Grape Tartaric Acid: Chemistry, Function, Metabolism, and Regulation
Abstract
1. Introduction
2. Characteristics of TA
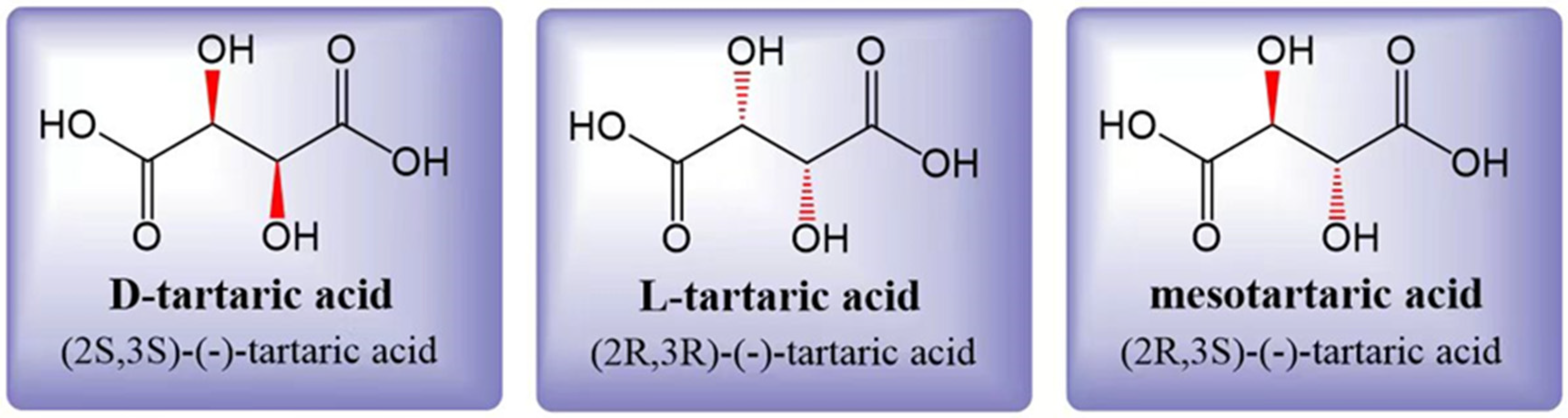
2.1. Structure and Properties
2.2. Distribution and Transportation of TA
3. The Biosynthetic Pathway of TA
3.1. AsA Biosynthesis Stage
3.2. TA Biosynthesis Stage
4. Key Enzymes in TA Biosynthesis Pathway
4.1. Enzymes of the L-Galactose Pathway for AsA Biosynthesis
4.1.1. GDP-D-Mannose-3′,5′-Epimerase, GME
4.1.2. GDP-L-Galactose Phosphorylase, GGP/VTC2
4.1.3. L-Galactose Dehydrogenase, GalDH
4.1.4. L-Galactono-1,4-Lactone Dehydrogenase, GalLDH
4.2. Enzymes of the Alternative Pathway for AsA Biosynthesis
4.2.1. L-Gulono-1,4-Lactone Oxidase, GulLO
4.2.2. D-Galacturonate Reductase, GalUR
4.2.3. Aldonolactonase, Alase
4.2.4. Myo-Inositol Oxygenase, MIOX
4.3. TA Biosynthetic Enzymes
4.3.1. L-Idonate Dehydrogenase, L-IdnDH
4.3.2. 2-Keto-L-Gulonate Reductase, 2-KGR
4.3.3. Transketolase (TK) and Tartaric Semialdehyde Dehydrogenase (TSAD)
5. Metabolism of TA
6. Factors Affecting TA Content
6.1. Environmental Factors
6.2. Grape Training Systems and Rootstocks
6.3. Regulatory Effects of Transcription Factors
7. Future Perspectives
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mu, W.; Feng, J.; Tian, D.; Mu, X. The international trade and domestic demand of the table grape industry in China. China Fruits 2019, 196, 5–10. [Google Scholar] [CrossRef]
- Liu, F.; Wang, H.; Hu, C. Current situation of main fruit tree industry in China and it’s development countermeasure during the “14th five-year plan” period. China Fruits 2021, 1–5. [Google Scholar] [CrossRef]
- Zhu, L.; Chen, Y.; Hu, X.; Li, X.; Zhan, C.; Lyu, S. Research Progress of Organic Acids in Grape. Sino-Overseas Grapevine Wine 2022, 246, 88–95. [Google Scholar] [CrossRef]
- He, D.; Ma, X.; Kang, Q.; Sun, Y. Effects oforganic acids content on the fermentation of wine. China Brew. 2022, 41, 62–67. [Google Scholar]
- Liu, H. Study on Sugars and Acid Composition, Inheritance and Sucrose Metabolism Related Enzymes Activities in Grape Berries. Ph.D. Thesis, China Agricultural Univeisity, Beijing, China, 2005. [Google Scholar]
- Zhang, X.; Liu, C.; Liu, Q.; Fan, X.; Zhang, Y.; Sun, L.; Niu, S. Organic Acid Components and Content Characteristics of Grape Berry. Food Sci. 2021, 43, 228–234. [Google Scholar] [CrossRef]
- Mira de Orduña, R. Climate change associated effects on grape and wine quality and production. Food Res. Int. 2010, 43, 1844–1855. [Google Scholar] [CrossRef]
- Burbidge, C.A.; Ford, C.M.; Melino, V.J.; Wong, D.C.J.; Jia, Y.; Jenkins, C.L.D.; Soole, K.L.; Castellarin, S.D.; Darriet, P.; Rienth, M.; et al. Biosynthesis and Cellular Functions of Tartaric Acid in Grapevines. Front. Plant Sci. 2021, 12, 643024. [Google Scholar] [CrossRef]
- Frioni, T.; Bertoloni, G.; Squeri, C.; Garavani, A.; Ronney, L.; Poni, S.; Gatti, M. Biodiversity of Local Vitis vinifera L. Germplasm: A Powerful Tool Toward Adaptation to Global Warming and Desired Grape Composition. Front. Plant Sci. 2020, 11, 608. [Google Scholar] [CrossRef]
- Derewenda, Z.S. On wine, chirality and crystallography. Acta Crystallogr. A 2008, 64, 246–258. [Google Scholar] [CrossRef]
- Cholet, C.; Claverol, S.; Claisse, O.; Rabot, A.; Osowsky, A.; Dumot, V.; Ferrari, G.; Geny, L. Tartaric acid pathways in Vitis vinifera L. (cv. Ugni blanc): A comparative study of two vintages with contrasted climatic conditions. BMC Plant Biol. 2016, 16, 144. [Google Scholar] [CrossRef]
- Duchêne, É. How can grapevine genetics contribute to the adaptation to climate change? OENO One 2016, 50, 113–124. [Google Scholar] [CrossRef]
- Picariello, L.; Rinaldi, A.; Martino, F.; Petracca, F.; Moio, L.; Gambuti, A. Modification of the organic acid profile of grapes due to climate changes alters the stability of red wine phenolics during controlled oxidation. Vitis 2019, 58, 127–133. [Google Scholar] [CrossRef]
