Genetic Analysis of the Grapevine GATA Gene Family and Their Expression Profiles in Response to Hormone and Downy Mildew Infection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification of GATA Family Genes in Grapevine
2.2. Phylogenetic Analysis, Protein Structural Analysis, and Identification of Promoter Cis-Elements
2.3. Plant Material, Fungal Materials, and Inoculation
2.4. Callus Cells Treatments
2.5. RNA Extraction and Expression Pattern Analysis
2.6. Confocal Imaging and Transcriptional Assay
3. Results
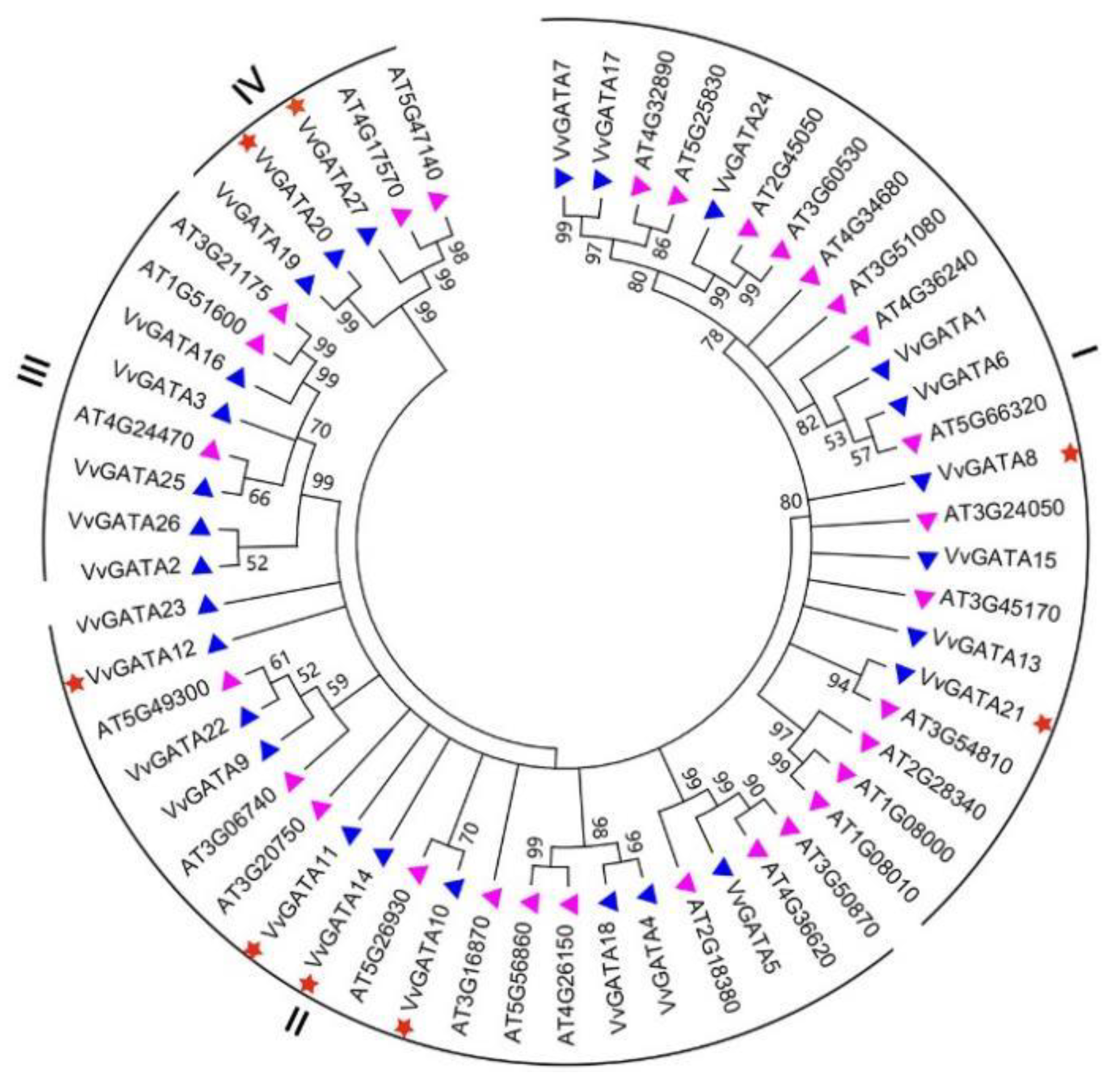
3.1. Identification of GATA Transcription Factor Genes in V. vinifera
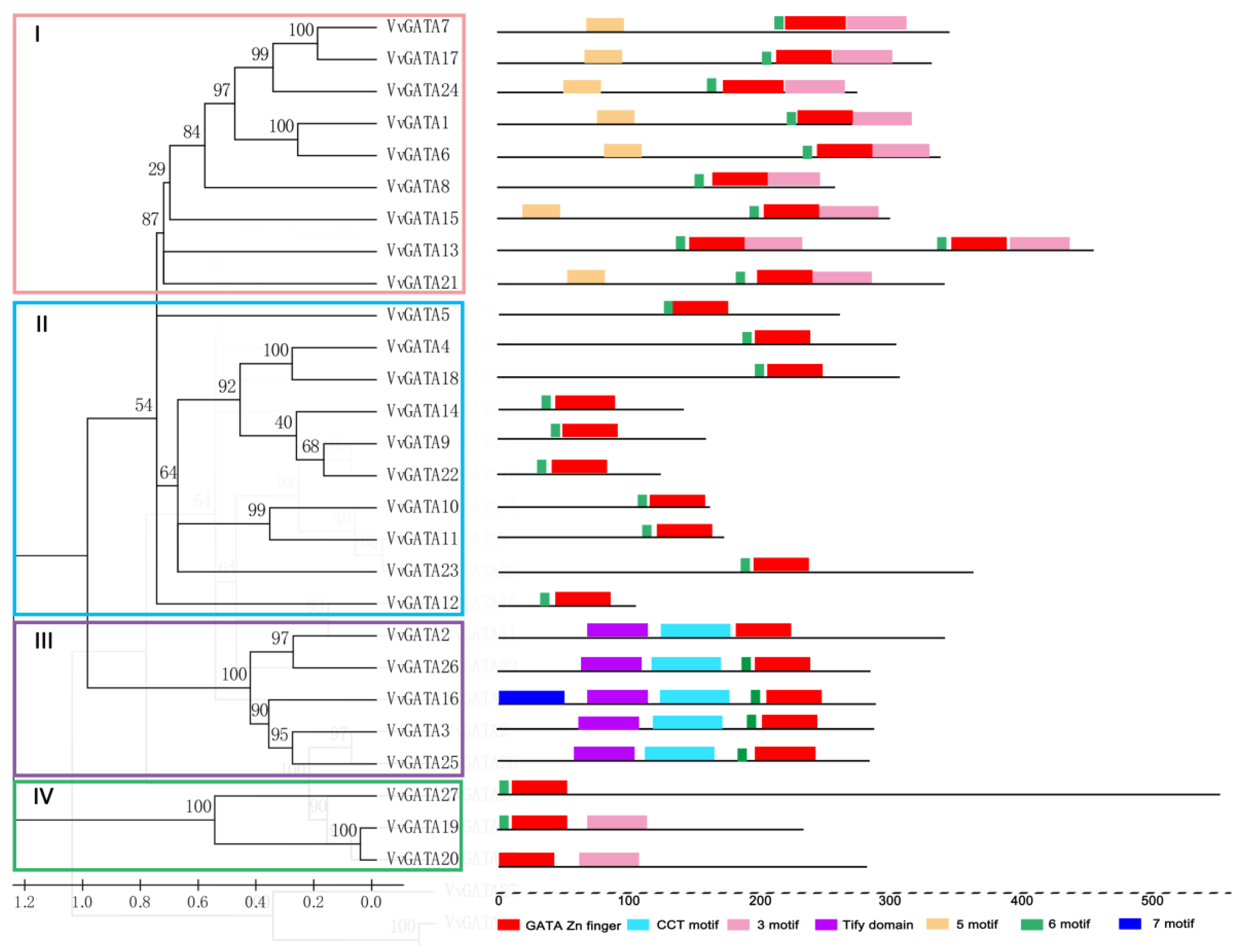
3.2. Phylogenetic Analysis, Conserved Motifs, and Structural Features of the V. vinifera GATA Gene Family
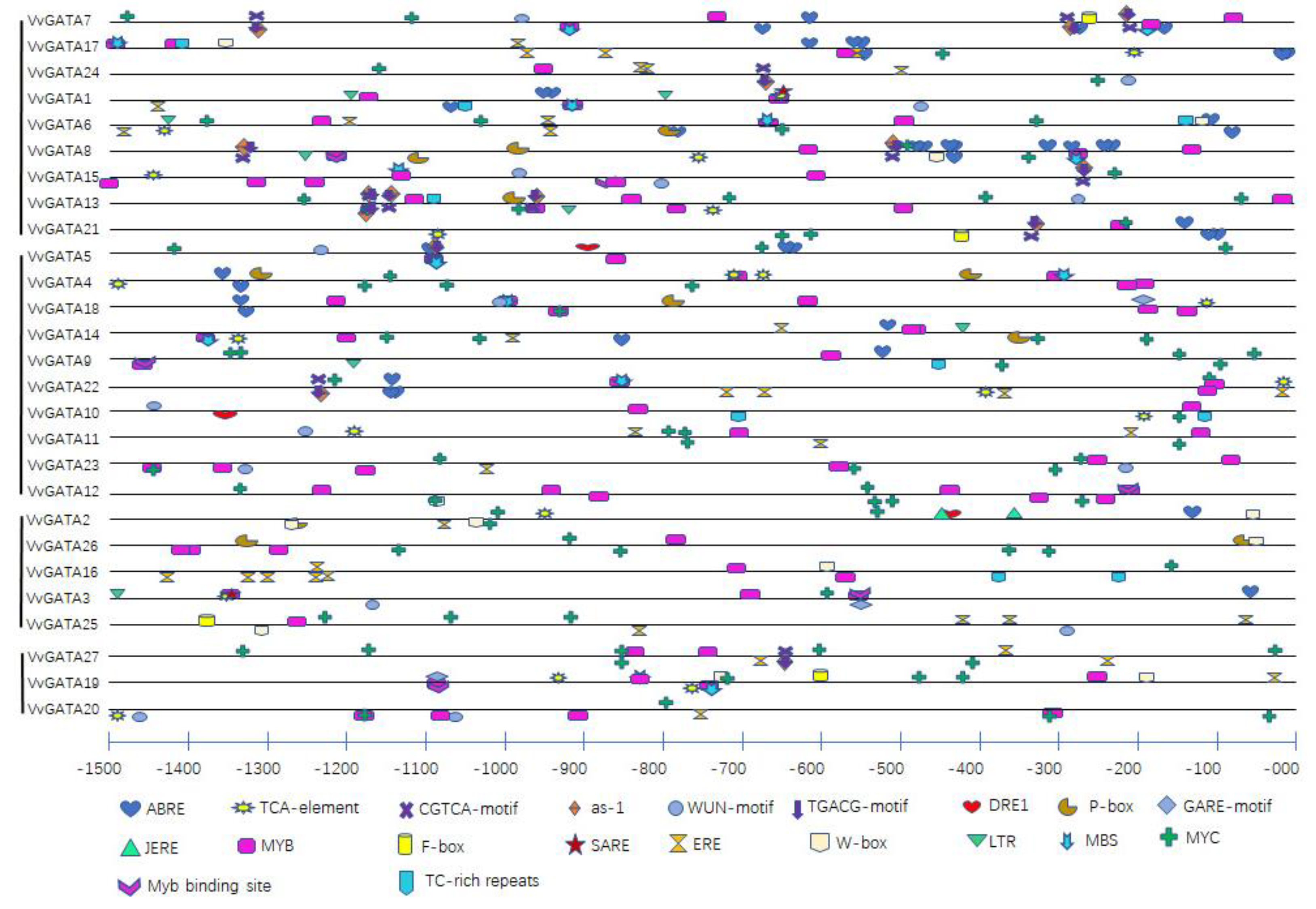
3.3. Cis-Elements in the Promoter Region of VvGATA Factors
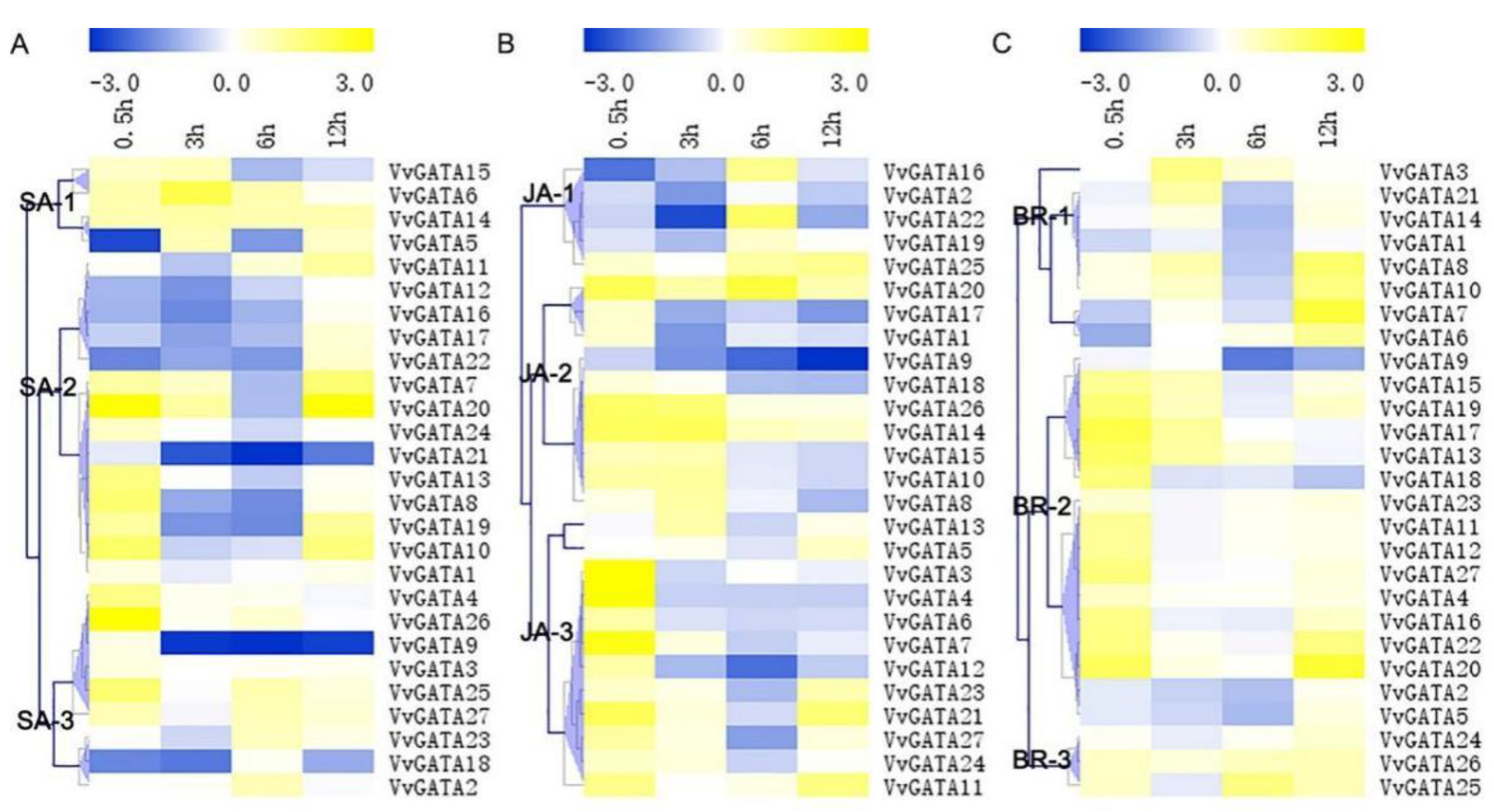
3.4. Expression Profiles of the VvGATA Genes in Response to Hormones
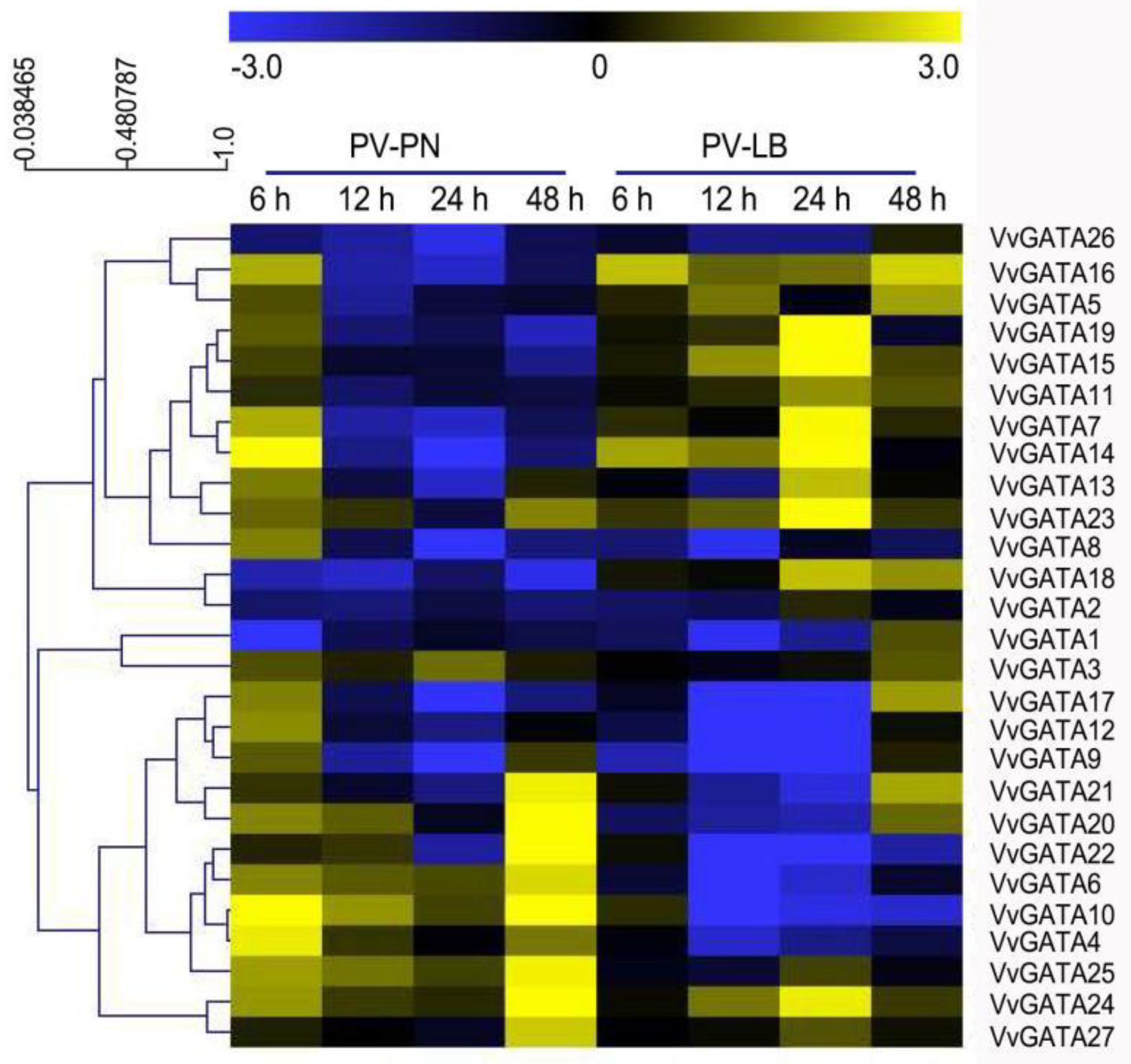
3.5. Expression Profiles of the VvGATA Genes in Response to P. viticola
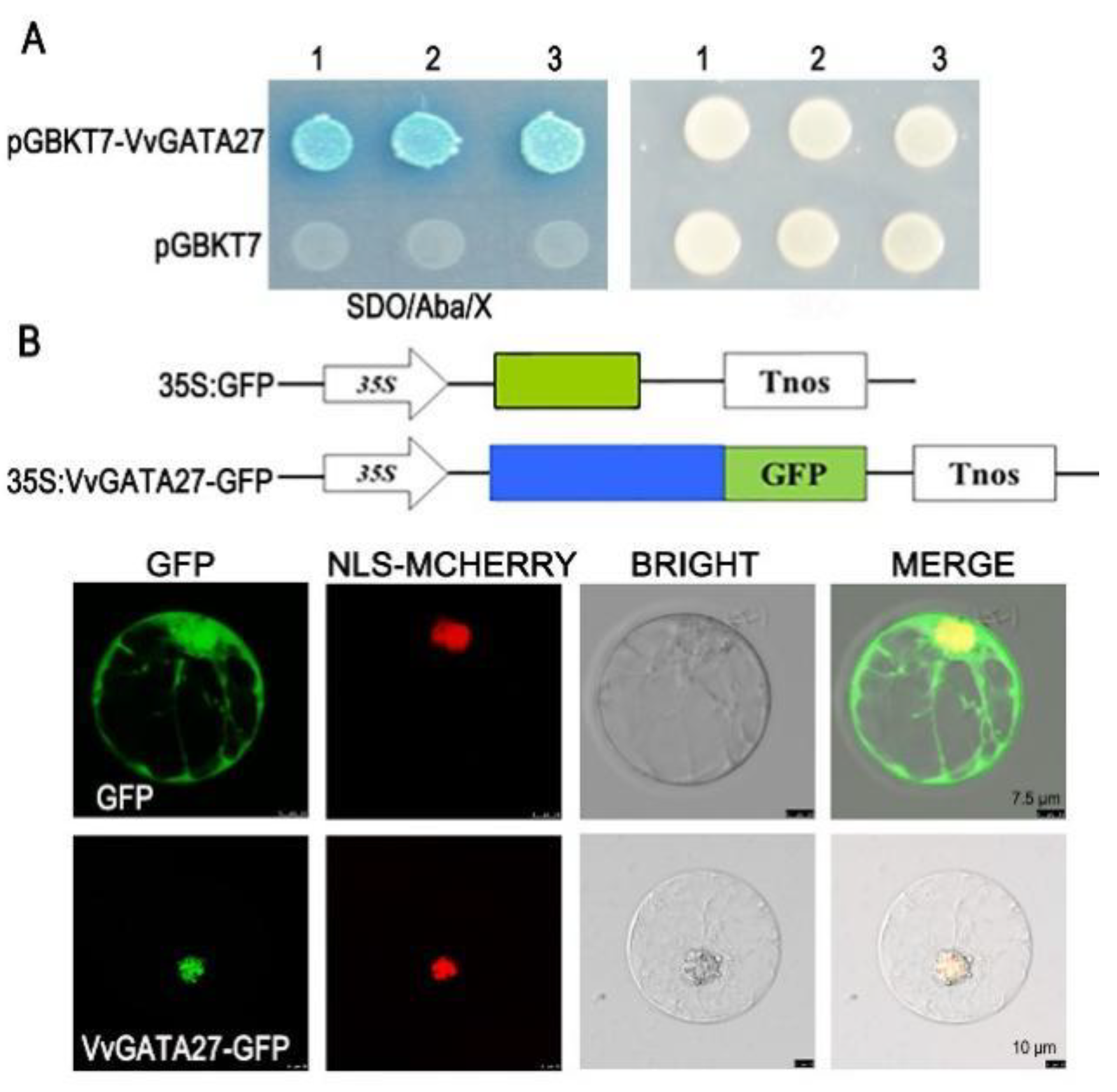
3.6. VvGATA27 Localizes to the Nucleus and Shows Transcriptional Activity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, R.; Wang, L.; Zhu, J.; Chen, T.; Wang, Y.; Xu, Y. Histological responses to downy mildew in resistant and susceptible grapevines. Protoplasma 2015, 252, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Weng, K.; Dou, M.; Chen, T.; Yin, X.; Li, Z.; Li, T.; Zhang, C.; Xiang, G.; Liu, G.; et al. Transcriptomic analysis of Chinese wild Vitis pseudoreticulata in response to Plasmopara viticola. Protoplasma 2019, 256, 1409–1424. [Google Scholar] [CrossRef] [PubMed]
- Pap, D.; Riaz, S.; Dry, I.B.; Jermakow, A.; Tenscher, A.C.; Cantu, D.; Oláh, R.; Walker, M.A. Identification of two novel powdery mildew resistance loci, Ren6 and Ren7, from the wild Chinese grape species Vitis piasezkii. BMC Plant Biol. 2016, 16, 170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, X.; Liu, R.Q.; Su, H.; Su, L.; Guo, Y.R.; Wang, Z.J.; Wei Du, W.; Li, M.J.; Zhang, X.; Wang, Y.J.; et al. Pathogen development and host responses to Plasmopara viticola in resistant and susceptible grapevines, an ultrastructural study. Hortic. Res. 2017, 4, 17033. [Google Scholar] [CrossRef]
