Genome-Wide Identification of Strawberry Metal Tolerance Proteins and Their Expression under Cadmium Toxicity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification of MTP Gene from Strawberry
2.2. Physicochemical Properties, Sequence Alignment, and Phylogenetic Analysis
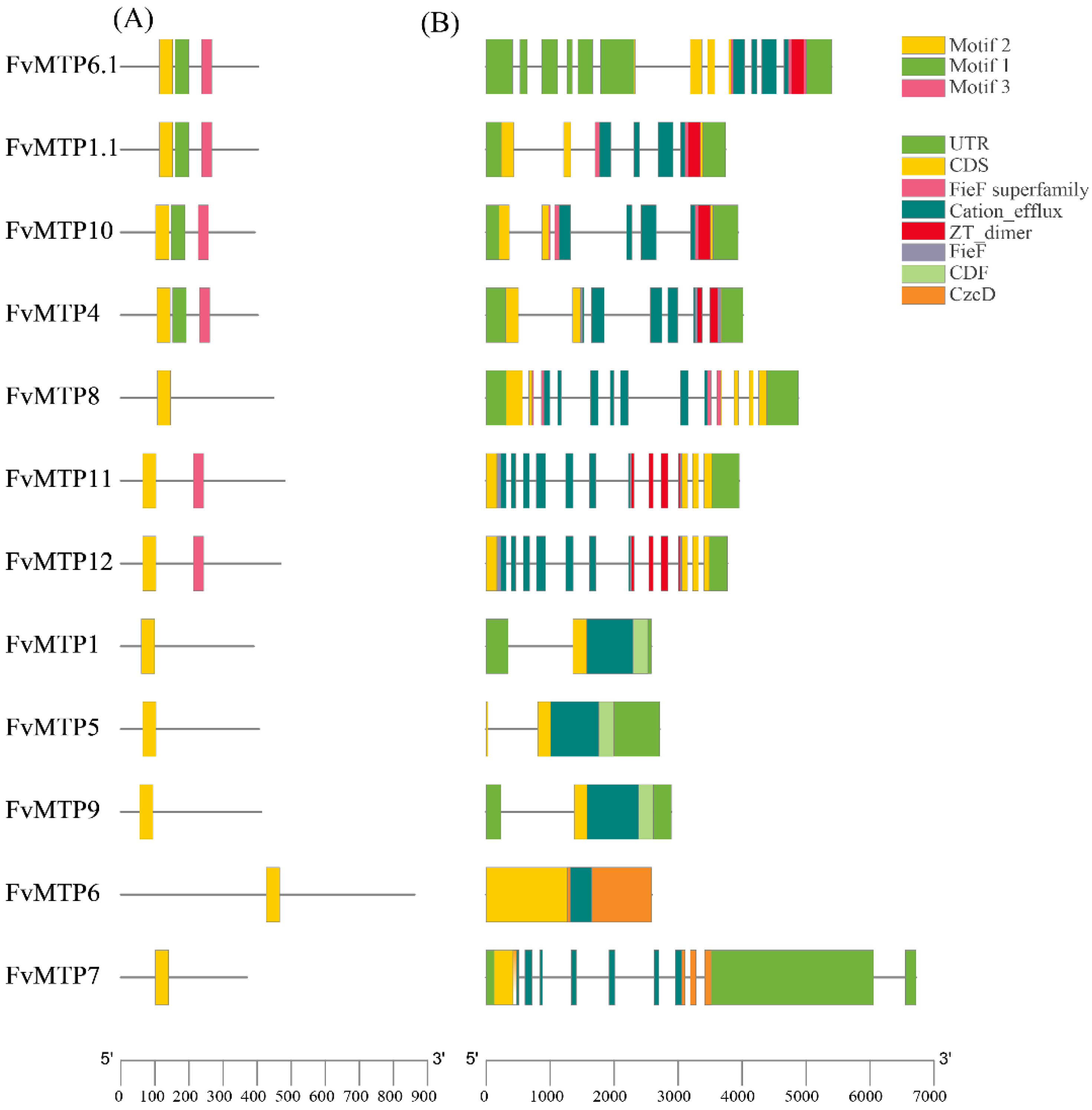
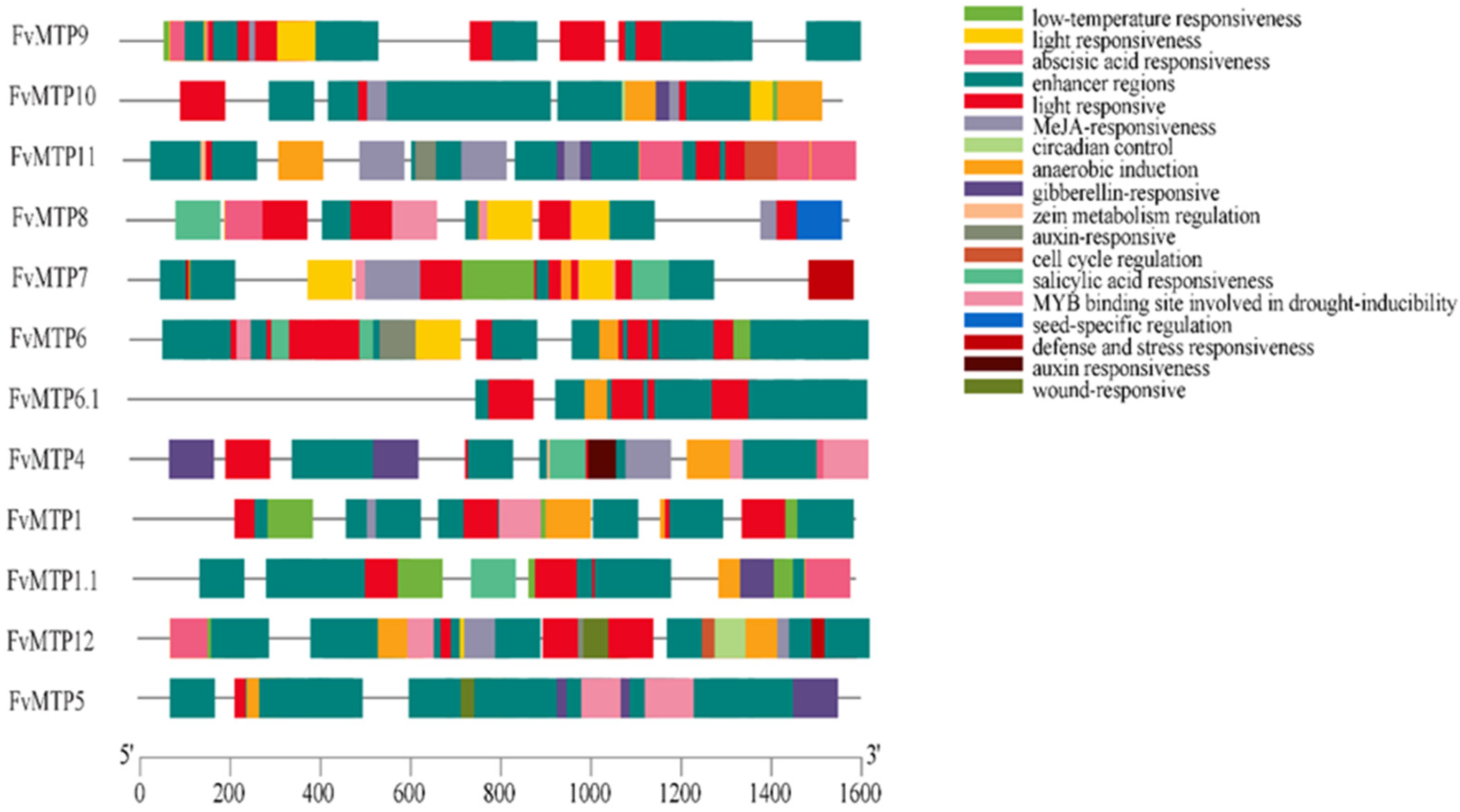
2.3. Conserved Motifs, Gene Structure, and Promoter Analysis
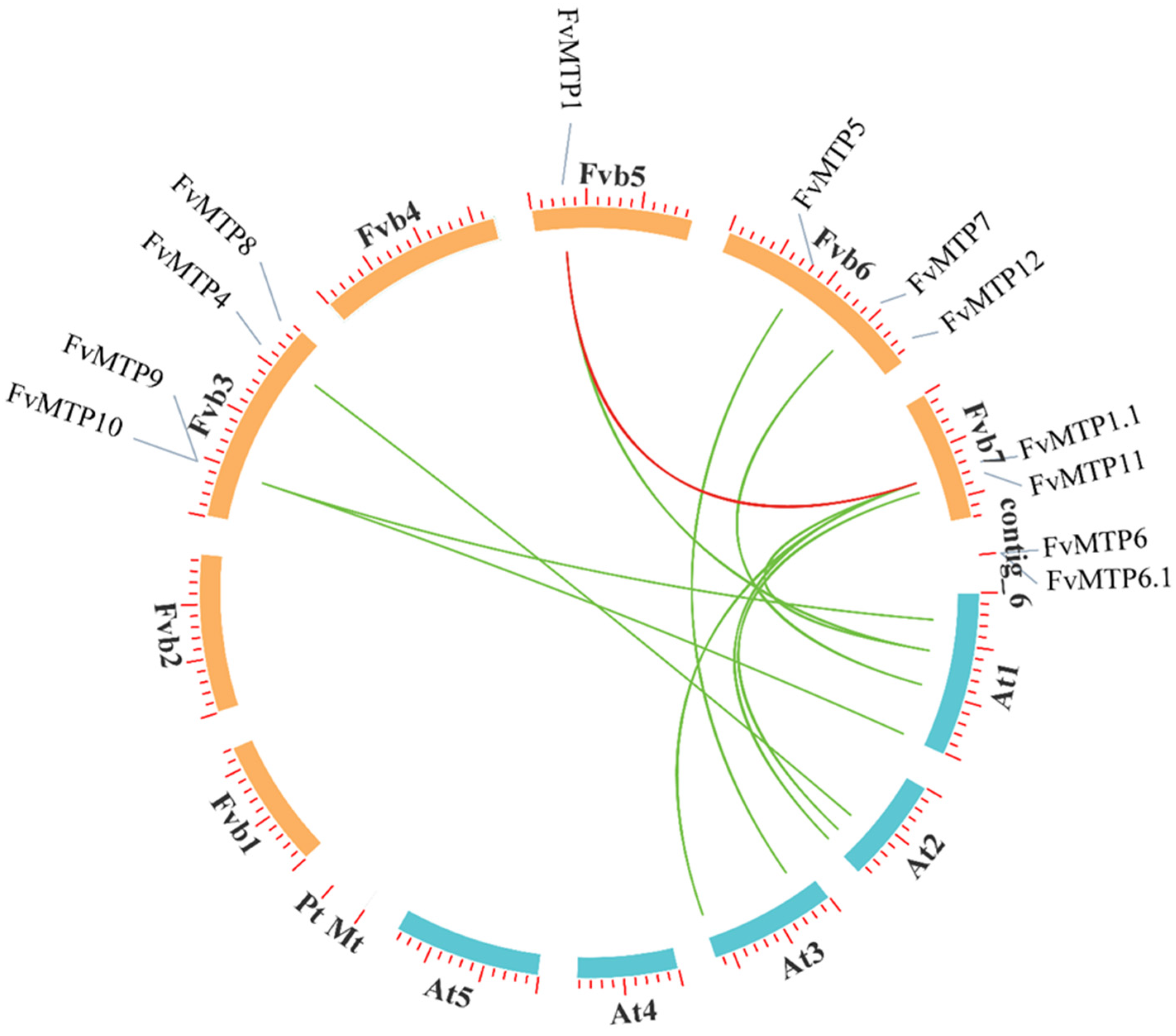
2.4. Chromosomal Positioning and Collinearity Analysis
2.5. Gene Ontology (GO) and Kyoto-Encyclopedia of Genes and Genomes (KEGG) Analysis
2.6. Plant Material and Cd Stress
2.7. RNA Extraction and qRT-PCR Analysis
2.8. H2O2 Accumulation, and Cd Quantification
2.9. Statistical Analysis
3. Results
3.1. Identification and Sequence Properties of FvMTP Genes
3.2. Phylogenetic Characterization, Motif Composition, and Gene Structure Analysis of FvMTPs
3.3. Chromosomal Orientation, Promoters and Functional Enrichment Analysis of FvMTPs
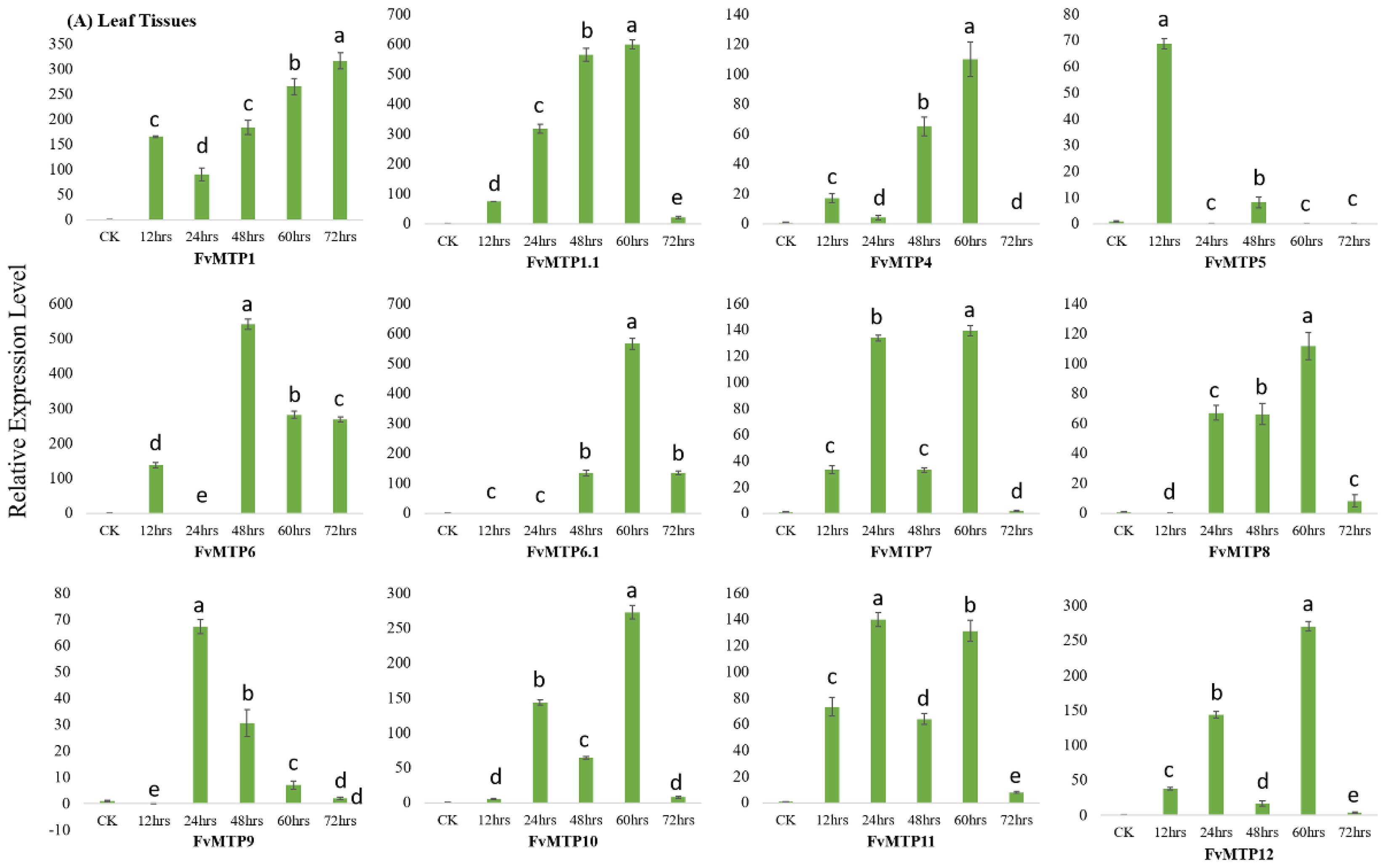
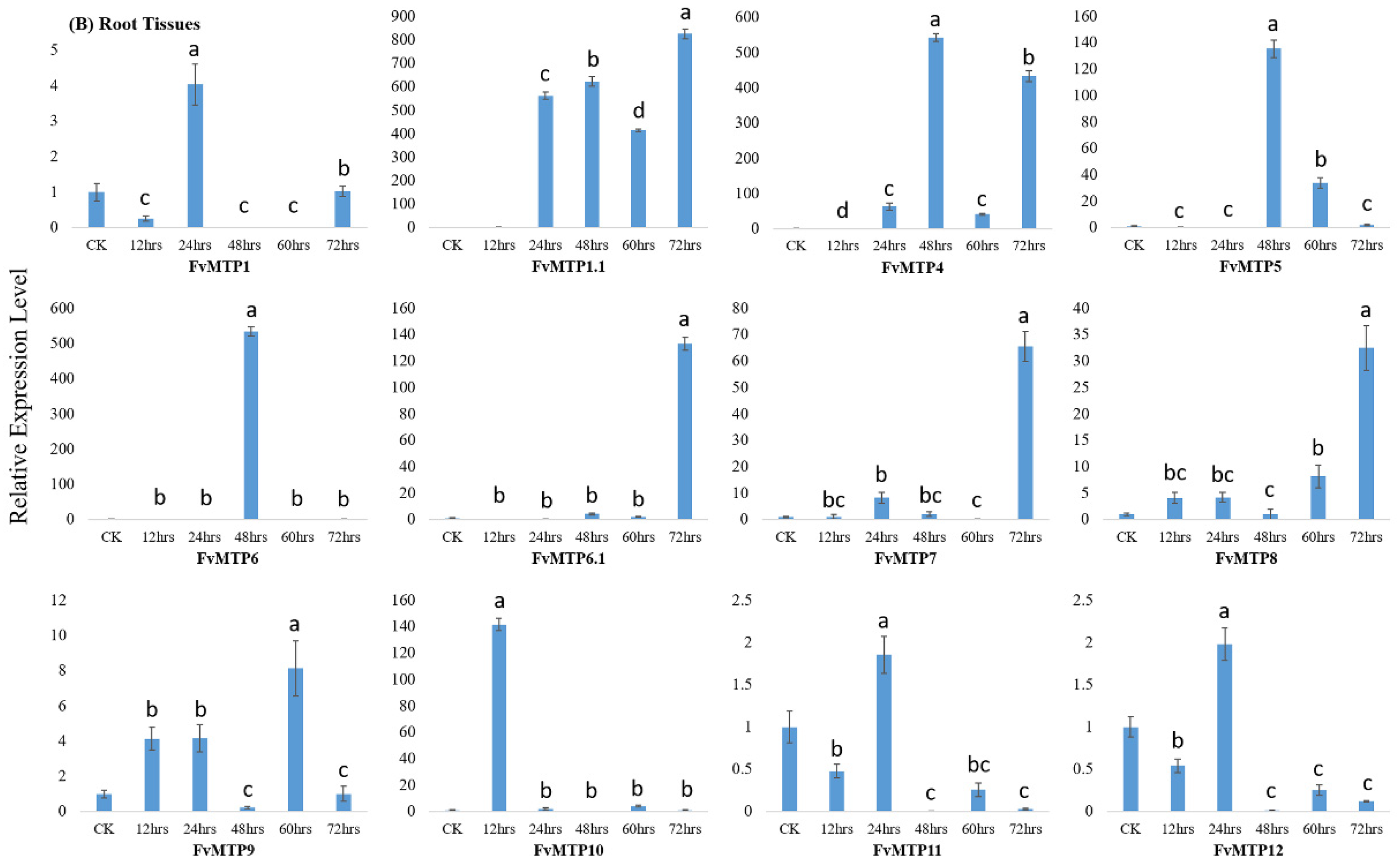
3.4. Cd Treatment and qRT-PCR Analysis
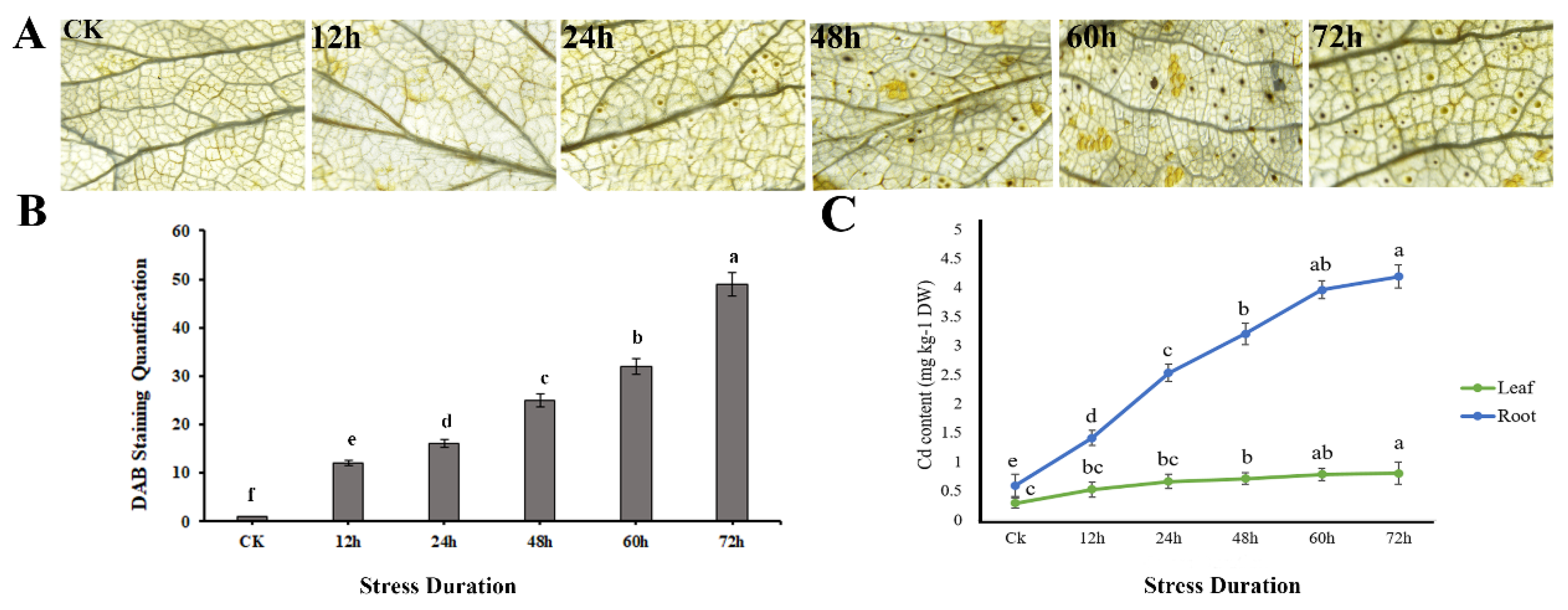
3.5. H2O2 Accumulation, and Cd Quantification
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Yuan, L.; Yang, S.; Liu, B.; Zhang, M.; Wu, K. Molecular characterization of a rice metal tolerance protein, OsMTP1. Plant Cell Rep. 2012, 31, 67–79. [Google Scholar] [CrossRef] [PubMed]
