Mining Genes Related to Single Fruit Weight of Peach (Prunus persica) Based on WGCNA and GSEA
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Phenotype Identification of Single Fruit Weight
2.3. RNA Extracting, Database Building and Data Filtering
2.4. Comparative Analysis of Reference Sequences and Differential Expression
2.5. Analysis of WGCNA
2.6. GSEA Analysis
2.7. Fluorescence Quantitative PCR Analysis
3. Results
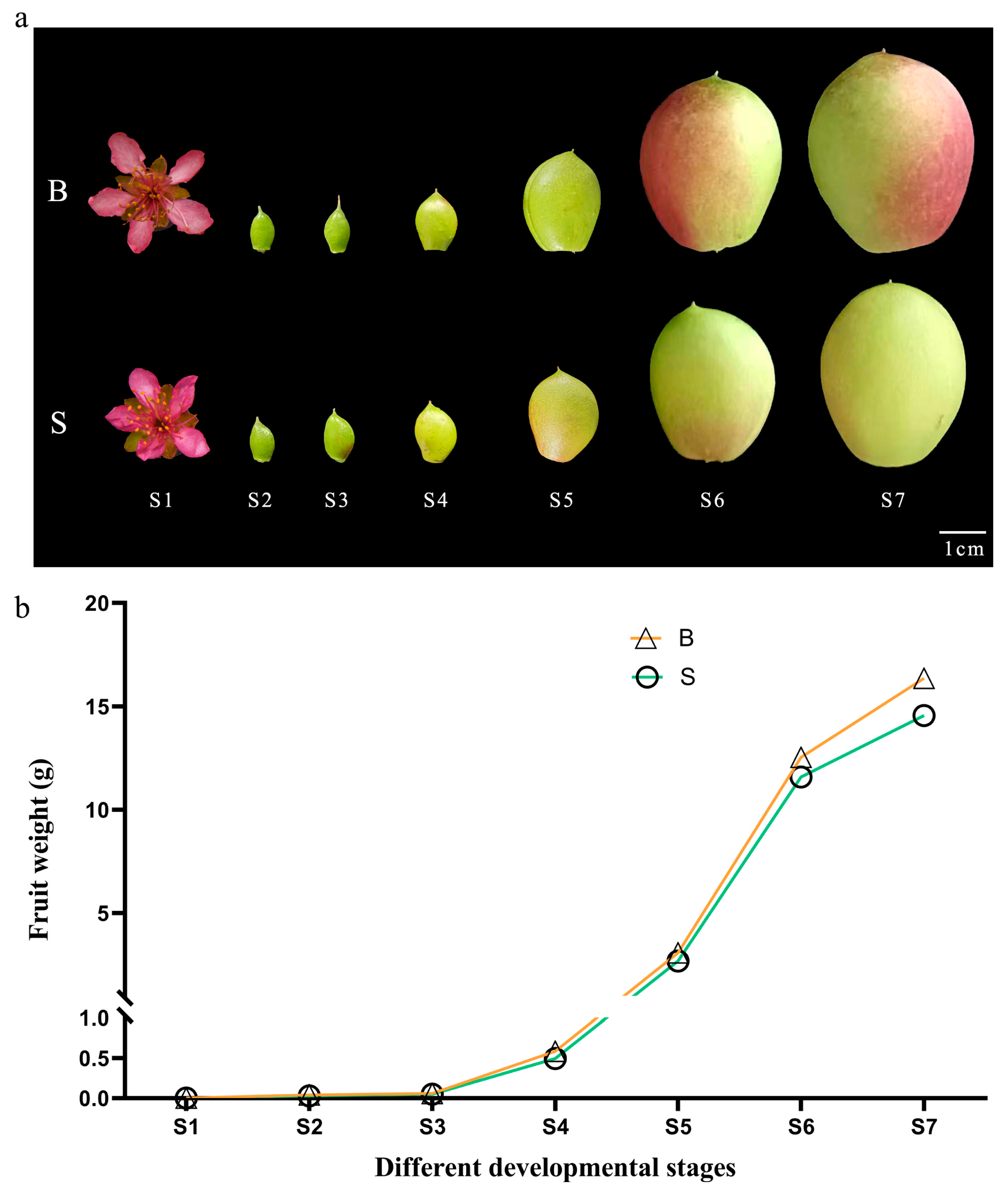
3.1. Changes of Single Fruit Weight Traits during Peach Fruit Development
3.2. The Quality Evaluation of RNA-Seq Sequencing Data
3.3. Statistics of Differentially Expressed Genes
3.4. Screening Key Candidate Genes Based on WGCNA
3.5. Gene Set Enrichment Analysis GSEA
3.6. Screening of Key Genes for Single Fruit Weight by Three Methods
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Biscarini, F.; Nazzicari, N.; Bink, M.; Arús, P.; Aranzana, M.J.; Verde, I.; Micali, S.; Pascal, T.; Quilot-Turion, B.; Lambert, P.; et al. Genome-enabled predictions for fruit weight and quality from repeated records in European peach progenies. BMC Genom. 2017, 18, 432. [Google Scholar] [CrossRef]
- Yu, Y.; Guan, J.; Xu, Y.; Ren, F.; Zhang, Z.; Yan, J.; Fu, J.; Guo, J.; Shen, Z.; Zhao, J.; et al. Population-scale peach genome analyses unravel selection patterns and biochemical basis underlying fruit flavor. Nat. Commun. 2021, 12, 3604. [Google Scholar] [CrossRef]
- Chalmers, D.J.; Ende, B.V.D. Productivity of peach trees: Factors affecting dry-weight distribution during tree growth. Ann. Bot. 1975, 39, 423–432. [Google Scholar] [CrossRef]
- Horiguchi, G.; Ferjani, A.; Fujikura, U.; Tsukaya, H. Coordination of cell proliferation and cell expansion in the control of leaf size in Arabidopsis thaliana. J. Plant Res. 2006, 119, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Si, L.; Chen, J.; Huang, X.; Gong, H.; Luo, J.; Hou, Q.; Zhou, T.; Lu, T.; Zhu, J.; Shangguan, Y.; et al. OsSPL13 controls grain size in cultivated rice. Nat. Genet. 2016, 48, 447–456. [Google Scholar] [CrossRef] [PubMed]
- Masami, Y.; Takashi, H.; Masanori, M.; Hideaki, Y. Varietal differences in cell division and enlargement periods during peach (Prunus persica Batsch) fruit development. J. Jpn. Soc. Hortic. Sci. 2002, 71, 155–163. [Google Scholar]
- Bradley, M.V. Mean cell size in the mesocarp of mature peaches of different sizes. Proc. Am. Soc. Hortic. Sci. 1959, 73, 120–124. [Google Scholar]
