwin.cirad.fr/ (accessed on 10 November 2021).
- Perrier, X.; Jacquemoud-Collet, J. Darwin. Dissimilarity Analysis and Representation for Windows, Version 6; CIRAD: Montpellier, France, 2014. [Google Scholar]
- Bedjaoui, H.; Benbouza, H. Assessment of phenotypic diversity of local Algerian date palm (Phoenix dactylifera L.) cultivars. J. Saudi Soc. Agric. Sci. 2020, 19, 65–75. [Google Scholar] [CrossRef]
- Vardareli, N.; Doğaroğlu, T.; Doğaç, E.; Taşkın, V.; Taşkın, B.G. Genetic characterization of tertiary relict endemic Phoenix theophrasti populations in Turkey and phylogenetic relations of the species with other palm species revealed by SSR markers. Plant Syst. Evol. 2019, 305, 415–429. [Google Scholar] [CrossRef]
- Pintaud, J.C.; Zehdi, S.; Couvreur, T.; Barrow, S.; Henderson, S.; Aberlenc-Bertossi, F.; Tregear, J.; Billotte, N. Species delimitation in the genus Phoenix (Arecaceae) based on SSR markers, with emphasis on the identity of the date palm (Phoenix dactylifera L.). In Diversity, Phylogeny, and Evolution in the Monocotyledons; Seberg, O., Petersen, G., Barfod, A.S., Davis, J.I., Eds.; Aarhus University Press: Aarhus, Denmark, 2010; pp. 267–286. [Google Scholar]
- Ballardini, M.; Mercuri, A.; Littardi, C.; Abbas, S.; Couderc, M.; Ludeña, B.; Pintaud, J.C. The chloroplast DNA locus psbZ-trnfM as a potential barcode marker in Phoenix, L. (Arecaceae). ZooKeys 2013, 365, 71–82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saro-Hernandez, I. Variabilidad Genética y Dispersión Polínica del Endemismo Canario Phoenix canariensis. Ph.D. Thesis, Universidad de Las Palmas de Gran Canaria, Las Palmas, Spain, 2016. [Google Scholar]
- Rivera, D.; Obón, C.; García-Arteaga, J.; Egea, T.; Alcaraz, F.; Laguna, E.; Carreño, E.; Johnson, D.; Krueger, R.; Delgadillo, J.; et al. Carpological analysis of Phoenix (Arecaceae): Contributions to the taxonomy and evolutionary history of the genus. Bot. J. Linn. Soc. 2014, 175, 74–122. [Google Scholar] [CrossRef] [Green Version]
- Martínez-Rico, M. El Género Phoenix en Jardinería y Paisajismo: El Caso de Phoenix canariensis. Ph.D. Thesis, Universidad Miguel Hernández, Alicante, Spain, 2017. Available online: http://dspace.umh.es/handle/11000/4504 (accessed on 15 December 2021).
- Gardner, E. The Pleistocene fauna and flora of Kharga Oasis, Egypt. Q. J. Geol. Soc. Lond. 1935, 91, 479–518. [Google Scholar] [CrossRef]
- Mai, D. Fossile Früchte und Samen aus dem Mitteleozän des Geiseltales. Abh. Zent. Geol. Inst. 1976, 26, 93–149. [Google Scholar]