- Staudt, G.; Kassemeyer, H.H. Evaluation of downy mildew resistance in various accessions of wild Vitis species. Vitis 1995, 34, 225–228. [Google Scholar]
- Wan, Y.Z.; Schwaniniger, H.; He, P.C.; Wang, Y.J. Comparison of resistance to powdery mildew and downy mildew in Chinese wild grapes. Vitis 2007, 46, 132–136. [Google Scholar]
- Riaz, S.; Boursiquot, J.M.; Dangl, G.S.; Lacombe, T.; Laucou, V.; Tenscher, A.C.; Walker, M.A. Identification of mildew resistance in wild and cultivated Central Asian grape germplasm. BMC Plant Biol. 2013, 13, 149. [Google Scholar] [CrossRef] [Green Version]
- Vergnes, S.; Ladouce, N.; Fournier, S.; Ferhou, H.; Attia, F.; Dumas, B. Foliar treatments with Gaultheria procumbens essential oil induce defense responses and resistance against a fungal pathogen in Arabidopsis. Front. Plant Ence. 2014, 5, 477. [Google Scholar] [CrossRef]
- Cheong, Y.H.; Chang, H.S.; Gupta, R.; Wang, X.; Zhu, T.; Luan, S. Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis. Plant Physiol. 2002, 129, 661–677. [Google Scholar] [CrossRef] [Green Version]
- Fan, C.; Yao, H.; Qiu, Z.; Ma, H.; Zeng, B. Genome-wide analysis of Eucalyptus grandis WRKY genes family and their expression profiling in response to hormone and abiotic stress treatment. Gene 2018, 678, 38–48. [Google Scholar] [CrossRef]
- Li, T.; Cheng, X.; Wang, Y.; Yin, X.; Li, Z.; Liu, R.; Liu, G.; Xu, Y. Genome-wide analysis of glyoxalase-like gene families in grape (vitis vinifera l.) and their expression profiling in response to downy mildew infection. BMC Genom. 2019, 20, 362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lowry, J.A.; Atchley, W.R. Molecular evolution of the GATA family of transcription factors, conservation within the DNA-binding Domain. J. Mol. Evol. 2000, 50, 103–115. [Google Scholar] [CrossRef] [PubMed]