- Poni, S.; Gatti, M.; Palliotti, A.; Dai, Z.; Duchêne, E.; Truong, T.-T.; Ferrara, G.; Matarrese, A.M.S.; Gallotta, A.; Bellincontro, A.; et al. Grapevine quality: A multiple choice issue. Sci. Hortic. 2018, 234, 445–462. [Google Scholar] [CrossRef]
- DeBolt, S.; Cook, D.R.; Ford, C.M. L-tartaric acid synthesis from vitamin C in higher plants. Proc. Natl. Acad. Sci. USA 2006, 103, 5608–5613. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Guo, Y.; Guo, X. Dynamic Changes of Sugar and Acid Content in Grape Fruits during Development. Xinjiang Agric. Sci. 2022, 59, 1680–1689. [Google Scholar]
- Amerine, M.A. The maturation of wine grapes. Wines Vines 1956, 37, 1–11. [Google Scholar]
- Stafford, H.A. Distribution of Tartaric Acid in the Leaves of Certain Angiosperms. Am. J. Bot. 1959, 46, 347–352. [Google Scholar] [CrossRef]
- Stafford, H.A.; Loewus, F.A. The Fixation of CO(2) into Tartaric and Malic Acids of Excised Grape Leaves. Plant Physiol. 1958, 33, 194–199. [Google Scholar] [CrossRef]
- Williams, M.; Loewus, F.A. Biosynthesis of (+)-Tartaric Acid from l-[4-C]Ascorbic Acid in Grape and Geranium. Plant Physiol. 1978, 61, 672–674. [Google Scholar] [CrossRef]
- Kirikoi, Y.T.; Sokolov, O.A. The localization of anabolism and the possibility of the conversion of organic acids into carbohydrates during grape ripening. Fiziol. Rastenii 1974, 21, 780–787. [Google Scholar]
- Etienne, A.; Génard, M.; Lobit, P.; Mbeguié, A.M.D.; Bugaud, C. What controls fleshy fruit acidity? A review of malate and citrate accumulation in fruit cells. J. Exp. Bot. 2013, 64, 1451–1469. [Google Scholar] [CrossRef] [PubMed]
- Sweetman, C.; Deluc, L.G.; Cramer, G.R.; Ford, C.M.; Soole, K.L. Regulation of malate metabolism in grape berry and other developing fruits. Phytochemistry 2009, 70, 1329–1344. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.Y.; Wang, C.K.; Zhao, Y.W.; Sun, C.H.; Hu, D.G. Mechanisms and regulation of organic acid accumulation in plant vacuoles. Hortic. Res. 2021, 8, 227. [Google Scholar] [CrossRef] [PubMed]
- Davey, M.W.; Montagu, M.V.; Inze, D.; Sanmartin, M.; Kanellis, A.; Smirnoff, N.; Benzie, I.J.J.; Strain, J.J.; Favell, D.; Fletcher, J. Plant L-ascorbic acid: Chemistry, function, metabolism, bioavailability and effects of processing. J. Sci. Food Agric. 2000, 80, 825–860. [Google Scholar] [CrossRef]
- Hancock, R.D.; McRae, D.; Haupt, S.; Viola, R. Synthesis of L-ascorbic acid in the phloem. BMC Plant Biol. 2003, 3, 7. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pignocchi, C.; Foyer, C.H. Apoplastic ascorbate metabolism and its role in the regulation of cell signalling. Curr. Opin. Plant Biol. 2003, 6, 379–389. [Google Scholar] [CrossRef]
- Wen, Y.Q.; Li, J.M.; Zhang, Z.Z.; Zhang, Y.F.; Pan, Q.H. Antibody preparation, gene expression and subcellular localization of L-idonate dehydrogenase in grape berry. Biosci. Biotechnol. Biochem. 2010, 74, 2413–2417. [Google Scholar] [CrossRef][Green Version]
- Ford, C.M. The Biochemistry of Organic Acids in the Grape. Biochem. Grape Berry 2012, 22, 67–88. [Google Scholar] [CrossRef]
- Melino, V.J.; Soole, K.L.; Ford, C.M. Ascorbate metabolism and the developmental demand for tartaric and oxalic acids in ripening grape berries. BMC Plant Biol. 2009, 9, 145. [Google Scholar] [CrossRef]
- Narnoliya, L.K.; Sangwan, R.S.; Singh, S.P. Transcriptome mining and in silico structural and functional analysis of ascorbic acid and tartaric acid biosynthesis pathway enzymes in rose-scanted geranium. Mol. Biol. Rep. 2018, 45, 315–326. [Google Scholar] [CrossRef]