- Chandler, M. The Lower Tertiary floras of Southern England I. Palaeocene Floras, London Clay Flora (Supplement); Trustees of the British Museum (Natural History): London, UK, 1961. [Google Scholar]
- Ortega, F.; Bardet, N.; Barroso Barcenilla, F.; Callapez, P.M.; Domingo Martínez, L. The biota of the Upper Cretaceous site of Lo Hueco (Cuenca, Spain). J. Iber. Geol. 2015, 41, 83–99. [Google Scholar] [CrossRef] [Green Version]
- Berry, E. Fruits of a date palm in the Tertiary deposits of eastern Texas. Am. J. Sci. 1914, 37, 403–406. [Google Scholar] [CrossRef] [Green Version]
- Smith, T.; Quesnel, F.; De Plöeg, G.; De Franceschi, D.; Metais, G.; De Bast, E.; Solé, F.; Folie, A.; Boura, A.; Claude, J.; et al. First Clarkforkian equivalent land mammal age in the latest Paleocene basal Sparnacian facies of Europe: Fauna, flora, paleoenvironment and (bio) stratigraphy. PLoS ONE 2014, 9, e86229. [Google Scholar] [CrossRef] [Green Version]
- Billotte, N.; Marseillac, N.; Brottier, P.; Noyer, J.L.; Jacquemoud-Collet, J.P.; Moreau, C.; Couvreur, T.; Chevallier, M.H.; Pintaud, J.C.; Risterucci, A.M. Nuclear microsatellite markers for the date palm (Phoenix dactylifera L.): Characterization and utility across the genus Phoenix and in other palm genera. Mol. Ecol. Notes 2004, 4, 256–258. [Google Scholar] [CrossRef]
- Elshibli, S.; Korpelainen, H. Microsatellite markers reveal high genetic diversity in date palm (Phoenix dactylifera L.) germplasm from Sudan. Genetica 2008, 134, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Chaluvadi, S.R.; Young, P.; Thompson, K.; Bahri, B.A.; Gajera, B.; Narayanan, S.; Krueger, R.; Bennetzen, J.L. Phoenix phylogeny, and analysis of genetic variation in a diverse collection of date palm (Phoenix dactylifera) and related species. Plant Divers. 2019, 41, 330–339. [Google Scholar] [CrossRef] [PubMed]
- Naqvi, S.A. Date Palm Diversity in Pakistan and Its Relationship with World Dates Germplasm for Exploring the Center of Origin of Phoenix dactylifera L. Ph.D. Thesis, University of Agriculture, Faisalabad, Pakistan, 2015. [Google Scholar]
- Moussouni, S.; Pintaud, J.C.; Vigouroux, Y.; Bouguedoura, N. Diversity of Algerian oases date palm (Phoenix dactylifera L., Arecaceae): Heterozygote excess and cryptic structure suggest farmer management had a major impact on diversity. PLoS ONE 2017, 12, 0175232. [Google Scholar] [CrossRef] [Green Version]
