Efficient Editing of SoCSLD2 by CRISPR/Cas9 Affects Morphogenesis of Root Hair in Spinach
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials, Growth, and Culture Conditions
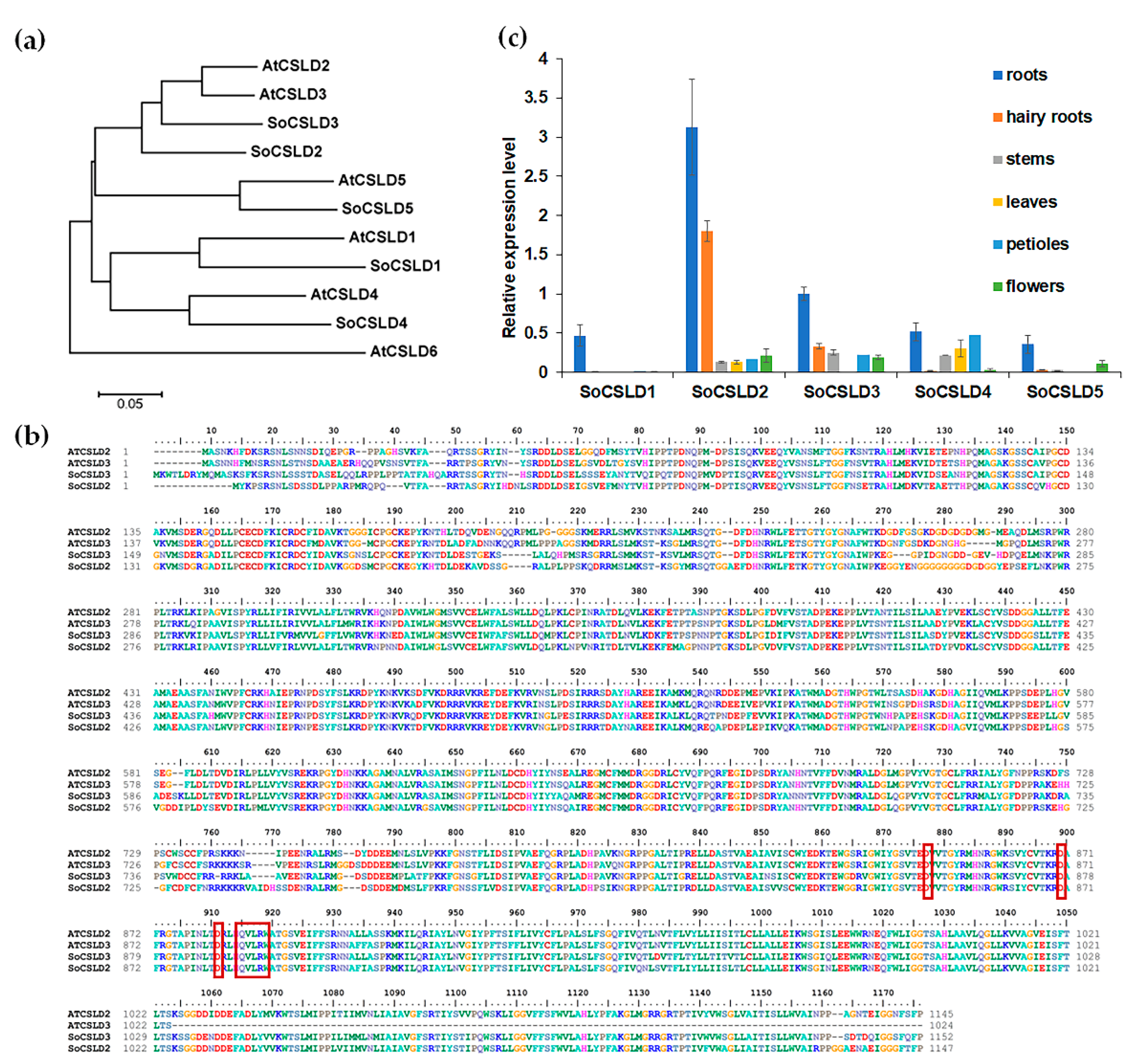
2.2. Bioinformation Analysis of CSLD Protein Sequence
2.3. Quantitative Real-Time PCR
2.4. Target Sites Design and Vector Construction
2.5. Induction and Identification of Transgenic Hairy Roots of Spinach
2.6. Mutation Type Analysis
2.7. Phenotype Observation
2.8. Off-Target Analysis
2.9. Transcriptome Sequencing of Root Hairs
3. Results
3.1. SoCSLD2 and SoCSLD3 Were Strongly Expressed in Roots and Hairy Roots
3.2. CRISPR/Cas9 Vector Construction and Co-Transformation Efficiency in Hairy Roots
3.3. Genotyping and Phenotyping of Edited Hairy Root Lines of SoCSLD2
3.4. Genotyping and Phenotyping of Edited Hairy Root Lines of SoCSLD3
3.5. Mutation Variety and Frequency of CRISPR/Cas9-Edited Hairy Roots
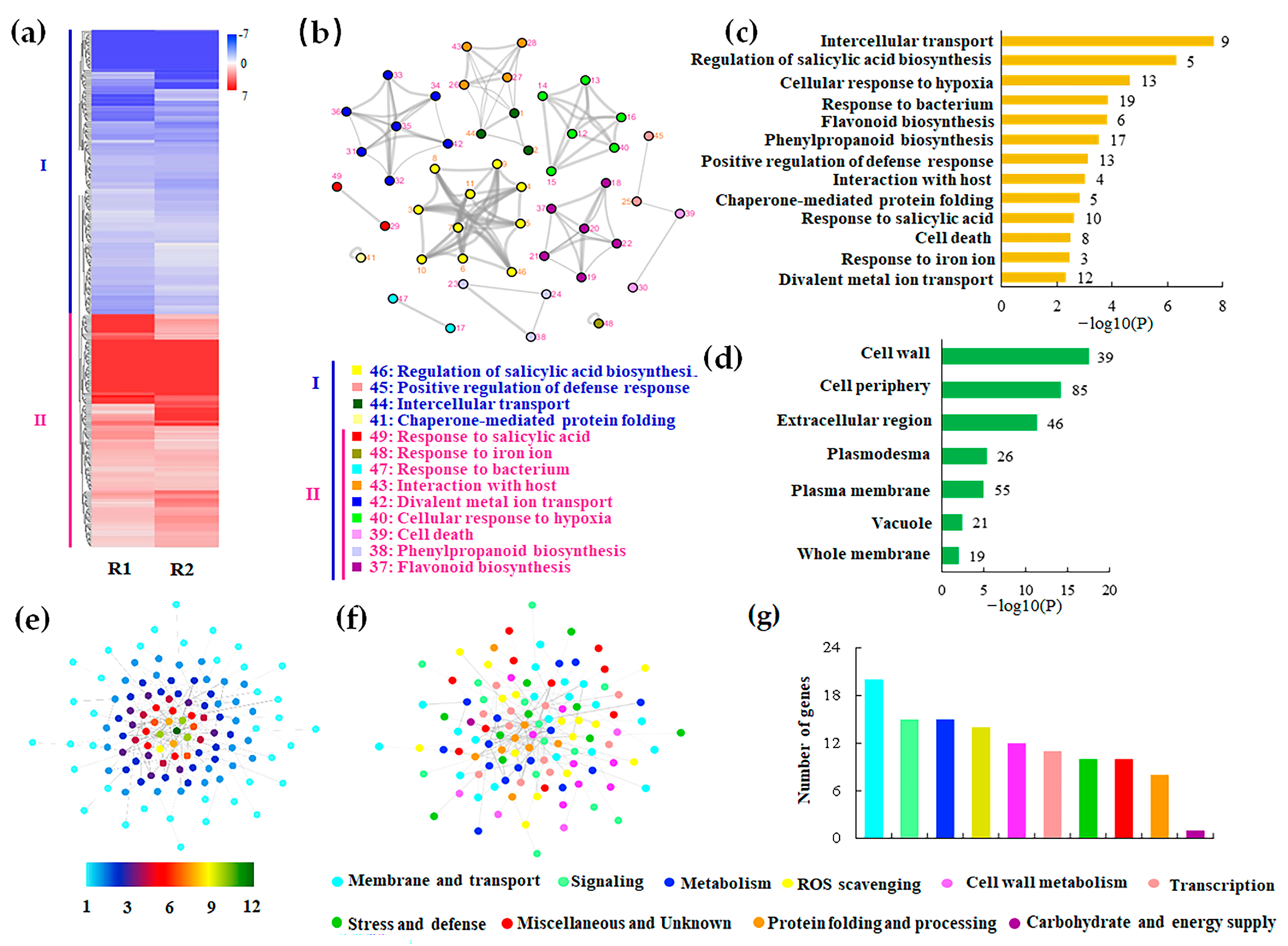
3.6. Transcriptomic Analysis of CRISPR/Cas9 Edited Hairy Roots
4. Discussion
4.1. The tRNA–gRNA Cassette Expression System Is an Efficient Tool for Multiplex Targeted Mutagenesis in Spinach
4.2. SoCSLD2 Regulates the Spinach Root-Hair Growth
4.3. SoCSLD2 Is Involved in Cell-Wall Remodulation in Hairy Roots of Spinach
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xu, C.; Jiao, C.; Sun, H.; Cai, X.; Wang, X.; Ge, C.; Zheng, Y.; Liu, W.; Sun, X.; Xu, Y.; et al. Draft genome of spinach and transcriptome diversity of 120 Spinacia accessions. Nat. Commun. 2017, 8, 15275. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Sun, X.; Xu, C.; Sun, H.; Wang, X.; Ge, C.; Zhang, Z.; Wang, Q.; Fei, Z.; Jiao, C.; et al. Genomic analyses provide insights into spinach domestication and the genetic basis of agronomic traits. Nat. Commun. 2021, 12, 7246. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Cao, Y.P.; Wang, Y.; Fu, C.; Dai, S. Agrobacterium rhizogenes-mediated transformation system of Spinacia oleracea. Chin. Bull. Bot. 2019, 54, 515–521. [Google Scholar] [CrossRef]
- Ge, C.; Cai, X.; Xu, C.; Wang, X.; Deng, J.; Liu, S.; Zhao, Q.; Dai, S.; Wang, Q. A new heat-resistant spinach cultivar ‘Hubo 1’. Acta Hortic. Sin. 2015, 42, 399–400. [Google Scholar] [CrossRef]