- Cambrollé, J.; García, J.L.; Figueroa, M.E.; Cantos, M. Evaluating wild grapevine tolerance to copper toxicity. Chemosphere 2015, 120, 171–178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomine, S.; Vert, G. Iron transport in plants: Better be safe than sorry. Curr. Opin. Plant Biol. 2013, 16, 322–327. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.S.; Hasanuzzaman, M.; Nahar, K.; Macovei, A.; Tuteja, N. Importance of nitric oxide in cadmium stress tolerance in crop plants. Plant Physiol. Biochem. 2013, 63, 254–261. [Google Scholar] [CrossRef]
- Clemens, S. Toxic metal accumulation, responses to exposure and mechanisms of tolerance in plants. Biochimie 2006, 88, 1707–1719. [Google Scholar] [CrossRef]
- Hall, J.L. Cellular mechanisms for heavy metal detoxification and tolerance. J. Exp. Bot. 2002, 53, 1–11. [Google Scholar] [CrossRef]
- Gaither, L.A.; Eide, D.J. Eukaryotic zinc transporters and their regulation. Biometals: An international journal on the role of metal ions. Biol. Biochem. Med. 2001, 14, 251–270. [Google Scholar]
- Hall, J.L.; Williams, L.E. Transition metal transporters in plants. J. Exp. Bot. 2003, 54, 2601–2613. [Google Scholar] [CrossRef]
- Gustin, J.L.; Zanis, M.J.; Salt, D.E. Structure and evolution of the plant cation diffusion facilitator family of ion transporters. BMC Evol. Biol. 2011, 11, 76. [Google Scholar] [CrossRef] [Green Version]
- Migocka, M.; Kosieradzka, A.; Papierniak, A.; Maciaszczyk-Dziubinska, E.; Posyniak, E.; Garbiec, A.; Filleur, S. Two metal-tolerance proteins, MTP1 and MTP4, are involved in Zn homeostasis and Cd sequestration in cucumber cells. J. Exp. Bot. 2015, 66, 1001–1015. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Gao, Y.; Tang, Y.; Wang, D.; Chen, X.; Yao, Y.; Guo, Y. Genome-wide identification, comprehensive gene feature, evolution, and expression analysis of plant metal tolerance proteins in tobacco under heavy metal toxicity. Front. Genet. 2019, 10, 345. [Google Scholar] [CrossRef] [PubMed]