- Reyes, J.C.; Muro-Pastor, M.I.; Florencio, F.J. The GATA Family of Transcription Factors in Arabidopsis and Rice. Plant Physiol. 2004, 134, 1718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villamayor, L.; Cano, D.A.; Rojas, A. GATA factors in pancreas development and disease. Int. Union Biochem. Mol. Biol. Life 2019, 72, 80–88. [Google Scholar] [CrossRef] [PubMed]
- Klermund, C.; Ranftl, Q.L.; Diener, J.; Bastakis, E.; Richter, R.; Schwechheimer, C. LLM-domain B-GATA transcription factors promote stomatal development downstream of light signaling pathways in Arabidopsis thaliana hypocotyls. Plant Cell 2016, 28, 646–660. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Zhou, Y.; Ding, L.; Wu, Z.; Liu, R.; Meyerowitz, E.M. Transcription repressor HANABA TARANU controls flower development by integrating the actions of multiple hormones, floral organ specification genes, and GATA3 family genes in Arabidopsis. Plant Cell 2013, 25, 83. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z. The Role of Anthocyanins and The GATA Transcription Factors GNC and CGA1 in The Plant Response to Stress. Ph.D. Thesis, The University of Guelph, Guelph, Canada, 2016. [Google Scholar]
- Chiang, Y.H.; Zubo, Y.O.; Tapken, W.; Kim, H.J.; Lavanway, A.M.; Howard, L.; Pilon, M.; Kieber, J.J.; Schaller, E.G. Functional characterization of the GATA transcription factors GNC and CGA1 reveals their key role in chloroplast development, growth, and division in Arabidopsis. Plant Physiol. 2012, 160, 332. [Google Scholar] [CrossRef] [Green Version]
- Hudson, D.; Guevara, D.R.; Hand, A.J.; Xu, Z.; Hao, L.; Chen, X.; Zhu, T.; Bi, Y.M.; Rothstein, S.J. Rice Cytokinin GATA Transcription Factor1 Regulates Chloroplast Development and Plant Architecture. Plant Physiol. 2013, 162, 132–144. [Google Scholar] [CrossRef] [Green Version]
- An, Y.; Han, X.; Tang, S.; Xia, X.; Yin, W. Poplar GATA transcription factor PdGNC, is capable of regulating chloroplast ultrastructure, photosynthesis, and vegetative growth in Arabidopsis, under varying nitrogen levels. Plant Cell Tissue Organ Cult. 2014, 119, 313–327. [Google Scholar] [CrossRef]