- Zhao, Q.; Chen, W.; Bian, J.; Xie, H.; Li, Y.; Xu, C.; Ma, J.; Guo, S.; Chen, J.; Cai, X.; et al. Proteomics and Phosphoproteomics of Heat Stress-Responsive Mechanisms in Spinach. Front. Plant Sci. 2018, 9, 800. [Google Scholar] [CrossRef]
- Li, S.; Yu, J.; Li, Y.; Zhang, H.; Bao, X.; Bian, J.; Xu, C.; Wang, X.; Cai, X.; Wang, Q.; et al. Heat-Responsive Proteomics of a Heat-Sensitive Spinach Variety. Int. J. Mol. Sci. 2019, 20, 3872. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, H.; Zhang, Y.; Liu, Y.; Li, Y.; Tian, H.; Guo, S.; Sun, M.; Qin, Z.; Dai, S. Genome-wide identification and expression analysis reveals spinach brassinosteroid-signaling kinase (BSK) gene family functions in temperature stress response. BMC Genom. 2022, 23, 453. [Google Scholar] [CrossRef]
- Barrangou, R.; Marraffini, L.A. CRISPR-Cas Systems: Prokaryotes Upgrade to Adaptive Immunity. Mol. Cell 2014, 54, 234–244. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Ma, X.; Liu, Y. CRISPR/Cas9-Based Multiplex Genome Editing in Monocot and Dicot Plants. Curr. Protoc. Mol. Biol. 2016, 115, 31.6.1–31.6.21. [Google Scholar] [CrossRef]
- Dong, F.; Xie, K.; Chen, Y.; Yang, Y.; Mao, Y. Polycistronic tRNA and CRISPR guide-RNA enables highly efficient multiplexed genome engineering in human cells. Biochem. Biophys. Res. Commun. 2017, 482, 889–895. [Google Scholar] [CrossRef]
- Wang, P.; Zhang, J.; Sun, L.; Ma, Y.; Xu, J.; Liang, S.; Deng, J.; Tan, J.; Zhang, Q.; Tu, L.; et al. High efficient multisites genome editing in allotetraploid cotton (Gossypium hirsutum) using CRISPR/Cas9 system. Plant Biotechnol. J. 2018, 16, 137–150. [Google Scholar] [CrossRef]
- Gao, C. Genome engineering for crop improvement and future agriculture. Cell 2021, 184, 1621–1635. [Google Scholar] [CrossRef]
- Zhu, H.; Li, C.; Gao, C. Applications of CRISPR–Cas in agriculture and plant biotechnology. Nat. Rev. Mol. Cell Biol. 2020, 11, 661–677. [Google Scholar] [CrossRef]
- Georgiev, M.I.; Agostini, E.; Ludwig-Müller, J.; Xu, J. Genetically transformed roots: From plant disease to biotechnological resource. Trends Biotechnol. 2012, 30, 528–537. [Google Scholar] [CrossRef]
- Libault, M.; Brechenmacher, L.; Cheng, J.; Xu, D.; Stacey, G. Root hair systems biology. Trends Plant Sci. 2010, 15, 641–650. [Google Scholar] [CrossRef]
- Yin, L.; Verhertbruggen, Y.; Oikawa, A.; Manisseri, C.; Knierim, B.; Prak, L.; Jensen, J.K.; Knox, J.P.; Auer, M.; Willats, W.G.; et al. The Cooperative Activities of CSLD2, CSLD3, and CSLD5 Are Required for Normal Arabidopsis Development. Mol. Plant 2011, 4, 1024–1037. [Google Scholar] [CrossRef]