- Montanini, B.; Blaudez, D.; Jeandroz, S.; Sanders, D.; Chalot, M. Phylogenetic and functional analysis of the cation diffusion facilitator (CDF) family: Improved signature and prediction of substrate specificity. BMC Genom. 2007, 8, 107. [Google Scholar] [CrossRef] [Green Version]
- Shirazi, Z.; Abedi, A.; Kordrostami, M.; Burritt, D.J.; Hossain, M.A.J.B. Genome-wide identification and characterization of the metal tolerance protein (MTP) family in grape (Vitis vinifera L.). 3 Biotech 2019, 9, 199. [Google Scholar] [CrossRef]
- Desbrosses-Fonrouge, A.-G.; Voigt, K.; Schröder, A.; Arrivault, S.; Thomine, S.; Krämer, U. Arabidopsis thaliana MTP1 is a Zn transporter in the vacuolar membrane which mediates Zn detoxification and drives leaf Zn accumulation. FEBS Lett. 2005, 579, 4165–4174. [Google Scholar] [CrossRef]
- Arrivault, S.; Senger, T.; Krämer, U. The Arabidopsis metal tolerance protein AtMTP3 maintains metal homeostasis by mediating Zn exclusion from the shoot under Fe deficiency and Zn oversupply. Plant J. 2006, 46, 861–879. [Google Scholar] [CrossRef] [PubMed]
- Delhaize, E.; Gruber, B.D.; Pittman, J.K.; White, R.G.; Leung, H.; Miao, Y.; Jiang, L.; Ryan, P.R.; Richardson, A.E. A role for the AtMTP11 gene of Arabidopsis in manganese transport and tolerance. Plant J. 2007, 51, 198–210. [Google Scholar] [CrossRef] [PubMed]