- Bohner, J.; Bangerth, F. Cell number, cell size and hormone levels in semi-isogenic mutants of lycopersicon pimpinellifolium differing in fruit size. Physiol. Plant. 1988, 72, 316–320. [Google Scholar] [CrossRef]
- Higashi, K.; Hosoya, K.; Ezura, H. Histological analysis of fruit development between two melon (Cucumis melo L. reticulatus) genotypes setting a different size of fruit. J. Exp. Bot. 1999, 50, 1593–1597. [Google Scholar] [CrossRef]
- Cheng, G.W.; Breen, P.J. Cell Count and Size in Relation to Fruit Size Among Strawberry Cultivars. J. Am. Soc. Hortic. Sci. 1992, 117, 946–950. [Google Scholar] [CrossRef]
- Goffinet, M.C.; Robinson, T.L.; Lakso, A.N. A comparison of ‘Empire’ apple fruit size and anatomy in unthinned and hand-thinning trees. J. Pomol. Hortic. Sci. 1995, 70, 375–387. [Google Scholar] [CrossRef]
- Cowan, A.K.; Moore-Gordon, C.S.; Bertling, I.; Wolstenholme, B.N. Metabolic Control of Avocado Fruit Growth (Isoprenoid Growth Regulators and the Reaction Catalyzed by 3-Hydroxy-3-Methylglutaryl Coenzyme A Reductase). Plant Physiol. 1997, 114, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Cao, K.; Zhou, Z.; Wang, Q.; Guo, J.; Zhao, P.; Zhu, G.; Fang, W.; Chen, C.; Wang, X.; Wang, X.; et al. Genome-wide association study of 12 agronomic traits in peach. Nat. Commun. 2016, 7, 13246. [Google Scholar] [CrossRef] [PubMed]
- Cao, K.; Li, Y.; Deng, C.H.; Gardiner, S.E.; Zhu, G.; Fang, W.; Chen, C.; Wang, X.; Wang, L. Comparative population genomics identified genomic regions and candidate genes associated with fruit domestication traits in peach. Plant Biotechnol. J. 2019, 17, 1954–1970. [Google Scholar] [CrossRef] [PubMed]
- Ge, H.; Song, W.; Liu, X.; Liang, H. Study on branch color and transcriptome of Salix caprea var. aurea. For. Ecol. Sci. 2022, 37, 370–377. [Google Scholar]
- Fan, Y.; Zheng, Y.; Chen, L.; Xu, L.; da Silva, J.A.T.; Wu, B.; Yu, X. Transcriptomic and proteomic analyses reveal changes in the metabolic pathways of Paeonia lactiflora petaloid stamens. Sci. Hortic. 2023, 312, 111859. [Google Scholar] [CrossRef]
- MacWilliams, J.R.; DNabity, P.; Mauck, K.E.; Kaloshian, I. Transcriptome analysis of aphid-resistant and susceptible near isogenic lines reveals candidate resistance genes in cowpea (Vigna unguiculata). BMC Plant Biol. 2023, 23, 22. [Google Scholar] [CrossRef]
- Hollender, C.A.; Kang, C.; Darwish, O.; Geretz, A.; Matthews, B.F.; Slovin, J.; Alkharouf, N.; Liu, Z. Floral Transcriptomes in Woodland Strawberry Uncover Developing Receptacle and Anther Gene Networks. Plant Physiol. 2014, 165, 1062–1075. [Google Scholar] [CrossRef]