- Wheeler, G.L.; Jones, M.A.; Smirnoff, N. The biosynthetic pathway of vitamin C in higher plants. Nature 1998, 393, 365–369. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Rus, E.; Botella, M.A.; Valpuesta, V.; Gomez-Jimenez, M.C. Analysis of genes involved in L-ascorbic acid biosynthesis during growth and ripening of grape berries. J. Plant Physiol. 2010, 167, 739–748. [Google Scholar] [CrossRef] [PubMed]
- Wolucka, B.A.; Van Montagu, M. GDP-mannose 3′,5′-epimerase forms GDP-L-gulose, a putative intermediate for the de novo biosynthesis of vitamin C in plants. J. Biol. Chem. 2003, 278, 47483–47490. [Google Scholar] [CrossRef] [PubMed]
- Agius, F.; Gonzalez-Lamothe, R.; Caballero, J.L.; Munoz-Blanco, J.; Botella, M.A.; Valpuesta, V. Engineering increased vitamin C levels in plants by overexpression of a D-galacturonic acid reductase. Nat. Biotechnol. 2003, 21, 177–181. [Google Scholar] [CrossRef] [PubMed]
- Lorence, A.; Chevone, B.I.; Mendes, P.; Nessler, C.L. myo-inositol oxygenase offers a possible entry point into plant ascorbate biosynthesis. Plant Physiol. 2004, 134, 1200–1205. [Google Scholar] [CrossRef] [PubMed]
- Laing, W.A.; Bulley, S.; Wright, M.; Cooney, J.; Jensen, D.; Barraclough, D.; MacRae, E. A highly specific L-galactose-1-phosphate phosphatase on the path to ascorbate biosynthesis. Proc. Natl. Acad. Sci. USA 2004, 101, 16976–16981. [Google Scholar] [CrossRef]
- Cao, H.; Shu, H.; Shao, J.; Zhang, H.; Ma, C. Research progress on biosynthesis of tartaric acid in grape berries. China Fruits 2021, 210, 8–13. [Google Scholar] [CrossRef]
- Narnoliya, L.K.; Kaushal, G.; Singh, S.P. Long noncoding RNAs and miRNAs regulating terpene and tartaric acid biosynthesis in rose-scented geranium. FEBS Lett. 2019, 593, 2235–2249. [Google Scholar] [CrossRef]
- Jia, Y.; Burbidge, C.A.; Sweetman, C.; Schutz, E.; Soole, K.; Jenkins, C.; Hancock, R.D.; Bruning, J.B.; Ford, C.M. An aldo-keto reductase with 2-keto-l-gulonate reductase activity functions in L-tartaric acid biosynthesis from vitamin C in Vitis vinifera. J. Biol. Chem. 2019, 294, 15932–15946. [Google Scholar] [CrossRef]
- Gilbert, L.; Alhagdow, M.; Nunes-Nesi, A.; Quemener, B.; Guillon, F.; Bouchet, B.; Faurobert, M.; Gouble, B.; Page, D.; Garcia, V.; et al. GDP-D-mannose 3,5-epimerase (GME) plays a key role at the intersection of ascorbate and non-cellulosic cell-wall biosynthesis in tomato. Plant J. 2009, 60, 499–508. [Google Scholar] [CrossRef]
- Tao, J.; Wu, H.; Li, Z.; Huang, C.; Xu, X. Molecular Evolution of GDP-D-Mannose Epimerase (GME), a Key Gene in Plant Ascorbic Acid Biosynthesis. Front. Plant Sci. 2018, 9, 1293. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Liu, J.; Zhang, Y.; Cai, X.; Gong, P.; Zhang, J.; Wang, T.; Li, H.; Ye, Z. Overexpression of SlGMEs leads to ascorbate accumulation with enhanced oxidative stress, cold, and salt tolerance in tomato. Plant Cell Rep. 2011, 30, 389–398. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Wang, Y.; Liu, W.; Liu, Z. Overexpression of an alfalfa GDP-mannose 3, 5-epimerase gene enhances acid, drought and salt tolerance in transgenic Arabidopsis by increasing ascorbate accumulation. Biotechnol. Lett. 2014, 36, 2331–2341. [Google Scholar] [CrossRef] [PubMed]
- Conklin, P.L.; Saracco, S.A.; Norris, S.R.; Last, R.L. Identification of ascorbic acid-deficient Arabidopsis thaliana mutants. Genetics 2000, 154, 847–856. [Google Scholar] [CrossRef] [PubMed]
- Linster, C.L.; Gomez, T.A.; Christensen, K.C.; Adler, L.N.; Young, B.D.; Brenner, C.; Clarke, S.G. Arabidopsis VTC2 encodes a GDP-L-galactose phosphorylase, the last unknown enzyme in the Smirnoff-Wheeler pathway to ascorbic acid in plants. J. Biol. Chem. 2007, 282, 18879–18885. [Google Scholar] [CrossRef]
- Linster, C.L.; Clarke, S.G. L-Ascorbate biosynthesis in higher plants: The role of VTC2. Trends Plant Sci. 2008, 13, 567–573. [Google Scholar] [CrossRef]