- Lampugnani, E.R.; Khan, G.A.; Somssich, M.; Persson, S. Building a plant cell wall at a glance. J. Cell Sci. 2018, 131, jcs207373. [Google Scholar] [CrossRef]
- Lampugnani, E.R.; Sandoval, E.F.; Tan, Q.W.; Mutwil, M.; Bowman, J.L.; Persson, S. Cellulose Synthesis—Central Components and Their Evolutionary Relationships. Trends Plant Sci. 2019, 24, 402–412. [Google Scholar] [CrossRef]
- Lerouxel, O.; Cavalier, D.M.; Liepman, A.; Keegstra, K. Biosynthesis of plant cell wall polysaccharides—A complex process. Curr. Opin. Plant Biol. 2006, 9, 621–630. [Google Scholar] [CrossRef]
- Richmond, T.A.; Somerville, C.R. The Cellulose Synthase Superfamily. Plant Physiol. 2000, 124, 495–498. [Google Scholar] [CrossRef] [PubMed]
- Bernal, A.J.; Yoo, C.-M.; Mutwil, M.; Jensen, J.K.; Hou, G.; Blaukopf, C.; Sørensen, I.; Blancaflor, E.B.; Scheller, H.V.; Willats, W.G. Functional Analysis of the Cellulose Synthase-Like Genes CSLD1, CSLD2, and CSLD4 in Tip-Growing Arabidopsis Cells. Plant Physiol. 2008, 148, 1238–1253. [Google Scholar] [CrossRef] [PubMed]
- Qi, G.; Hu, R.; Yu, L.; Chai, G.; Cao, Y.; Zuo, R.; Kong, Y.; Zhou, G. Two poplar cellulose synthase-like D genes, PdCSLD5 and PdCSLD6, are functionally conserved with Arabidopsis CSLD3. J. Plant Physiol. 2013, 170, 1267–1276. [Google Scholar] [CrossRef] [PubMed]
- Favery, B.; Ryan, E.; Foreman, J.; Linstead, P.; Boudonck, K.; Steer, M.; Shaw, P.; Dolan, L. KOJAK encodes a cellulose synthase-like protein required for root hair cell morphogenesis in Arabidopsis. Genes Dev. 2001, 15, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Yoo, C.-M.; Quan, L.; Blancaflor, E.B. Divergence and Redundancy in CSLD2 and CSLD3 Function During Arabidopsis Thaliana Root Hair and Female Gametophyte Development. Front. Plant Sci. 2012, 3, 111. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Zhang, R.; Dong, S.; Li, Y.; Fan, C.; Wang, Y.; Xia, T.; Chen, P.; Wang, L.; Feng, S.; et al. AtCSLD3 and GhCSLD3 mediate root growth and cell elongation in downstream of ethylene response pathway in Arabidopsis. J. Exp. Bot. 2018, 69, 1065–1080. [Google Scholar] [CrossRef]