- Lu, G.; Casaretto, J.A.; Ying, S.; Mahmood, K.; Liu, F.; Bi, Y.M.; Rothstein, S.J. Overexpression of OsGATA12 regulates chlorophyll content, delays plant senescence and improves rice yield under high density planting. Plant Mol. Biol. 2017, 94, 215–227. [Google Scholar] [CrossRef]
- Wang, T.; Yang, Y.; Lou, S.; Wei, W.; Zhao, Z.; Ren, Y.; Lin, C.; Ma, L. Genome-Wide Characterization and Gene Expression Analyses of GATA Transcription Factors in Moso Bamboo (Phyllostachys edulis). Int. J. Mol. Sci. 2020, 21, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhardwaj, A.R.; Joshi, G.; Kukreja, B. Global insights into high temperature and drought stress regulated genes by RNA-Seq in economically important oilseed crop Brassica juncea. BMC Plant Biol. 2015, 15, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, X.M.; Lin, W.H.; Zhu, S.; Zhu, J.Y.; Sun, Y.; Fan, X.Y.; Cheng, M.; Hao, Y.; Oh, E.; Tian, M.; et al. Integration of Light- and Brassinosteroid-Signaling Pathways by a GATA Transcription Factor in Arabidopsis. Dev. Cell 2010, 19, 872–883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, K.; Wang, X.; Zhu, W.; Qin, X.; Xu, J.; Cheng, C.; Lou, Q.; Li, J.; Chen, J. Complete resistance to powdery mildew and partial resistance to downy mildew in a Cucumis hystrix, introgression line of cucumber were controlled by a co-localized locus. Theor. Appl. Genet. 2018, 131, 2229–2243. [Google Scholar] [CrossRef]
- Yu, Y.H.; Bian, L.; Yu, K.K.; Yang, S.D.; Zhang, G.H.; Guo, D.L. Grape (Vitis davidii) VdGATA2 functions as a transcription activator and enhances powdery mildew resistance via the active oxygen species pathway. Sci. Hortic. 2020, 267, 109327. [Google Scholar] [CrossRef]
- Zhang, Z.; Ren, C.; Zou, L.; Wang, Y.; Li, S.; Liang, Z. Characterization of the GATA gene family in Vitis vinifera, genome-wide analysis, expression profiles, and involvement in light and phytohormone response. Genome 2018, 61, 713–723. [Google Scholar] [CrossRef] [Green Version]
- Jin, J.; Tian, F.; Yang, D.C.; Meng, Y.Q.; Gao, G. PlantTFDB 4.0, Toward a central hub for transcription factors and regulatory interactions in plants. Nuclc Acids Res. 2016, 1040–1045. [Google Scholar] [CrossRef] [Green Version]