- Dowdle, J.; Ishikawa, T.; Gatzek, S.; Rolinski, S.; Smirnoff, N. Two genes in Arabidopsis thaliana encoding GDP-L-galactose phosphorylase are required for ascorbate biosynthesis and seedling viability. Plant J. 2007, 52, 673–689. [Google Scholar] [CrossRef]
- Bulley, S.; Wright, M.; Rommens, C.; Yan, H.; Rassam, M.; Lin-Wang, K.; Andre, C.; Brewster, D.; Karunairetnam, S.; Allan, A.C.; et al. Enhancing ascorbate in fruits and tubers through over-expression of the L-galactose pathway gene GDP-L-galactose phosphorylase. Plant Biotechnol. J. 2012, 10, 390–397. [Google Scholar] [CrossRef]
- Smirnoff, N.; Conklin, P.L.; Loewus, F.A. BIOSYNTHESIS OF ASCORBIC ACID IN PLANTS: A Renaissance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 437–467. [Google Scholar] [CrossRef]
- Li, M.; Gao, J.; Ma, F.; Liang, D.; Hou, C. Relationship Between Expressions of GalDH and GalLDH and Ascorbate Content in Apple Fruits. Sci. Agric. Sin. 2010, 43, 351–357. [Google Scholar] [CrossRef]
- Liao, G.; Chen, L.; He, Y.; Li, X.; Lv, Z.; Yi, S.; Zhong, M.; Huang, C.; Jia, D.; Qu, X.; et al. Three metabolic pathways are responsible for the accumulation and maintenance of high AsA content in kiwifruit (Actinidia eriantha). BMC Genom. 2021, 22, 13. [Google Scholar] [CrossRef] [PubMed]
- Lu, N.; Wu, L.; Zhang, X.; Zhang, Y.; Shan, C. Selenium improves the content of vitamin C in the fruit of strawberry by regulating the enzymes responsible for vitamin C metabolism. Plant Soil Environ. 2022, 68, 205–211. [Google Scholar] [CrossRef]
- Maruta, T.; Ichikawa, Y.; Mieda, T.; Takeda, T.; Tamoi, M.; Yabuta, Y.; Ishikawa, T.; Shigeoka, S. The contribution of Arabidopsis homologs of L-gulono-1,4-lactone oxidase to the biosynthesis of ascorbic acid. Biosci. Biotechnol. Biochem. 2010, 74, 1494–1497. [Google Scholar] [CrossRef] [PubMed]
- Aboobucker, S.I.; Suza, W.P.; Lorence, A. Characterization of Two Arabidopsis L-Gulono-1,4-lactone Oxidases, AtGulLO3 and AtGulLO5, Involved in Ascorbate Biosynthesis. React. Oxyg. Species (Apex) 2017, 4, 389–417. [Google Scholar] [CrossRef]
- Amaya, I.; Osorio, S.; Martinez-Ferri, E.; Lima-Silva, V.; Doblas, V.G.; Fernandez-Munoz, R.; Fernie, A.R.; Botella, M.A.; Valpuesta, V. Increased antioxidant capacity in tomato by ectopic expression of the strawberry D-galacturonate reductase gene. Biotechnol. J. 2015, 10, 490–500. [Google Scholar] [CrossRef]
- Oba, K.; Ishikawa, S.; Nishikawa, M.; Mizuno, H.; Yamamoto, T. Purification and properties of L-galactono-gamma-lactone dehydrogenase, a key enzyme for ascorbic acid biosynthesis, from sweet potato roots. J. Biochem. 1995, 117, 120–124. [Google Scholar] [CrossRef]
- Mapson, L.W.; Breslow, E. Biological synthesis of ascorbic acid: L-galactono-gamma-lactone dehydrogenase. Biochem. J. 1958, 68, 395–406. [Google Scholar] [CrossRef]
- Cruz-Rus, E.; Amaya, I.; Sanchez-Sevilla, J.F.; Botella, M.A.; Valpuesta, V. Regulation of L-ascorbic acid content in strawberry fruits. J. Exp. Bot. 2011, 62, 4191–4201. [Google Scholar] [CrossRef]
- Badejo, A.A.; Wada, K.; Gao, Y.; Maruta, T.; Sawa, Y.; Shigeoka, S.; Ishikawa, T. Translocation and the alternative D-galacturonate pathway contribute to increasing the ascorbate level in ripening tomato fruits together with the D-mannose/L-galactose pathway. J. Exp. Bot. 2012, 63, 229–239. [Google Scholar] [CrossRef]
- Yao, Y.; Liu, Y.; Liu, M. Cloning and expression analysis of Alase family genes in kiwifruit (Actinidia chinensis). Chin. J. Appl. Environ. Biol. 2017, 23, 0209–0214. [Google Scholar] [CrossRef]
- Endres, S.; Tenhaken, R. Myoinositol oxygenase controls the level of myoinositol in Arabidopsis, but does not increase ascorbic acid. Plant Physiol. 2009, 149, 1042–1049. [Google Scholar] [CrossRef] [PubMed]
- Munir, S.; Mumtaz, M.A.; Ahiakpa, J.K.; Liu, G.; Chen, W.; Zhou, G.; Zheng, W.; Ye, Z.; Zhang, Y. Genome-wide analysis of Myo-inositol oxygenase gene family in tomato reveals their involvement in ascorbic acid accumulation. BMC Genom. 2020, 21, 284. [Google Scholar] [CrossRef] [PubMed]