- Gao, C.; Ju, Z.; Li, S.; Zuo, J.; Fu, D.; Tian, H.; Luo, Y.; Zhu, B. Deciphering Ascorbic Acid Regulatory Pathways in Ripening Tomato Fruit Using a Weighted Gene Correlation Network Analysis Approach. J. Integr. Plant Biol. 2013, 55, 1080–1091. [Google Scholar] [CrossRef]
- Bai, Y.; Dougherty, L.; Cheng, L.; Zhong, G.-Y.; Xu, K. Uncovering co-expression gene network modules regulating fruit acidity in diverse apples. BMC Genom. 2015, 16, 612. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, Y.; Shi, C.; Huang, Z.; Zhang, Y.; Li, S.; Li, Y.; Ye, J.; Yu, C.; Li, Z.; et al. SOAPnuke: A MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience 2018, 7, gix120. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B. Aligning Short Sequencing Reads with Bowtie. Curr. Protoc. Bioinform. 2010, 32, 11.7.1–11.7.14. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, 480–484. [Google Scholar] [CrossRef]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Zhang, M.; Zhang, S. Mitogen-activated protein kinase cascades in plant signaling. J. Integr. Plant Biol. 2022, 64, 301–341. [Google Scholar] [CrossRef]
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic Acid: Emergence of a Core Signaling Network. Annu. Rev. Plant Biol. 2010, 61, 651–679. [Google Scholar] [CrossRef] [PubMed]
- Liao, X.; Li, M.; Liu, B.; Yan, M.; Yu, X.; Zi, H.; Liu, R.; Yamamuro, C. Interlinked regulatory loops of ABA catabolism and biosynthesis coordinate fruit growth and ripening in woodland strawberry. Proc. Natl. Acad. Sci. USA 2018, 115, E11542–E11550. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Liu, J.; Zhao, T.; Du, C.; Nie, S.; Zhang, Y.; Lv, S.; Huang, S.; Wang, X. Modification of Threonine-1050 of SlBRI1 regulates BR Signalling and increases fruit yield of tomato. BMC Plant Biol. 2019, 19, 256. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.Q.; Wang, H.H.; Shi, C.H.; Zhang, X.Y.; Duan, K.; Luo, J. Morphological, cytological and fertility consequences of a spontaneous tetraploid of the diploid pear (Pyrus pyrifolia Nakai) cultivar ‘Cuiguan’. Sci. Hortic. 2015, 189, 59–65. [Google Scholar] [CrossRef]
- Xue, H.; Zhang, B.; Tian, J.R.; Chen, M.M.; Zhang, Y.Y.; Zhang, Z.H.; Ma, Y. Comparison of the morphology, growth and development of diploid and autotetraploid ‘Hanfu’ apple trees. Sci. Hortic. 2017, 225, 277–285. [Google Scholar] [CrossRef]