- Eddy, S.R. Profile hidden Markov models. Bioinformatics 1998, 14, 755–763. [Google Scholar] [CrossRef]
- Wang, L.; Li, Y.; Dai, W.; Yan, J.; Zhang, C. Establishment and optimization of grape cell suspension culture system. Biotechnol. Bull. 2018, 34, 86–92. [Google Scholar]
- Sun, H.; Jiang, X.; Sun, M.; Cong, H.; Qiao, F. Evaluation of reference genes for normalizing RT-qPCR in leaves and suspension cells of Cephalotaxus hainanensis under various stimuli. Plant Methods 2019, 15, 31. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Zhang, C.; Guo, Y.; Niu, W.; Wang, Y. Evolution and expression analysis reveal the potential role of the HD-Zip gene family in regulation of embryo abortion in grapes (Vitis vinifera L.). BMC Genom. 2017, 18, 744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, K. Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. Plant J. 2009, 59, 150–162. [Google Scholar] [CrossRef]
- Yu, C.; Li, N.; Yin, Y.; Wang, F.; Gao, S.; Jiao, C. Genome-wide identification and function characterization of GATA transcription factors during development and in response to abiotic stresses and hormone treatments in pepper. J. Appl. Genet. 2021, 7, 3019. [Google Scholar] [CrossRef] [PubMed]
- Timothy, L.B.; Gribskov, M. Combining evidence using p-values, application to sequence homology searches. Bioinformatics 1998, 14, 48–54. [Google Scholar]
- Zhao, Y.; Medrano, L.; Ohashi, K.; Fletcher, J.C.; Yu, H.; Sakai, H.; Meyerowitz, E.M. HANABA TARANU is a GATA transcription factor that regulates shoot apical meristem and flower development in Arabidopsis. Plant Cell 2004, 16, 2586–2600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chattopadhyay, S.; Puente, P.; Deng, X.W.; Wei, N. Combinatorial interaction of light-responsive elements plays a critical role in determining the response characteristics of light-regulated promoters in Arabidopsis. Plant J. 1998, 15, 69–77. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, X.; Wang, X.; Zhou, M.; Zhou, X.; Ye, X.; Wei, X. An R2R3 MYB transcription factor in wheat, TaPIMP1, mediates host resistance to Bipolaris sorokiniana and drought stresses through regulation of defenseand stress-related genes. New Phytol. 2012, 196, 1155–1170. [Google Scholar] [CrossRef] [PubMed]