- Duan, J.; Zhang, M.; Zhang, H.; Xiong, H.; Liu, P.; Ali, J.; Li, J.; Li, Z. OsMIOX, a myo-inositol oxygenase gene, improves drought tolerance through scavenging of reactive oxygen species in rice (Oryza sativa L.). Plant Sci 2012, 196, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Sweetman, C.; Wong, D.C.; Ford, C.M.; Drew, D.P. Transcriptome analysis at four developmental stages of grape berry (Vitis vinifera cv. Shiraz) provides insights into regulated and coordinated gene expression. BMC Genom. 2012, 13, 691. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.-Q.; Cui, J.; Zhang, Y.; Duan, C.-Q.; Pan, Q.-H. Comparison of organic acid levels and L-IdnDH expression in Chinese-type and European-type grapes. Euphytica 2013, 196, 63–76. [Google Scholar] [CrossRef]
- Jia, Y.; Wong, D.C.J.; Sweetman, C.; Bruning, J.B.; Ford, C.M. New insights into the evolutionary history of plant sorbitol dehydrogenase. BMC Plant Biol. 2015, 15, 101. [Google Scholar] [CrossRef]
- Saito, K.; Kasai, Z. Conversion of L-Ascorbic Acid to L-Idonic Acid, L-Idono-γ-lactoneane 2-Keto-L-idonic Acid in Slices of Immature Grapes. Plant Cell Physiol. 1982, 23, 499–507. [Google Scholar] [CrossRef]
- Yum, D.Y.; Bae, S.S.; Pan, J.G. Purification and characterization of the 2-ketoaldonate reductase from Brevibacterium ketosoreductum ATCC21914. Biosci. Biotechnol. Biochem. 1998, 62, 154–156. [Google Scholar] [CrossRef]
- Burbidge, C.A. Identification and Characterisation of the Enzymes Involved in the Biosynthetic Pathway of Tartaric Acid in Vitis vinifera. Ph.D. Thesis, Flinders University of South Australia, Adelaide, Australia, 2011. [Google Scholar]
- Truesdell, S.J.; Sims, J.C.; Boerman, P.A.; Seymour, J.L.; Lazarus, R.A. Pathways for metabolism of ketoaldonic acids in an Erwinia sp. J. Bacteriol. 1991, 173, 6651–6656. [Google Scholar] [CrossRef][Green Version]
- Yum, D.Y.; Lee, B.Y.; Hahm, D.H.; Pan, J.G. The yiaE gene, located at 80.1 minutes on the Escherichia coli chromosome, encodes a 2-ketoaldonate reductase. J. Bacteriol. 1998, 180, 5984–5988. [Google Scholar] [CrossRef]
- Saito, K.; Kasai, Z. Synthesis of l-(+)-Tartaric Acid from l-Ascorbic Acid via 5-Keto-d-Gluconic Acid in Grapes. Plant Physiol. 1984, 76, 170–174. [Google Scholar] [CrossRef]
- Salusjarvi, T.; Povelainen, M.; Hvorslev, N.; Eneyskaya, E.V.; Kulminskaya, A.A.; Shabalin, K.A.; Neustroev, K.N.; Kalkkinen, N.; Miasnikov, A.N. Cloning of a gluconate/polyol dehydrogenase gene from Gluconobacter suboxydans IFO 12528, characterisation of the enzyme and its use for the production of 5-ketogluconate in a recombinant Escherichia coli strain. Appl. Microbiol. Biotechnol. 2004, 65, 306–314. [Google Scholar] [CrossRef]
- Wang, J.; Li, W.; Xin, Z.; Feng, W.; Sun, X.; Yuan, J. Molecular engineering of transketolase from Escherichia coli and tartaric semialdehyde biosynthesis. Chin. J. Biotechnol. 2022, 38, 4615–4629. [Google Scholar] [CrossRef]
- Chidi, B.S.; Bauer, F.F.; Rossouw, D. Organic Acid Metabolism and the Impact of Fermentation Practices on Wine Acidity: A Review. S. Afr. J. Enol. Vitic. 2018, 39. [Google Scholar] [CrossRef]
- Hrazdina, G.; Parsons, G.F.; Mattick, L.R. Physiological and Biochemical Events During Development and Maturation of Grape Berries. Am. J. Enol. Vitic. 1984, 35, 220–227. [Google Scholar] [CrossRef]
- Ruffner, H.P.; Brem, S.; Malipiero, U. The Physiology of Acid Metabolism in Grape Berry Ripening. Acta Hortic. 1983, 139, 123–128. [Google Scholar] [CrossRef]
- Saito, K.; Kasai, Z. Accumulation of tartaric acid in the ripening process of grapes. Plant Cell Physiol. 1968, 9, 529–537. [Google Scholar] [CrossRef]