- Peiter, E.; Montanini, B.; Gobert, A.; Pedas, P.; Husted, S.; Maathuis, F.J.M.; Blaudez, D.; Chalot, M.; Sanders, D. A secretory pathway-localized cation diffusion facilitator confers plant manganese tolerance. Proc. Nat. Acad. Sci. USA 2007, 104, 8532–8537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujiwara, T.; Kawachi, M.; Sato, Y.; Mori, H.; Kutsuna, N.; Hasezawa, S.; Maeshima, M. A high molecular mass zinc transporter MTP12 forms a functional heteromeric complex with MTP5 in the Golgi in Arabidopsis thaliana. FEBS J. 2015, 282, 1965–1979. [Google Scholar] [CrossRef]
- Topcu, H.; Degirmenci, I.; Sonmez, D.A.; Paizila, A.; Karci, H.; Kafkas, S.; Kafkas, E.; Ercisli, S.; Alatawi, A. Sugar, invertase enzyme activities and invertase gene expression in different developmental stages of strawberry fruits. Plants 2022, 11, 509. [Google Scholar] [CrossRef]
- Das, N.; Bhattacharya, S.; Maiti, M.K.J.P.P. Enhanced cadmium accumulation and tolerance in transgenic tobacco overexpressing rice metal tolerance protein gene OsMTP1 is promising for phytoremediation. Plant Physiol. Biochem. 2016, 105, 297–309. [Google Scholar] [CrossRef]
- Juric, S.; Vlahovicek-Kahlina, K.; Duralija, B.; Maslov Bandic, L.; Nekic, P.; Vincekovic, M. Stimulation of plant secondary metabolites synthesis in soilless cultivated strawberries (Fragaria × ananassa Duchesne) using zinc-alginate microparticles. Turk. J. Agric. For. 2021, 45, 324–334. [Google Scholar] [CrossRef]
- Kilic, N.; Burgut, A.; Gündesli, M.A.; Nogay, G.; Ercisli, S.; Kafkas, N.E.; Ekiert, H.; Elansary, H.O.; Szopa, A. The effect of organic, inorganic fertilizers and their combinations on fruit quality parameters in strawberry. Horticulturae 2021, 7, 354. [Google Scholar] [CrossRef]