- Zehdi-Azouzi, S.; Cherif, E.; Moussouni, S.; Gros-Balthazard, M.; Abbas Naqvi, S.; Ludeña, B.; Castillo, K.; Chabrillange, N.; Bouguedoura, N.; Bennaceur, M.; et al. Genetic structure of the date palm (Phoenix dactylifera) in the Old World reveals a strong differentiation between eastern and western populations. Ann. Bot. 2015, 116, 101–112. [Google Scholar] [CrossRef] [Green Version]
- Mathew, L.S.; Seidel, M.A.; George, B.; Mathew, S.; Spannagl, M.; Haberer, G.; Torres, M.F.; Al-Dous, E.K.; Al-Azwani, E.K.; Diboun, I.; et al. A genome-wide survey of date palm cultivars supports two major subpopulations in Phoenix dactylifera. Genes Genomes Genet. 2015, 5, 1429–1438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hazzouri, K.M.; Flowers, J.M.; Visser, H.J.; Khierallah, H.S.; Rosas, U.; Pham, G.M.; Meyer, R.S.; Johansen, C.K.; Fresquez, Z.A.; Masmoudi, K.; et al. Whole genome re-sequencing of date palms yields insights into diversification of a fruit tree crop. Nat. Commun. 2015, 6, 8824. [Google Scholar] [CrossRef] [Green Version]
- Mohamoud, Y.A.; Mathew, L.S.; Torres, M.F.; Younuskunju, S.; Krueger, R.; Suhre, K.; Malek, J.A. Novel subpopulations in date palm (Phoenix dactylifera) identified by population-wide organellar genome sequencing. BMC Genom. 2019, 20, 498. [Google Scholar] [CrossRef] [Green Version]
- Flowers, J.M.; Hazzouri, K.M.; Gros-Balthazard, M.; Mo, Z.; Koutroumpa, K.; Perrakis, A.; Ferrand, S.; Khierallah, H.S.; Fuller, D.Q.; Aberlenc, F.; et al. Cross-species hybridization and the origin of North African date palms. Proc. Natl. Acad. Sci. USA 2019, 116, 1651–1658. [Google Scholar] [CrossRef] [Green Version]
- Zango, O.; Cherif, E.; Chabrillange, N.; Zehdi-Azouzi, S.; Gros-Balthazard, M.; Naqvi, S.A.; Lemansour, A.; Rey, H.; Bakasso, Y.; Aberlenc, F. Genetic diversity of Southeastern Nigerien date palms reveals a secondary structure within Western populations. Tree Genet. Genomes 2017, 13, 75. [Google Scholar] [CrossRef]
- Vavilov, N.I. The law of homologous series in variation. J. Genet. 1922, 12, 47–89. [Google Scholar] [CrossRef]
- Carreño, E.; Rivera, D.; Obón, C.; Alcaraz, F.; Johnson, D.; Bartual, J. What are candits? Study of a date palm landrace in Spain belonging to the western cluster of Phoenix dactylifera L. Genet. Resour. Crop Evol. 2021, 68, 135–149. [Google Scholar] [CrossRef]
- Johnson, D.V.; Al-Khayri, J.M.; Jain, S.M. Seedling date palms (Phoenix dactylifera L.) as genetic resources. Emir. J. Food Agric. 2013, 24, 809–830. [Google Scholar] [CrossRef] [Green Version]
- Al-Khayri, J.M.; Jain, S.M.; Johnson, D.V. (Eds.) Date Palm Genetic Resources and Utilization; Springer: Dordrecht, The Netherlands; Berlin/Heidelberg, Germany; London, UK; New York, NY, USA, 2015; Volumes 1 and 2. [Google Scholar]