| Gene Name | Gene Cribi ID | Genome Location | GeneBank Accession No. | Motif | Length (aa) | E-Value | Used Name (NCBI, Vitis Vinifera) | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Subgroup | znF-GATA | Tify | Another | |||||||
| VvGATA1 | VIT_03s0038g00490 | chr3, 452898..453933 | XM_002274836.3 | I | 1 | 317 | 4.33 × 10−13 | GATA transcription factor 5 | ||
| VvGATA2 | VIT_03s0038g00580 | chr3_random, 512571..559934 | XM_019217517.1 | III | 1 | 1 | CCT | 353 | 4.10 × 10−15 | GATA transcription factor 19 |
| VvGATA3 | VIT_03s0038g00480 | chr3_random, 452201..461888 | XM_002270325.4 | III | 1 | 1 | CCT | 302 | 6.56 × 10−13 | GATA transcription factor 24 |
| VvGATA4 | VIT_04s0008g01290 | chr4, 1062865..1063577 | XM_002279247.3 | II | 1 | 306 | 4.49 × 10−20 | GATA transcription factor 22 | ||
| VvGATA5 | VIT_04s0023g01840 | chr4, 18421693..18422139 | XM_010650837.2 | II | 1 | 249 | 2.00 × 10−17 | GATA transcription factor 18 | ||
| VvGATA6 | VIT_04s0023g02880 | chr4, 19468180..19469275 | XM_002272726.3 | I | 1 | 338 | 1.28 × 10−15 | GATA transcription factor 5 | ||
| VvGATA7 | VIT_04s0008g03270 | chr4, 2730051..2730368 | XM_002283709.3 | I | 1 | 342 | 1.60 × 10−16 | GATA transcription factor 9-like | ||
| VvGATA8 | VIT_05s0051g00450 | chr5, 11067051..11068106 | XM_010651913.1 | I | 1 | 258 | 5.13 × 10−15 | GATA transcription factor 1 | ||
| VvGATA9 | VIT_05s0077g01450 | chr5, 1164009..1164539 | XM_010651243.2 | II | 1 | 153 | 6.12 × 10−13 | GATA transcription factor 16 | ||
| VvGATA10 | VIT_206s0004g01275 | chr6, 1521644..1522690 | XM_019220522.1 | II | 1 | 162 | 5.08 × 10−19 | GATA transcription factor 12-like | ||
| VvGATA11 | none | chr6, 1526283..1528520 | XM_010653013.2 | II | 1 | 171 | 6.22 × 10−14 | GATA transcription factor 13 | ||
| VvGATA12 | VIT_06s0004g01280 | chr6, 1535752..1535895 | XM_019220525.1 | II | 1 | 104 | 1.50 × 10−16 | GATA transcription factor 23-like | ||
| VvGATA13 | VIT_06s0004g02740 | chr6, 3428913..3432090 | XM_010652872.2 | I | 2 | 464 | 3.52 × 10−13 | GATA transcription factor 8 | ||