- Hu, H.; Zhang, R.; Tang, Y.; Peng, C.; Wu, L.; Feng, S.; Chen, P.; Wang, Y.; Du, X.; Peng, L. Cotton CSLD3 restores cell elongation and cell wall integrity mainly by enhancing primary cellulose production in the Arabidopsis cesa6 mutant. Plant Mol. Biol. 2019, 101, 389–401. [Google Scholar] [CrossRef]
- Peng, X.; Pang, H.; Abbas, M.; Yan, X.; Dai, X.; Li, Y.; Li, Q. Characterization of Cellulose synthase-like D (CSLD) family revealed the involvement of PtrCslD5 in root hair formation in Populus trichocarpa. Sci. Rep. 2019, 9, 1452. [Google Scholar] [CrossRef]
- Karas, B.J.; Ross, L.; Novero, M.; Amyot, L.; Shrestha, A.; Inada, S.; Nakano, M.; Sakai, T.; Bonetta, D.; Sato, S.; et al. Intragenic complementation at the Lotus japonicus CELLULOSE SYNTHASE-LIKE D1 locus rescues root hair defects. Plant Physiol. 2021, 186, 2037–2050. [Google Scholar] [CrossRef]
- Xie, K.; Minkenberg, B.; Yang, Y. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc. Natl. Acad. Sci. USA 2015, 112, 3570–3575. [Google Scholar] [CrossRef]
- Li, B.; Cui, G.; Shen, G.; Zhan, Z.; Huang, L.; Chen, J.; Qi, X. Targeted mutagenesis in the medicinal plant Salvia miltiorrhiza. Sci. Rep. 2017, 7, srep43320. [Google Scholar] [CrossRef]
- Gupta, D.; Bhattacharjee, O.; Mandal, D.; Sen, M.K.; Dey, D.; Dasgupta, A.; Kazi, T.A.; Gupta, R.; Sinharoy, S.; Acharya, K.; et al. CRISPR-Cas9 system: A new-fangled dawn in gene editing. Life Sci. 2019, 232, 116636. [Google Scholar] [CrossRef]
- Li, Y.; Li, S.; Wang, J.; Liu, G. CRISPR/Cas Systems towards Next-Generation Biosensing. Trends Biotechnol. 2019, 37, 730–743. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, X.; Xie, X.; Deng, L.; Zheng, H.; Pan, H.; Li, D.; Li, L.; Zhong, C. ABE8e with Polycistronic tRNA-gRNA Expression Cassette Sig-Nificantly Improves Adenine Base Editing Efficiency in Nicotiana benthamiana. Int. J. Mol. Sci. 2021, 22, 5663. [Google Scholar] [CrossRef]
- Qi, W.; Zhu, T.; Tian, Z.; Li, C.; Zhang, W.; Song, R. High-efficiency CRISPR/Cas9 multiplex gene editing using the glycine tRNA-processing system-based strategy in maize. BMC Biotechnol. 2016, 16, 58. [Google Scholar] [CrossRef]
- Gagnon, J.A.; Valen, E.; Thyme, S.B.; Huang, P.; Ahkmetova, L.; Pauli, A.; Montague, T.G.; Zimmerman, S.; Richter, C.; Schier, A.F. Efficient Mutagenesis by Cas9 Protein-Mediated Oligonucleotide Insertion and Large-Scale Assessment of Single-Guide RNAs. PLoS ONE 2014, 9, e98186. [Google Scholar] [CrossRef]