- Tas, A.; Berk, S.K.; Orman, E.; Gundogdu, M.; Ercisli, S.; Karatas, N.; Jurikova, T.; Adamkova, A.; Nedomova, S.; Mlcek, J. Influence of pre-harvest gibberellic acid and post-harvest 1-Methyl Cyclopropane treatments on phenolic compounds, vitamin C and organic acid contents during the shelf life of strawberry fruits. Plants 2021, 10, 121. [Google Scholar] [CrossRef] [PubMed]
- Urün, I.; Attar, S.H.; Sönmez, D.A.; Gündeşli, M.A.; Ercişli, S.; Kafkas, N.E.; Bandić, L.M.; Duralija, B. Comparison of polyphenol, sugar, organic acid, volatile compounds, and antioxidant capacity of commercially grown strawberry cultivars in Turkey. Plants 2021, 10, 1654. [Google Scholar] [CrossRef] [PubMed]
- Haider, M.S.; Khan, N.; Pervaiz, T.; Zhongjie, L.; Nasim, M.; Jogaiah, S.; Mushtaq, N.; Jiu, S.; Jinggui, F. Genome-wide identification, evolution, and molecular characterization of the PP2C gene family in woodland strawberry. Gene 2019, 702, 27–35. [Google Scholar] [CrossRef]
- Jiu, S.; Haider, M.S.; Kurjogi, M.M.; Zhang, K.; Zhu, X.; Fang, J. Genome-wide characterization and expression analysis of sugar transporter family genes in woodland strawberry. Plant Genome 2018, 11, 170103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fierascu, R.C.; Temocico, G.; Fierascu, I.; Ortan, A.; Babeanu, N.E. Fragaria genus: Chemical composition and biological activities. Molecules 2020, 25, 498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bebek Markovinovic, A.; Putnik, P.; Duralija, B.; Krivohlavek, A.; Ivešic, M.; Mandic Andacic, I.; Palac Bešlic, I.; Pavlic, B.; Lorenzo, J.M.; Bursac