- Pattison, R.J.; Catalá, C. Evaluating auxin distribution in tomato (Solanum lycopersicum) through an analysis of the PIN and AUX/LAX gene families. Plant J. 2012, 70, 585–598. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wu, T.; Liu, J.; Cong, L.; Zhu, Y.; Zhai, R.; Yang, C.; Wang, Z.; Ma, F.; Xu, L. PbGA20ox2 Regulates Fruit Set and Induces Parthenocarpy by Enhancing GA4 Content. Front. Plant Sci. 2020, 11, 113. [Google Scholar] [CrossRef]
- Pang, Y.; Zang, X.Y.; Pang, F.T.; Zhou, T.H.; Tian, F.Z. Changes of CTK and few nitrogen index during development of flower and fruit in Zhanhua jujube. J. N. China Agric. 2017, 5, 101–104. [Google Scholar]
- Zhao, Z.H.; Liu, M.J.; Zhao, J.; Dai, L.; Liu, P. Study on changes of endogenous hormone content in fruit development of ‘Dongzao’ and ‘Linyilizao’. Acta Hortic. Sin. 2014, 41, 2628. [Google Scholar]
- Feng, C.Z.; Chen, Y.; Wang, C.; Kong, Y.H.; Wu, W.H.; Chen, Y.F. Arabidopsis RAV1 transcription factor, phosphorylated by SnRK2 kinases, regulates the expression of ABI3, ABI4, and ABI5 during seed germination and early seedling development. Plant J. 2014, 80, 654–668. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Chen, J.; Xiao, K.; Yang, W.C. Origin of the domesticated horticultural species and molecular bases of fruit shape and size changes during the domestication, taking tomato as an example. Hortic. Plant J. 2017, 3, 125–132. [Google Scholar] [CrossRef]
- Liu, M.-J.; Zhao, J.; Cai, Q.-L.; Liu, G.-C.; Wang, J.-R.; Zhao, Z.-H.; Liu, P.; Dai, L.; Yan, G.; Wang, W.-J.; et al. The complex jujube genome provides insights into fruit tree biology. Nat. Commun. 2014, 5, 5315. [Google Scholar] [CrossRef] [PubMed]
- Jiao, C.; Sun, X.; Yan, X.; Xu, X.; Yan, Q.; Gao, M.; Fei, Z.; Wang, X. Grape Transcriptome Response to Powdery Mildew Infection: Comparative Transcriptome Profiling of Chinese Wild Grapes Provides Insights Into Powdery Mildew Resistance. Phytopathology 2021, 111, 2041–2051. [Google Scholar] [CrossRef] [PubMed]
- Kamber, T.; Buchmann, J.P.; Pothier, J.F.; Smits, T.H.M.; Wicker, T.; Duffy, B. Fire blight disease reactome: RNA-seq transcriptional profile of apple host plant defense responses to Erwinia amylovora pathogen infection. Sci. Rep. 2016, 6, 21600. [Google Scholar] [CrossRef]
- Nardozza, S.; Cooney, J.; Boldingh, H.L.; Hewitt, K.G.; Trower, T.; Jones, D.; Thrimawithana, A.H.; Allan, A.C.; Richardson, A.C. Phytohormone and Transcriptomic Analysis Reveals Endogenous Cytokinins Affect Kiwifruit Growth under Restricted Carbon Supply. Metabolites 2020, 10, 23. [Google Scholar] [CrossRef]