- Ren, X.; Yang, Z.; Xu, J.; Sun, J.; Mao, D.; Hu, Y.; Yang, S.-J.; Qiao, H.-H.; Wang, X.; Hu, Q.; et al. Enhanced Specificity and Efficiency of the CRISPR/Cas9 System with Optimized sgRNA Parameters in Drosophila. Cell Rep. 2014, 9, 1151–1162. [Google Scholar] [CrossRef]
- Liu, X.; Homma, A.; Sayadi, J.; Yang, S.; Ohashi, J.; Takumi, T. Sequence features associated with the cleavage efficiency of CRISPR/Cas9 system. Sci. Rep. 2016, 6, 19675. [Google Scholar] [CrossRef]
- Zhou, Z.; Tan, H.; Li, Q.; Chen, J.; Gao, S.; Wang, Y.; Chen, W.; Zhang, L. CRISPR/Cas9-mediated efficient targeted mutagenesis of RAS in Salvia miltiorrhiza. Phytochemistry 2018, 148, 63–70. [Google Scholar] [CrossRef]
- Johnson, R.A.; Gurevich, V.; Filler, S.; Samach, A.; Levy, A.A. Comparative assessments of CRISPR-Cas nucleases’ cleavage efficiency in planta. Plant Mol. Biol. 2015, 87, 143–156. [Google Scholar] [CrossRef]
- Shan, Q.; Wang, Y.; Li, J.; Zhang, Y.; Chen, K.; Liang, Z.; Zhang, K.; Liu, J.; Xi, J.J.; Qiu, J.-L.; et al. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 2013, 31, 686–688. [Google Scholar] [CrossRef] [PubMed]
- Oh, Y.; Lee, B.; Kim, H.; Kim, S.-G. A multiplex guide RNA expression system and its efficacy for plant genome engineering. Plant Methods 2020, 16, 37. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Zhang, B.; Ding, W.; Liu, X.; Yang, D.-L.; Wei, P.; Cao, F.; Zhu, S.; Zhang, F.; Mao, Y.; et al. Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 2013, 23, 1229–1232. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Chen, L.; Liu, X.; Sun, S.; Wu, C.; Jiang, B.; Han, T.; Hou, W. CRISPR/Cas9-Mediated Genome Editing in Soybean Hairy Roots. PLoS ONE 2015, 10, e0136064. [Google Scholar] [CrossRef]
- Fauser, F.; Schiml, S.; Puchta, H. Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J. 2014, 79, 348–359. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, J.; Wei, P.; Zhang, B.; Gou, F.; Feng, Z.; Mao, Y.; Yang, L.; Zhang, H.; Xu, N.; et al. The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol. J. 2014, 12, 797–807. [Google Scholar] [CrossRef]
- Doench, J.G.; Hartenian, E.; Graham, D.B.; Tothova, Z.; Hegde, M.; Smith, I.; Sullender, M.; Ebert, B.L.; Xavier, R.J.; Root, D.E. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat. Biotechnol. 2014, 32, 1262–1267. [Google Scholar] [CrossRef]