Kovacevic, D. Chemometric valorization of strawberry (Fragaria × ananassa Duch.) cv. ‘Albion’ for the production of functional juice: The impact of physicochemical, toxicological, sensory, and bioactive value. Foods 2022, 11, 640. [Google Scholar] [CrossRef]
- Palma, G.; Freer, J.; Baeza, J. Removal of metal ions by modified Pinus radiata bark and tannins from water solutions. Water Res. 2003, 37, 4974–4980. [Google Scholar] [CrossRef]
- Muradoglu, F.; Gundogdu, M.; Ercisli, S.; Encu, T.; Balta, F.; Jaafar, H.Z.; Zia-Ul-Haq, M. Cadmium toxicity affects chlorophyll a and b content, antioxidant enzyme activities and mineral nutrient accumulation in strawberry. Biol. Res. 2015, 48, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Treder, W.; Cieslinski, G. Cadmium uptake and distribution in strawberry plants as affected by its concentration in soil. J. Fruit Ornam. Plant Res. 2000, 8, 127–135. [Google Scholar]
- Migeon, A.; Blaudez, D.; Wilkins, O.; Montanini, B.; Campbell, M.M.; Richaud, P.; Thomine, S.; Chalot, M.J.C.; Sciences, M.L. Genome-wide analysis of plant metal transporters, with an emphasis on poplar. Cell. Mol. Life Sci. 2010, 67, 3763–3784. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Bae, D.W.; Kim, S.H.; Han, H.J.; Liu, X.; Park, H.C.; Lim, C.O.; Lee, S.Y.; Chung, W.S. Comparative proteomic analysis of the short-term responses of rice roots and leaves to cadmium. J. Plant Physiol. 2010, 167, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Xingzheng, F.; Yahua, T.; Xue, Z.; Lili, L.; Changpin, C.; Li, C.; Ming, Z.; Liangzhi, P. Genome-wide identification of sweet orange (Citrus sinensis) metal tolerance proteins and analysis of their expression patterns under zinc, manganese, copper, and cadmium toxicity. Gene 2017, 629, 1–8. [Google Scholar]