- Shimazu, Y.; Uehara, M.; Watanabe, M. Decomposition of l-Tartaric and l-Malic Acids in Grape Must by Botrytis cinerea. Agric. Biol. Chem. 2014, 48, 1565–1573. [Google Scholar] [CrossRef]
- Hurlbert, R.E.; Jakoby, W.B. Tartaric Acid Metabolism. I. Subunits of L(+)-Tartaric Acid Dehydrase. J. Biol. Chem. 1965, 240, 2772–2777. [Google Scholar] [CrossRef]
- Martin, W.R.; Foster, J.W. Production of trans-L-epoxysuccinic acid by fungi and its microbiological conversion to meso-tartartic acid. J. Bacteriol. 1955, 70, 405–414. [Google Scholar] [CrossRef]
- Rosenberger, R.F.; Shilo, M. Diauxie in tartrate-utilising strains of Pseudomonas and its control by oxaloacetate. Biochem. Biophys. Res. Commun. 1961, 4, 414–419. [Google Scholar] [CrossRef] [PubMed]
- Kun, E. Enzymatic mechanism of oxidation of tartrate. J. Biol. Chem. 1956, 221, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Kohn, L.D.; Jakoby, W.B. Tartaric acid metabolism II. Crystalline protein converting meso-tartrate and dihydroxyfumarate to glycerate. Biochem. Biophys. Res. Commun. 1966, 22, 33–37. [Google Scholar] [CrossRef]
- Crouzet, P.; Otten, L. Sequence and mutational analysis of a tartrate utilization operon from Agrobacterium vitis. J. Bacteriol. 1995, 177, 6518–6526. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ruffner, H.P. Metabolism of tartaric and malic acids in Vitis: A review—Part A. Vitis 1982, 21, 247–259. [Google Scholar] [CrossRef]
- Kliewer, W.M.; Lider, L.A.; Schultz, H.B. Influence of Artificial Shading of Vineyards on the Concentration of Sugar and Organic Acid in Grapes. Am. J. Enol. Vitic. 1967, 18, 78–86. [Google Scholar] [CrossRef]
- Kiiewer, W.M. Effect of Day Temperature and Light Intensity on Concentration of Malic and Tartaric Acids in Vitis vinifera L. Grapes. J. Am. Soc. Hortic. Sci. 1971, 96, 372–377. [Google Scholar] [CrossRef]
- Ristic, R.; Iland, P.G.; Ford, C.M. Altered light interception reduces grape berry weight and modulates organic acid biosynthesis during development. J. Am. Soc. Hortic. Sci. 2008, 43, 957–961. [Google Scholar] [CrossRef]
- Kliewer, W.M. Influence of Environment on Metabolism of Organic Acids and Carbohydrates in Vitis Vinifera. I. Temperature. Plant Physiol. 1964, 39, 869–880. [Google Scholar] [CrossRef]
- Melino, V.J.; Hayes, M.A.; Soole, K.L.; Ford, C.M. The role of light in the regulation of ascorbate metabolism during berry development in the cultivated grapevine Vitis vinifera L. J. Sci. Food Agric. 2011, 91, 1712–1721. [Google Scholar] [CrossRef]
- Ziliotto, F.; Corso, M.; Rizzini, F.M.; Rasori, A.; Botton, A.; Bonghi, C. Grape berry ripening delay induced by a pre-véraison NAA treatment is paralleled by a shift in the expression pattern of auxin- and ethylene-related genes. BMC Plant Biol. 2012, 12, 185. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Qian, Y.; Wu, W.; Zhao, M.; Zhou, B.; Wang, Z.; Wu, J. Effect of 6-BA on Organic Acid Content and Related Genes Expression in Grape Berry. Acta Agric. Boreali-Sin. 2017, 32, 149–153. [Google Scholar]
- Niu, Y.; Dong, Y.; Zhang, P.; Niu, T.; Wen, P. Effect of Foliar Spraying SA on the Organic Acids Content of Grape Jizaomi during Berry Development. J. Shanxi Agric. Sci. 2017, 45, 1426–1429. [Google Scholar] [CrossRef]
- Chi, M.; Li, M.; Zhang, Z. Effect of Different Training Systems on Quality of ‘Cabernet Sauvignon’ Grape Berries. North. Hortic. 2014, 321, 50–53. [Google Scholar]
- Niu, D. Effects of Orthopedics and Rootstock on Sugarandacid Metabolism and Organic Acid-Related Gene Expression in Grape Fruits. Master’s Thesis, Shanxi Agricultural University, Taiyuan, China, 2019. [Google Scholar]
- Attia, F.; Garcia, F.; Garcia, M. Effect of Rootstock on Organic Acids in Leaves and Berries and on Must and Wine Acidity of Two Red Wine Grape Cultivars ‘Malbec’ and ‘Négrette’ (Vitis vinifera L.) Grown Hydroponically. Acta Hortic. 2007, 754, 473–482. [Google Scholar] [CrossRef]