- Wang, W.; Wang, L.; Chen, C.; Xiong, G.; Tan, X.-Y.; Yang, K.-Z.; Wang, Z.-C.; Zhou, Y.; Ye, D.; Chen, L.-Q. Arabidopsis CSLD1 and CSLD4 are required for cellulose deposition and normal growth of pollen tubes. J. Exp. Bot. 2011, 62, 5161–5177. [Google Scholar] [CrossRef]
- Park, S.; Szumlanski, A.L.; Gu, F.; Guo, F.; Nielsen, E. A role for CSLD3 during cell-wall synthesis in apical plasma membranes of tip-growing root-hair cells. Nat. Cell Biol. 2011, 13, 973–980. [Google Scholar] [CrossRef]
- Wang, X.; Cnops, G.; Vanderhaeghen, R.; De Block, S.; Van Montagu, M.; Van Lijsebettens, M. AtCSLD3, A Cellulose Synthase-Like Gene Important for Root Hair Growth in Arabidopsis. Plant Physiol. 2001, 126, 575–586. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 2005, 6, 850–861. [Google Scholar] [CrossRef] [PubMed]
- Thomas, B.R.; Inouhe, M.; Simmons, C.R.; Nevins, D.J. Endo-1,3;1,4-β-glucanase from coleoptiles of rice and maize: Role in the regulation of plant growth. Int. J. Biol. Macromol. 2000, 27, 145–149. [Google Scholar] [CrossRef]
- Li, J.; Cao, C.; Jiang, Y.; Huang, Q.; Shen, Y.; Ni, J. A Novel Digestive GH16 β-1,3(4)-Glucanase from the Fungus-Growing Termite Macrotermes barneyi. Appl. Biochem. Biotechnol. 2020, 192, 1284–1297. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Eberhard, S.; Pattathil, S.; Warder, C.; Glushka, J.; Yuan, C.; Hao, Z.; Zhu, X.; Avci, U.; Miller, J.S.; et al. An Arabidopsis Cell Wall Proteoglycan Consists of Pectin and Arabinoxylan Covalently Linked to an Arabinogalactan Protein. Plant Cell 2013, 25, 270–287. [Google Scholar] [CrossRef]
- Showalter, A.M. Arabinogalactan-proteins: Structure, expression and function. Cell Mol. Life Sci. 2001, 58, 1399–1417. [Google Scholar] [CrossRef]
- Kong, Y.; Zhou, G.; Yin, Y.; Xu, Y.; Pattathil, S.; Hahn, M.G. Molecular Analysis of a Family of Arabidopsis Genes Related to Galacturonosyltransferases. Plant Physiol. 2011, 155, 1791–1805. [Google Scholar] [CrossRef]
- Philippe, F.; Pelloux, J.; Rayon, C. Plant pectin acetylesterase structure and function: New insights from bioinformatic analysis. BMC Genom. 2017, 18, 456. [Google Scholar] [CrossRef]
- Van Acker, R.; Déjardin, A.; Desmet, S.; Hoengenaert, L.; Vanholme, R.; Morreel, K.; Laurans, F.; Kim, H.; Santoro, N.; Foster, C.; et al. Different Routes for Conifer- and Sinapaldehyde and Higher Saccharification upon Deficiency in the Dehydrogenase CAD1. Plant Physiol. 2017, 175, 1018–1039. [Google Scholar] [CrossRef]