- Kolaj-Robin, O.; Russell, D.; Hayes, K.A.; Pembroke, J.T.; Soulimane, T. Cation diffusion facilitator family: Structure and function. FEBS Lett. 2015, 589, 1283–1295. [Google Scholar] [CrossRef]
- Sahin, U.; Anapali, O.; Ercisli, S. Physico-chemical and physical properties of some substrates used in horticulture. Gartenbauwissenschaft 2002, 67, 55–60. [Google Scholar]
- Nosek, M.; Kaczmarczyk, A.; Jędrzejczyk, R.J.; Supel, P.; Kaszycki, P.; Miszalski, Z. Expression of genes involved in heavy metal trafficking in plants exposed to salinity stress and elevated Cd concentrations. Plants 2020, 9, 475. [Google Scholar] [CrossRef] [Green Version]
- Lu, M.; Fu, D. Structure of the zinc transporter YiiP. Science 2007, 317, 1746–1748. [Google Scholar] [CrossRef] [Green Version]
- Ricachenevsky, F.K.; Menguer, P.K.; Sperotto, R.A.; Williams, L.E.; Fett, J.P. Roles of plant metal tolerance proteins (MTP) in metal storage and potential use in biofortification strategies. Front. Plant Sci. 2013, 4, 144. [Google Scholar] [CrossRef] [Green Version]
- Khan, N.; Fatima, F.; Haider, M.S.; Shazadee, H.; Liu, Z.; Zheng, T.; Fang, J. Genome-Wide identification and expression profiling of the Polygalacturonase (PG) and Pectin Methylesterase (PME) genes in grapevine (Vitis vinifera L.). Int. J. Mol. Sci. 2019, 20, 3180. [Google Scholar] [CrossRef] [Green Version]