- Rivera, D.; Obón, C. Hybridization between Sideritis serrata Lag. and Sideritis bourgaeana Boiss. (Lamiaceae) in their hybrid zone in Spain. Ann. Bot. 1990, 66, 147–154. [Google Scholar]
- Bergman, P. Phoenix hybrids: Those promiscuous Phoenix! Palms 2005, 181, 21–23. [Google Scholar]
- Gros-Balthazard, M. Hybridization in the genus Phoenix: A review. Emir. J. Food Agric. 2013, 25, 831–842. [Google Scholar] [CrossRef] [Green Version]
- Hannachi, S.; Khitri, D.; Benkhalifa, A.; Brac De La Perriere, R.A. Inventaire Variétal de la Palmeraie Algérienne; ANEP: Algiers, Algeria, 1998. [Google Scholar]
- Rekis, A. Conservation des Ressources Phytogénétiques en Algérie. Cas des Palmiers Dattiers Cultivés et Sub-Spontanés (Phoenix dactylifera L.). Ph.D. Thesis, Université Mohamed Khider Biskra, Biskra, Algeria, 2021. Available online: http://thesis.univ-biskra.dz/5485/ (accessed on 20 December 2021).









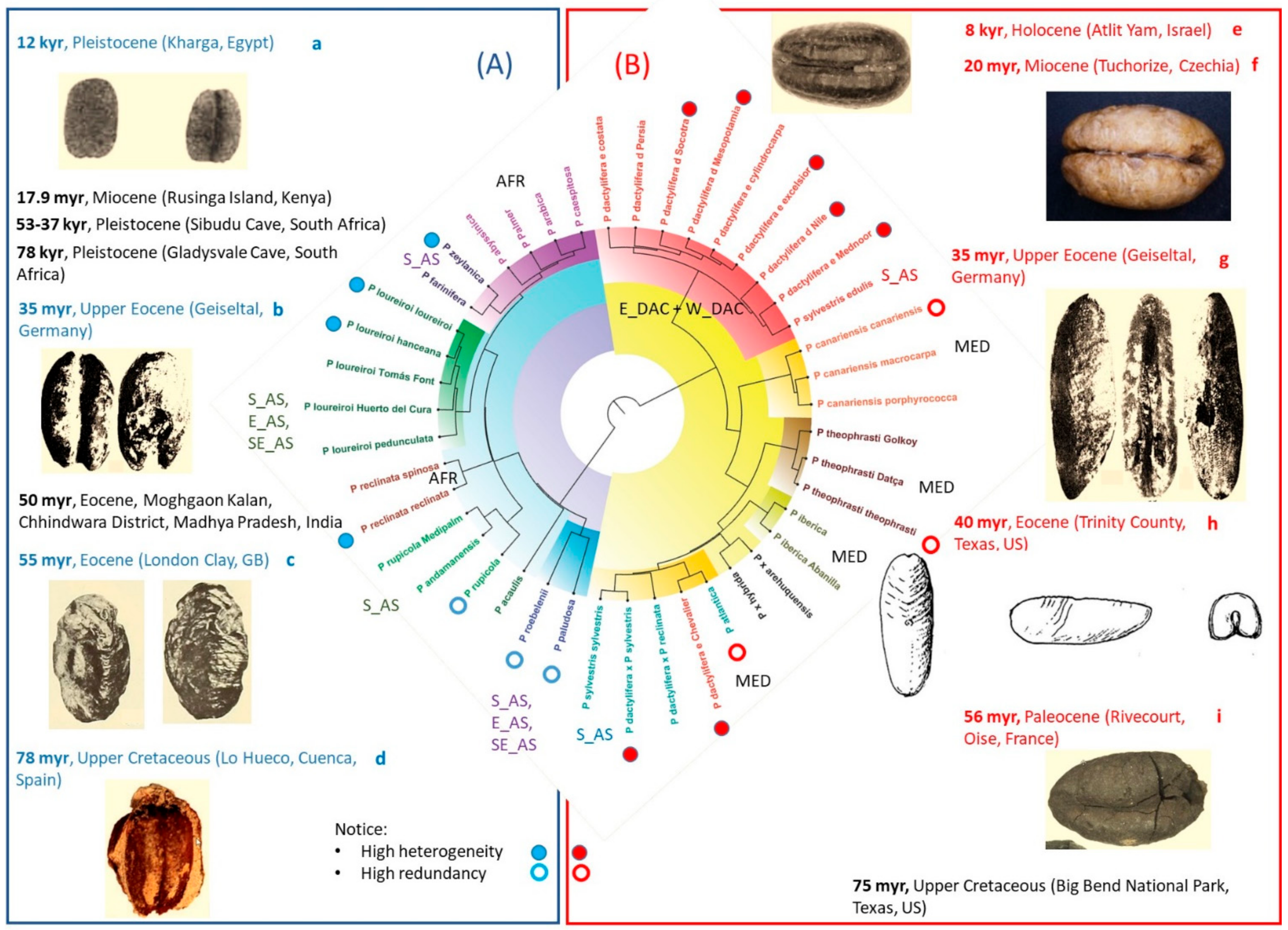

| Recognised Species and Major Groups | OTUs that Include Typical and Related Taxa still under Study and Groups of Landraces |
|---|---|
| Phoenix acaulis Roxb. | P. acaulis (10) 1 |
| Phoenix canariensis H.Wildpret | P. canariensis var. canariensis (24) 1, P. canariensis “Macrocarpa” (20) 1, P. canariensis var. porphyrococca Vasc. & Franco (32) 1 |
| Phoenix dactylifera L. Eastern Group | P. dactylifera “Mesopotamia” (32) 1, P. dactylifera “Nile” (12) 1, P. dactylifera “Persia” (4) 1, P. dactylifera “Socotra” (18) 1 |
| Phoenix dactylifera L. Western Group | P. atlantica A.Chev. (15) 1, P. dactylifera “Chevalier” (8) 1, P. dactylifera var. costata Becc. (10) 1, P. dactylifera var. cylindrocarpa Mart. (66) 1, P. dactylifera “Mednoor” (10) 1, P. excelsior Cav. (61) 1, P. iberica D.Rivera, S.Ríos & Obón (10) 1, P. iberica “Abanilla” (6) 1 |
| Phoenix loureiroi Kunth | P. andamanensis W.Mill., J.G.Sm. & N.Taylor (8) 1, P. hanceana Naudin (8) 1, P. loureiroi “Huerto del Cura” (4) 1, P. loureiroi var. loureiroi (10) 1, P. loureiroi var. pedunculata (Griff.) Govaerts (4) 1, P loureiroi “Tomás Font” (8) 1 |
| Phoenix paludosa Roxb. | P. paludosa Roxb. (15) 1 |
| Phoenix pusilla Gaertn. | P. farinifera Roxb. (10) 1, P. zeylanica Trimen (12) 1 |
| Phoenix reclinata Jacq. | P. abyssinica Drude (10) 1, P. arabica Burret (10) 1, P. caespitosa Chiov. (11) 1, P. reclinata var. reclinata (11) 1, P. “Palmer” (10) 1, P. spinosa Schumach. & Thonn. (8) 1 |
| Phoenix roebelenii O’Brien | P. roebelenii O’Brien (20) 1 |