- Bonawitz, N.D.; Chapple, C. The Genetics of Lignin Biosynthesis: Connecting Genotype to Phenotype. Annu. Rev. Genet. 2010, 44, 337–363. [Google Scholar] [CrossRef]
- Irish, B.M.; Correll, J.C.; Koike, S.T.; Morelock, T.E. Three New Races of the Spinach Downy Mildew Pathogen Identified by a Modified Set of Spinach Differentials. Plant Dis. 2007, 91, 1392–1396. [Google Scholar] [CrossRef]



| Gene | Number of Hairy Root Lines | Target Site/Sequence | GC Content (%) | Number of Hairy Root Lines with Mutation | Mutation Rate (%) | Number of Hairy Root Lines with Each Mutation Type | |||
|---|---|---|---|---|---|---|---|---|---|
| i | r | d | c | ||||||
| SoCSLD2 | 15 | GTGTCAAACTCCCTCTTTAC | 45 | 3 | 20.0 | 0 | 2 | 1 | 0 |
| CTCCTACCCAGCAGAGACGA | 60 | 1 | 6.7 | 0 | 1 | 0 | 0 | ||
| AAACCAGAGTTCACAAACCA | 40 | 13 | 86.7 | 1 | 0 | 12 | 2 | ||
| SoCSLD3 | 15 | AACATCCAATGAGCAGGAGT | 45 | 13 | 86.7 | 6 | 2 | 5 | 7 |
| ACAAAGGGGACATATGGGTA | 45 | 9 | 60.0 | 0 | 0 | 4 | 5 | ||
| TTCTGGGATTCTTCCTCGTG | 50 | 8 | 53.3 | 0 | 0 | 5 | 5 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, Y.; Xu, Y.; Zhang, Y.; Zhang, H.; Qin, Z.; Bai, C.; Zhang, H.; Ma, D.; Wang, Q.; Fu, C.; et al. Efficient Editing of SoCSLD2 by CRISPR/Cas9 Affects Morphogenesis of Root Hair in Spinach. Horticulturae 2022, 8, 735. https://doi.org/10.3390/horticulturae8080735
Cao Y, Xu Y, Zhang Y, Zhang H, Qin Z, Bai C, Zhang H, Ma D, Wang Q, Fu C, et al. Efficient Editing of SoCSLD2 by CRISPR/Cas9 Affects Morphogenesis of Root Hair in Spinach. Horticulturae. 2022; 8(8):735. https://doi.org/10.3390/horticulturae8080735
Chicago/Turabian StyleCao, Yingping, Yue Xu, Yue Zhang, Heng Zhang, Zhi Qin, Chen Bai, Hailing Zhang, Dongmei Ma, Quanhua Wang, Chunxiang Fu, and et al. 2022. "Efficient Editing of SoCSLD2 by CRISPR/Cas9 Affects Morphogenesis of Root Hair in Spinach" Horticulturae 8, no. 8: 735. https://doi.org/10.3390/horticulturae8080735
APA StyleCao, Y., Xu, Y., Zhang, Y., Zhang, H., Qin, Z., Bai, C., Zhang, H., Ma, D., Wang, Q., Fu, C., Sun, M., & Dai, S. (2022). Efficient Editing of SoCSLD2 by CRISPR/Cas9 Affects Morphogenesis of Root Hair in Spinach. Horticulturae, 8(8), 735. https://doi.org/10.3390/horticulturae8080735