- Vatansever, R.; Filiz, E.; Eroglu, S. Genome-wide exploration of metal tolerance protein (MTP) genes in common wheat (Triticum aestivum): Insights into metal homeostasis and biofortification. Biometals 2017, 30, 217–235. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haider, M.S.; Sheikh, T.M.M.; Jiu, S.; Aleem, M.; Shafqat, W.; Shoukat, K.; Khan, N.; Jaskani, M.J.; Naqvi, S.A.; Ercisli, S.; et al. Genome-Wide Identification of Strawberry Metal Tolerance Proteins and Their Expression under Cadmium Toxicity. Horticulturae 2022, 8, 477. https://doi.org/10.3390/horticulturae8060477
Haider MS, Sheikh TMM, Jiu S, Aleem M, Shafqat W, Shoukat K, Khan N, Jaskani MJ, Naqvi SA, Ercisli S, et al. Genome-Wide Identification of Strawberry Metal Tolerance Proteins and Their Expression under Cadmium Toxicity. Horticulturae. 2022; 8(6):477. https://doi.org/10.3390/horticulturae8060477
Chicago/Turabian StyleHaider, Muhammad Salman, Taha Majid Mahmood Sheikh, Songtao Jiu, Muqaddas Aleem, Waqar Shafqat, Komal Shoukat, Nadeem Khan, Muhammad Jafar Jaskani, Summar A. Naqvi, Sezai Ercisli, and et al. 2022. "Genome-Wide Identification of Strawberry Metal Tolerance Proteins and Their Expression under Cadmium Toxicity" Horticulturae 8, no. 6: 477. https://doi.org/10.3390/horticulturae8060477
APA StyleHaider, M. S., Sheikh, T. M. M., Jiu, S., Aleem, M., Shafqat, W., Shoukat, K., Khan, N., Jaskani, M. J., Naqvi, S. A., Ercisli, S., Assouguem, A., Kara, M., Ullah, R., Aljabri, M., & Qari, S. H. (2022). Genome-Wide Identification of Strawberry Metal Tolerance Proteins and Their Expression under Cadmium Toxicity. Horticulturae, 8(6), 477. https://doi.org/10.3390/horticulturae8060477