| Phoenix rupicola T.Anderson | P. rupicola var. rupicola (10) 1, P. rupicola “Medipalm” (5) 1 |
| Phoenix sylvestris (L.) Roxb. | P. sylvestris var. sylvestris (15) 1, P. sylvestris “Edulis” (6) 1 |
| Phoenix theophrasti Greuter | P. theophrasti var. theophrasti (14) 1, P. theophrasti “Datça” (10) 1, P theophrasti “Gölköy” (10) 1 |
| Hybrids | |
| Phoenix canariensis × P. reclinata | P. canariensis × P. reclinata (7) 1 |
| Phoenix dactylifera × P. canariensis | P. dactylifera × P. canariensis (12) 1 |
| Phoenix dactylifera × P. reclinata | P. dactylifera × P. reclinata (4) 1 |
| Phoenix dactylifera × P. sylvestris | P. dactylifera × P. sylvestris (6) 1 |
| Group | Subgroups | Characters | States |
|---|---|---|---|
| Category | Evaluation | 2 | 7 |
| Phenology | 2 | 11 | |
| Vegetative | 42 | 163 | |
| Male reproductive | 11 | 31 | |
| Female reproductive | 13 | 46 | |
| Fruit and seed | 46 | 191 | |
| Type | Qualitative ordinal | 12 | 43 |
| Quantitative discrete | 7 | 37 | |
| Quantitative continuous discretized | 48 | 231 | |
| Qualitative nominal | 29 | 104 | |
| Binary qualitative | 6 | 6 | |
| Binary qualitative duplicate | 14 | 28 |
| Group | Heterogeneity | Normalized Heterogeneity |
|---|---|---|
| Total | 2.49 | 1 |
| Fruit and seed | 2.04 | 0.82 |
| Reproductive | 1.99 | 0.79 |
| Vegetative | 1.50 | 0.61 |
| Female inflorescence and flower | 1.04 | 0.42 |
| Male inflorescence and flower | 0.87 | 0.35 |
| Taxa | Geog. | AVarIntr (Average) 1 | Heter. 2 | Redund. 3 | Pond. Heter. | Accessions | SF/SM |
|---|---|---|---|---|---|---|---|
| P. abyssinica Drude | AFR | 0.32 | 2.03 | 0.39 | 1.72 | 10 | 1.00 |
| P. acaulis Roxb. | S_AS | 0.31 | 2.26 | 0.32 | 2.02 | 10 | 1.00 |
| P. andamanensis W.Mill., J.G.Sm. & N.Taylor | S_AS | 0.31 | 2.23 | 0.33 | 2.17 | 8 | 3.00 |
| P. arabica Burret | AFR | 0.33 | 2.16 | 0.35 | 2.00 | 10 | 1.00 |
| P. atlantica A.Chev. | MED | 0.31 | 2.48 | 0.25 | 1.96 | 15 | 4.00 |
| P. caespitosa Chiov. | AFR | 0.34 | 2.37 | 0.29 | 2.09 | 11 | 1.20 |
| P. canariensis var. canariensis | MED | 0.33 | 2.35 | 0.29 | 1.77 | 24 | 3.80 |
| P. canariensis “Macrocarpa” | MED | 0.32 | 2.48 | 0.25 | 1.89 | 20 | 3.00 |
| P. canariensis var. porphyrococca Vasc. & Franco | MED | 0.33 | 2.79 | 0.16 | 2.32 | 32 | 1.91 |
| P. dactylifera “Mesopotamia” | E_DAC | 0.33 | 2.86 | 0.14 | 2.89 | 32 | 1.00 |
| P. dactylifera “Nile” | E_DAC | 0.32 | 2.65 | 0.20 | 2.68 | 12 | 1.00 |
| P. dactylifera “Persia” | E_DAC | 0.33 | 1.53 | 0.54 | 2.01 | 4 | 1.00 |
| P. dactylifera “Socotra” | E_DAC | 0.33 | 2.81 | 0.15 | 2.99 | 18 | 1.00 |