| VvGATA14 | VIT_207s0005g01085 | chr7, 3630384..3630512 | XM_010653922.2 | II | 1 | 140 | 2.71 × 10−14 | GATA transcription factor 15 | ||
| VvGATA15 | VIT_08s0007g07550 | chr8, 21037563..21038886 | XM_002282189.3 | I | 1 | 299 | 3.63 × 10−15 | GATA transcription factor 2 | ||
| VvGATA16 | VIT_09s0054g00440 | chr9, 20974328..20988907 | XM_002263671.4 | III | 1 | 1 | CCT | 299 | 2.32 × 10−13 | GATA transcription factor 24 |
| VvGATA17 | VIT_09s0002g03750 | chr9, 3439127..3440408 | XM_002283992.3 | I | 1 | 329 | 3.95 × 10−16 | GATA transcription factor 9 | ||
| VvGATA18 | VIT_11s0016g02210 | chr11, 1818033..1819175 | XM_002282137.3 | II | 1 | 309 | 2.72 × 10−19 | GATA transcription factor 21 | ||
| VvGATA19 | VIT_12s0121g00120 | chr12, 11859275..11882585 | XM_019223388.1 | IV | 1 | 216 | 4.00 × 10−15 | GATA transcription factor 27 | ||
| VvGATA20 | VIT_14s0066g00150 | chr12, 11878739..11880211 | FN596758.1 | IV | 1 | 1 | BET | 259 | 4.00 × 10−12 | unnamed protein product |
| VvGATA21 | VIT_13s0019g04390 | chr13, 5760956..5762854 | XM_002273466.4 | I | 1 | 340 | 6.51 × 10−17 | GATA transcription factor 8 | ||
| VvGATA22 | VIT_14s0060g01520 | chr14, 1203496..1204229 | XM_002278333.4 | II | 1 | 125 | 8.46 × 10−16 | GATA transcription factor 16 | ||
| VvGATA23 | VIT_14s0066g00150 | chr14, 26717092..26720948 | XM_010662532.2 | II | 1 | 1 | 367 | 5.30 × 10−15 | GATA transcription factor 7 | |
| VvGATA24 | VIT_15s0021g02510 | chr15, 13505714..13506649 | XM_002277923.3 | I | 1 | 270 | 1.45 × 10−14 | GATA transcription factor 4 | ||
| VvGATA25 | VIT_18s0001g07720 | chr18, 6040264..6057788 | XM_002283717.3 | III | 1 | CCT | 294 | 2.52 × 10−14 | GATA transcription factor 25 | |
| VvGATA26 | VIT_18s0001g07730 | chr18, 6060621..6094190 | XM_010666122.2 | III | 1 | CCT | 368 | 8.90 × 10−17 | GATA transcription factor 24 | |
| VvGATA27 | VIT_200s2393g00010 | chrUn, 41800730..41802661 | AM449204.2 | IV | 1 | ASXH | 542 | 5.60 × 10−16 | GATA transcription factor 26 | |
| Cis-Element Name | Sequence | Response | Number | Gene Number |
|---|---|---|---|---|