- Wei, L.; Cheng, J.; Li, L.; Mei, J.; Wu, J. Effects of SO4 and Beta rootstocks on the growth and berry quality of Yinhong grapevine. Sino-Overseas Grapevine Wine 2012, 183, 23–25. [Google Scholar] [CrossRef]
- Liu, L.; White, M.J.; MacRae, T.H. Transcription factors and their genes in higher plants functional domains, evolution and regulation. Eur. J. Biochem. 1999, 262, 247–257. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, J.; Zhang, R.; Huang, R. The ethylene response factor AtERF98 enhances tolerance to salt through the transcriptional activation of ascorbic acid synthesis in Arabidopsis. Plant J. 2012, 71, 273–287. [Google Scholar] [CrossRef]
- Wang, Q. Identification and Functional Characterization of Regulatory Genes of Organic Acid Accumulation in Peach. Ph.D. Thesis, Huazhong Agricultural University, Wuhan, China, 2022. [Google Scholar]
- Zhang, L. Molecular Mechanism of MdWRKY126 in Regulating Organic Acid and Sugar Content in Apple Fruit. Ph.D. Thesis, Northwest A&F University, Yangling, China, 2022. [Google Scholar]
- Zhang, Q. Molecular Mechanism of Action of Nitrate-Responsive Gene MdBT2 Involved in the Regulation of Malate Accumulation in Apple. Ph.D. Thesis, Shandong Agricultural University, Tai’an, China, 2021. [Google Scholar]
- Liu, S.; Wang, Y.; Shi, M.; Maoz, I.; Gao, X.; Sun, M.; Yuan, T.; Li, K.; Zhou, W.; Guo, X.; et al. SmbHLH60 and SmMYC2 antagonistically regulate phenolic acids and anthocyanins biosynthesis in Salvia miltiorrhiza. J. Adv. Res. 2022, 42, 205–219. [Google Scholar] [CrossRef]
- Deng, C.; Hao, X.; Shi, M.; Fu, R.; Wang, Y.; Zhang, Y.; Zhou, W.; Feng, Y.; Makunga, N.P.; Kai, G. Tanshinone production could be increased by the expression of SmWRKY2 in Salvia miltiorrhiza hairy roots. Plant Sci. 2019, 284, 1–8. [Google Scholar] [CrossRef]
- Ye, J.; Li, W.; Ai, G.; Li, C.; Liu, G.; Chen, W.; Wang, B.; Wang, W.; Lu, Y.; Zhang, J.; et al. Genome-wide association analysis identifies a natural variation in basic helix-loop-helix transcription factor regulating ascorbate biosynthesis via D-mannose/L-galactose pathway in tomato. PLoS Genet. 2019, 15, e1008149. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Yan, M.; Dong, H.; Luo, J.; Ke, Y.; Guo, A.; Chen, Y.; Zhang, J.; Huang, X. Maize bHLH55 functions positively in salt tolerance through modulation of AsA biosynthesis by directly regulating GDP-mannose pathway genes. Plant Sci. 2021, 302, 110676. [Google Scholar] [CrossRef] [PubMed]
- Miura, K.; Sato, A.; Shiba, H.; Kang, S.W.; Kamada, H.; Ezura, H. Accumulation of antioxidants and antioxidant activity in tomato, Solanum lycopersicum, are enhanced by the transcription factor SlICE1. Plant Biotechnol. 2012, 29, 261–269. [Google Scholar] [CrossRef]
- Liu, L.; Jia, C.; Zhang, M.; Chen, D.; Chen, S.; Guo, R.; Guo, D.; Wang, Q. Ectopic expression of a BZR1-1D transcription factor in brassinosteroid signalling enhances carotenoid accumulation and fruit quality attributes in tomato. Plant Biotechnol. J. 2014, 12, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Sawake, S.; Tajima, N.; Mortimer, J.C.; Lao, J.; Ishikawa, T.; Yu, X.; Yamanashi, Y.; Yoshimi, Y.; Kawai-Yamada, M.; Dupree, P.; et al. KONJAC1 and 2 Are Key Factors for GDP-Mannose Generation and Affect l-Ascorbic Acid and Glucomannan Biosynthesis in Arabidopsis. Plant Cell 2015, 27, 3397–3409. [Google Scholar] [CrossRef] [PubMed]
- Hu, T.; Ye, J.; Tao, P.; Li, H.; Zhang, J.; Zhang, Y.; Ye, Z. The tomato HD-Zip I transcription factor SlHZ24 modulates ascorbate accumulation through positive regulation of the D-mannose/L-galactose pathway. Plant J. 2016, 85, 16–29. [Google Scholar] [CrossRef]
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef]