| P. dactylifera “Chevalier” | W_DAC | 0.33 | 2.10 | 0.37 | 2.56 | 8 | 1.00 |
| P. dactylifera var. costata Becc. | W_DAC | 0.32 | 2.41 | 0.27 | 2.46 | 10 | 1.00 |
| P. dactyliferavar. cylindrocarpa Mart. | W_DAC | 0.32 | 2.86 | 0.14 | 2.45 | 66 | 1.00 |
| P. excelsior Cav. | W_DAC | 0.33 | 3.00 | 0.10 | 2.58 | 61 | 3.36 |
| P. dactylifera “Mednoor” | W_DAC | 0.33 | 2.75 | 0.17 | 3.18 | 10 | 1.00 |
| P. farinifera Roxb. | S_AS | 0.35 | 2.36 | 0.29 | 2.37 | 10 | 1.00 |
| P. iberica D.Rivera, S.Ríos & Obón | MED | 0.35 | 2.49 | 0.25 | 2.49 | 10 | 1.00 |
| P. iberica “Abanilla” | MED | 0.32 | 1.97 | 0.41 | 2.09 | 6 | 2.00 |
| P. hanceana Naudin | E_AS | 0.39 | 2.24 | 0.33 | 2.65 | 8 | 1.00 |
| P. loureiroi “Huerto del Cura” | S_AS | 0.35 | 1.46 | 0.56 | 1.97 | 4 | 1.00 |
| P. loureiroivar. loureiroi | SE_AS | 0.34 | 2.77 | 0.16 | 2.96 | 10 | 1.00 |
| P.loureiroi var. pedunculata (Griff.) Govaerts | S_AS | 0.34 | 1.44 | 0.57 | 1.88 | 4 | 1.00 |
| P. loureiroi “Tomás Font” | S_AS | 0.39 | 1.00 | 0.70 | 0.96 | 8 | 1.00 |
| P. “Palmer” | AFR | 0.37 | 1.88 | 0.43 | 1.83 | 10 | 1.00 |
| P. paludosa Roxb. | S_AS, SE_AS | 0.31 | 2.42 | 0.27 | 1.86 | 15 | 2.00 |
| P. reclinatavar. reclinata | AFR | 0.32 | 2.75 | 0.17 | 2.55 | 11 | 1.75 |
| P. spinosa Schumach. & Thonn. | AFR | 0.31 | 2.02 | 0.39 | 1.89 | 8 | 3.00 |
| P. roebelenii O’Brien | SE_AS | 0.32 | 2.49 | 0.25 | 1.79 | 20 | 2.33 |
| P. rupícola var. rupícola | S_AS | 0.32 | 1.95 | 0.41 | 1.62 | 10 | 1.00 |
| P. rupicola “Medipalm” | S_AS | 0.34 | 1.37 | 0.59 | 1.48 | 5 | 1.50 |
| P. sylvestris “Edulis” | S_AS | 0.32 | 1.81 | 0.46 | 1.86 | 6 | 1.00 |
| P. sylvestris var. sylvestris | S_AS | 0.33 | 2.63 | 0.21 | 2.20 | 15 | 2.00 |
| P. theophrasti “Datça” | MED | 0.34 | 2.45 | 0.26 | 2.43 | 10 | 1.00 |
| P theophrasti “Gölköy” | MED | 0.34 | 2.29 | 0.31 | 2.00 | 10 | 1.00 |
| P. theophrasti var. theophrasti | MED | 0.36 | 2.30 | 0.31 | 2.05 | 14 | 1.00 |
| P. zeylanica Trimen | S_AS | 0.33 | 2.80 | 0.16 | 2.68 | 12 | 1.40 |
| Hybrids | |||||||
| P. canariensis × P. reclinata | - | 0.35 | 2.01 | 0.39 | 2.26 | 7 | 2.50 |
| P. dactylifera × P. canariensis | - | 0.35 | 2.20 | 0.34 | 1.99 | 12 | 5.00 |
| P. dactylifera × P. reclinata | - | 0.31 | 1.38 | 0.59 | 1.66 | 4 | 1.00 |
| P. dactylifera × P. sylvestris | - | 0.34 | 2.17 | 0.35 | 3.04 | 6 | 2.00 |
| Recognised Species and Major Groups | Geog. | AVarIntr (Range) 1 | Heterogeneity 2 | Redundancy | Pondered Heterogeneity 3 | SM 4 | SF 4 |
|---|---|---|---|---|---|---|---|