| ABRE | ACGTG | Abscisic acid | 44 | 14 |
| TCA-element | CCATCTTTTT | Salicylic acid | 21 | 17 |
| CGTCA-motif | CGTCA | Methyl jasmonate | 15 | 9 |
| as-1 | TGACG | Salicylic acid | 15 | 9 |
| WUN-motif | AATTT(A)C | Mechanical injury | 16 | 13 |
| TGACG-motif | TGACG | Methyl jasmonate | 15 | 9 |
| DRE1 | ACCGAGA | Abscisic acid | 3 | 3 |
| P-box | CCTTTTG | Gibberellin | 11 | 8 |
| GARE-motif | TCTGTTG | Gibberellin | 3 | 3 |
| JERE | AGACCGCC | Methyl jasmonate | 2 | 1 |
| MYB | T(C)AAC | Methyl jasmonate | 89 | 27 |
| F-box | CTATTCTCATT | Abscisic acid, Light, Drought | 4 | 4 |
| SARE | TTCGACCATCTT | Salicylic acid | 2 | 2 |
| ERE | ATTTTAAA | Ethylene | 38 | 14 |
| W-box | TTGACC | Defense | 12 | 9 |
| LTR | CCGAAA | Low-temperature | 8 | 7 |
| MBS | CAACTG | Drought | 14 | 12 |
| MYC | CAA(T)G(T)TG(A) | Abscisic acid | 84 | 26 |
| Myb binding site | CAACAG | Drought | 7 | 7 |
| TC-rich repeats | TTC(T)TCT | Defense and stress | 9 | 7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, T.; Peng, J.; Li, M.; Dou, M.; Lei, Y.; Wang, Y.; Xu, Y. Genetic Analysis of the Grapevine GATA Gene Family and Their Expression Profiles in Response to Hormone and Downy Mildew Infection. Horticulturae 2022, 8, 303. https://doi.org/10.3390/horticulturae8040303
Chen T, Peng J, Li M, Dou M, Lei Y, Wang Y, Xu Y. Genetic Analysis of the Grapevine GATA Gene Family and Their Expression Profiles in Response to Hormone and Downy Mildew Infection. Horticulturae. 2022; 8(4):303. https://doi.org/10.3390/horticulturae8040303
Chicago/Turabian StyleChen, Tingting, Jing Peng, Meijie Li, Mengru Dou, Yan Lei, Yuejing Wang, and Yan Xu. 2022. "Genetic Analysis of the Grapevine GATA Gene Family and Their Expression Profiles in Response to Hormone and Downy Mildew Infection" Horticulturae 8, no. 4: 303. https://doi.org/10.3390/horticulturae8040303
APA StyleChen, T., Peng, J., Li, M., Dou, M., Lei, Y., Wang, Y., & Xu, Y. (2022). Genetic Analysis of the Grapevine GATA Gene Family and Their Expression Profiles in Response to Hormone and Downy Mildew Infection. Horticulturae, 8(4), 303. https://doi.org/10.3390/horticulturae8040303