- Yuan, J.; Yu, Z.; Lin, T.; Wang, L.; Chen, X.; Liu, T.; Wang, J.; Hou, X.; Li, Y. BcERF070, a novel ERF (ethylene-response factor) transcription factor from non-heading Chinese cabbage, affects the accumulation of ascorbic acid by regulating ascorbic acid-related genes. Mol. Breed. 2019, 40, 2. [Google Scholar] [CrossRef]
- Chen, Y.; Shu, P.; Wang, R.; Du, X.; Xie, Y.; Du, K.; Deng, H.; Li, M.; Zhang, Y.; Grierson, D.; et al. Ethylene response factor AcERF91 affects ascorbate metabolism via regulation of GDP-galactose phosphorylase encoding gene (AcGGP3) in kiwifruit. Plant Sci. 2021, 313, 111063. [Google Scholar] [CrossRef]
- Lu, D.; Wu, Y.; Pan, Q.; Zhang, Y.; Qi, Y.; Bao, W. Identification of key genes controlling L-ascorbic acid during Jujube (Ziziphus jujuba Mill.) fruit development by integrating transcriptome and metabolome analysis. Front. Plant Sci. 2022, 13, 950103. [Google Scholar] [CrossRef]
- Liu, X.; Wu, R.; Bulley, S.M.; Zhong, C.; Li, D. Kiwifruit MYBS1-like and GBF3 transcription factors influence l-ascorbic acid biosynthesis by activating transcription of GDP-L-galactose phosphorylase 3. New Phytol. 2022, 234, 1782–1800. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Lorence, A.; Gruszewski, H.A.; Chevone, B.I.; Nessler, C.L. AMR1, an Arabidopsis gene that coordinately and negatively regulates the mannose/l-galactose ascorbic acid biosynthetic pathway. Plant Physiol. 2009, 150, 942–950. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yu, Y.; Zhang, Z.; Quan, R.; Zhang, H.; Ma, L.; Deng, X.W.; Huang, R. Arabidopsis CSN5B interacts with VTC1 and modulates ascorbic acid synthesis. Plant Cell 2013, 25, 625–636. [Google Scholar] [CrossRef]
- Gago, C.; Drosou, V.; Paschalidis, K.; Guerreiro, A.; Miguel, G.; Antunes, D.; Hilioti, Z. Targeted gene disruption coupled with metabolic screen approach to uncover the LEAFY COTYLEDON1-LIKE4 (L1L4) function in tomato fruit metabolism. Plant Cell Rep. 2017, 36, 1065–1082. [Google Scholar] [CrossRef]
- Castro, J.C.; Castro, C.G.; Cobos, M. Genetic and biochemical strategies for regulation of L-ascorbic acid biosynthesis in plants through the L-galactose pathway. Front. Plant Sci. 2023, 14, 1099829. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Hu, T.; Ye, J.; Wang, B.; Liu, G.; Wang, Y.; Yuan, L.; Li, J.; Li, F.; Ye, Z.; et al. A CCAAT-binding factor, SlNFYA10, negatively regulates ascorbate accumulation by modulating the D-mannose/L-galactose pathway in tomato. Hortic. Res. 2020, 7, 200. [Google Scholar] [CrossRef]
- Kakan, X.; Yu, Y.; Li, S.; Li, X.; Huang, R.; Wang, J. Ascorbic acid modulation by ABI4 transcriptional repression of VTC2 in the salt tolerance of Arabidopsis. BMC Plant Biol. 2021, 21, 112. [Google Scholar] [CrossRef]
- Tabata, K.; Takaoka, T.; Esaka, M. Gene expression of ascorbic acid-related enzymes in tobacco. Phytochemistry 2002, 61, 631–635. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, M.; Su, J.; Yang, H.; Feng, L.; Wang, M.; Xu, G.; Shao, J.; Ma, C. Grape Tartaric Acid: Chemistry, Function, Metabolism, and Regulation. Horticulturae 2023, 9, 1173. https://doi.org/10.3390/horticulturae9111173
Li M, Su J, Yang H, Feng L, Wang M, Xu G, Shao J, Ma C. Grape Tartaric Acid: Chemistry, Function, Metabolism, and Regulation. Horticulturae. 2023; 9(11):1173. https://doi.org/10.3390/horticulturae9111173
Chicago/Turabian StyleLi, Menghan, Jing Su, Huanqi Yang, Lei Feng, Minghui Wang, Gezhe Xu, Jianhui Shao, and Chunhua Ma. 2023. "Grape Tartaric Acid: Chemistry, Function, Metabolism, and Regulation" Horticulturae 9, no. 11: 1173. https://doi.org/10.3390/horticulturae9111173
APA StyleLi, M., Su, J., Yang, H., Feng, L., Wang, M., Xu, G., Shao, J., & Ma, C. (2023). Grape Tartaric Acid: Chemistry, Function, Metabolism, and Regulation. Horticulturae, 9(11), 1173. https://doi.org/10.3390/horticulturae9111173