| Phoenix acaulis Roxb. | S_AS | 0.31 | 2.26 | 0.32 | 2.02 | 5 | 5 |
| Phoenix canariensis H.Wildpret | MED | 0.32–0.33 | 2.35–2.79, mean 2.54 | 0.16–0.29, mean 0.23 | 1.77–2.32, mean 1.99 | 21 | 55 |
| Phoenix dactylifera L. Eastern Group | E_DAC | 0.32–0.32 | 1.53–2.86, mean 2.46 | 0.14–0.54, mean 0.26 | 2.01–2.99, mean 2.64 | 33 | 33 |
| Phoenix dactylifera L. Western Group | W_DAC | 0.31–0.35 | 2.10–3.0, mean 2.62 | 0.10–0.37, mean 0.21 | 2.45–3.18, mean 2.65 | 61 | 94 |
| Phoenix loureiroi Kunth | S_AS, E_AS, SE_AS | 0.31–0.39 | (1) 5 1.46–2.77, mean 2.17 | 0.16–0.56 (0.70) 5, mean 0.35 | (0.96) 5 1.97–2.96, mean 2.53 | 15 | 15 |
| Phoenix paludosa Roxb. | S_AS, SE_AS | 0.31 | 2.42 | 0.27 | 1.86 | 5 | 10 |
| Phoenix pusilla Gaertn. | S_AS | 0.33–0.35 | 2.36–2.80, mean 2.58 | 0.16–0.29, mean 0.23 | 2.37–2.68, mean 2.53 | 10 | 12 |
| Phoenix reclinata Jacq. | AFR | 0.31–0.37 | (1.88) 5 2.02–2.75, mean 2.26 | 0.17–0.39 (0.43) 5, mean 0.32 | (1.83) 5 1.72–2.55, mean 2.05 | 26 | 34 |
| Phoenix roebelenii O’Brien | SE_AS | 0.32 | 2.49 | 0.25 | 1.79 | 6 | 14 |
| Phoenix rupicola T.Anderson | S_AS | 0.32–0.34 | (1.37) 5 1.95 | 0.41 (0.59) 5 | (1.62) 5 1.82 | 7 | 8 |
| Phoenix sylvestris (L.) Roxb. | S_AS | 0.32–0.33 | 1.81–2.63, mean 2.22 | 0.21–0.46, mean 0.34 | 1.86–2.2, mean 2.03 | 8 | 13 |
| Phoenix theophrasti Greuter | MED | 0.34–0.36 | 2.1–2.45, mean 2.28 | 0.26–0.37, mean 0.33 | 2–2.43, mean 2.16 | 17 | 17 |
| Hybrids | |||||||
| Phoenix canariensis × P. reclinata | - | 0.35 | 2.01 | 0.39 | 2.26 | 2 | 5 |
| Phoenix dactylifera × P. canariensis | - | 0.35 | 2.20 | 0.34 | 1.99 | 2 | 10 |
| Phoenix dactylifera × P. reclinata | - | 0.31 | 1.36 | 0.59 | 1.66 | 2 | 2 |
| Phoenix dactylifera × P. sylvestris | - | 0.34 | 2.17 | 0.35 | 3.04 | 2 | 4 |
| Distinctive Characters | Western Syndrome | W Cultivars Examples | Eastern Syndrome | E Cultivars Examples |
|---|---|---|---|---|
| Leaf | ||||
| Basal neck length in the upper spines | 0–15 mm | Abel, Alig, Candits, Criollo, Deglet Nour, Ghars, Haziz | 15–30 (60) mm | Apdandon, Barhee, Dairy, Khalas, Koroch, Lulu |
| Leaf base width | 15–30 (35) cm | Candits, Medjool, Rhars, Thoory | 8–20 (25) cm | Apdandon, Korosh, Sarafana, Sayir, Sudra |
| Spines (acanthophylls) * | 10–30 | Abel, Abu Faqqus, Alig, Blonde Beauty, Criollo, Deglet Nour, Ghars, Maurs, Thoory | 2–17 | Khadrawy, Kustawy, Sudra, Sarafana |
| Fruits | ||||
| Fruiting Peduncle Colour | Orange, greenish orange or reddish orange | Beser Helou, Candits, Criollo, Medjool, Thoory | Greenish yellow, yellow or orange | Barhee, Braim, Dairy, Lulu, Sarafana, Sudra |
| Fruit Apex | Obtuse | Aziza, Candits, Maurs, Rhars, Tenats, Thoory | Obtuse, acute, truncate, mucronate, ovate-oblique, retuse emarginate | Barhee, Braim, Dairy, Kustawy, Maktoom, Piarom |
| Epicarp | Epicarp usually not adherent | Abel, Alig, Candits, Medjool, Thoory | Epicarp usually adherent | Barhee, Braim, Dairy, Piarom, Sayir |
| Seeds | ||||
| Seed width | 6–12 mm | Abel, Candits, Criollo, Ghars, Medjool, Tenats | 5–10 mm | Apdandon, Kessab, Maktoom, Muzawati, Sarafana, Sudra, Zahidi |
| Seed shape | Seed ovate to triangular, elliptical, elliptical-oblong, but also cylindrical-narrow | Blonde Beauty, Brunette Beauty, Candits, Criollo, Deglet Nour, Haziz, Medjool Thoory | Seed usually cylindrical-narrow | Apdandon, Dairy, Halawy, Khadrawy, Khalas, Muzawaty, Sarafana, Sayir, Sudra, Zahidi |
| Seed ventral furrow | Seed ventral furrow shallow or U-shaped, rarely deep V-shaped | Abel, Alig, Blonde Beauty, Brunette Beauty, Deglet Nour, Haziz, Medjool, San Ignacio | Seed ventral furrow usually deep V-shaped, less often U-shaped | Apdandon, Dairy, Halawy, Khadrawy, Khalas, Maktoom, Muzawaty, Sarafana, Sayir, Sudra, Zahidi |
| Seed surface | Rough or irregular, rarely smooth | Aziza, Beser Helou, Candits, Criollo, Medjool, Rhars, Tenats, Thoory | About 40% are smooth, the rest rough or irregular | Apdandon, Koroch, Lulu, Muzawati, Piarom |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rivera, D.; Alcaraz, F.; Rivera-Obón, D.J.; Obón, C. Phenotypic Diversity in Wild and Cultivated Date Palm (Phoenix, Arecaceae): Quantitative Analysis Using Information Theory. Horticulturae 2022, 8, 287. https://doi.org/10.3390/horticulturae8040287
Rivera D, Alcaraz F, Rivera-Obón DJ, Obón C. Phenotypic Diversity in Wild and Cultivated Date Palm (Phoenix, Arecaceae): Quantitative Analysis Using Information Theory. Horticulturae. 2022; 8(4):287. https://doi.org/10.3390/horticulturae8040287
Chicago/Turabian StyleRivera, Diego, Francisco Alcaraz, Diego J. Rivera-Obón, and Concepción Obón. 2022. "Phenotypic Diversity in Wild and Cultivated Date Palm (Phoenix, Arecaceae): Quantitative Analysis Using Information Theory" Horticulturae 8, no. 4: 287. https://doi.org/10.3390/horticulturae8040287
APA StyleRivera, D., Alcaraz, F., Rivera-Obón, D. J., & Obón, C. (2022). Phenotypic Diversity in Wild and Cultivated Date Palm (Phoenix, Arecaceae): Quantitative Analysis Using Information Theory. Horticulturae, 8(4), 287. https://doi.org/10.3390/horticulturae8040287