- Walsh, C.K.; Sadanandom, A. Ubiquitin chain topology in plant cell signaling: A new facet to an evergreen story. Front. Plant Sci. 2014, 5, 122. [Google Scholar] [CrossRef]
- Stone, S.L.; Williams, L.A.; Farmer, L.M.; Vierstra, R.D.; Callis, J. Keep on going, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell 2007, 18, 3415–3428. [Google Scholar] [CrossRef]
- Guo, M.; Zhang, Z.; Li, S.; Lian, Q.; Fu, P.; He, Y.; Qiao, J.; Xu, K.; Liu, L.; Wu, M.; et al. Genomic analyses of diverse wild and cultivated accessions provide insights into the evolutionary history of jujube. Plant Biotechnol. J. 2020, 19, 517–531. [Google Scholar] [CrossRef]
- Nemhauser, J.L.; Hong, F.; Chory, J. Different Plant Hormones Regulate Similar Processes through Largely Nonoverlapping Transcriptional Responses. Cell 2006, 126, 467–475. [Google Scholar] [CrossRef]
- Hagen, G.; Guilfoyle, T. Auxin-responsive gene expression: Genes, promoters and regulatory factors. Plant Mol. Biol. 2002, 49, 375–385. [Google Scholar] [CrossRef]
- Ljung, K.; Bhalerao, R.P.; Sandberg, G. Sites and homeostatic control of auxin biosynthesis in Arabidopsis during vegetative growth. Plant J. 2001, 28, 465–474. [Google Scholar] [CrossRef] [PubMed]
- Niek, S.; Marian, B. The SAUR gene family: The plant’s toolbox for adaptation of growth and development. J. Exp. Bot. 2018, 70, 17–27. [Google Scholar]
- McClure, B.A.; Guifoyle, T. Characterizaton of a class of small auxin-inducible soybean polyadenylated RNAs. Plant Mol. Biol. 1987, 9, 611–623. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.T.; Mori, H.; Imaseki, H. cDNA Cloning of Indole-3-Acetic Acid-Regulated Genes: Aux22 and SAUR from Mung Bean (Vigna radiata) Hypocotyl Tissue. Plant Cell Physiol. 1992, 33, 93–97. [Google Scholar] [CrossRef]
- Guilfoyle, T.J.; Hagen, G.; Li, Y.; Ulmasov, T.; Liu, Z.B.; Strabala, T.; Gee, M. Auxin-Regulated Transcription. Funct. Plant Biol. 1993, 20, 489–502. [Google Scholar] [CrossRef]
- Gil, P.; Liu, Y.; Orbović, V.; Verkamp, E.; Poff, K.L.; Green, P.J. Characterization of the Auxin-Inducible SAUR-AC1 Gene for Use as a Molecular Genetic Tool in Arabidopsis. Plant Physiol. 1994, 104, 777–784. [Google Scholar] [CrossRef] [PubMed]
- Roux, C.; Bilang, J.; Theunissen, B.H.; Perrot-Rechenmann, C. Identification of new early auxin markers in tobacco by mRNA differential display. Plant Mol. Biol. 1998, 37, 385–389. [Google Scholar] [CrossRef]
- Jain, M.; Tyagi, A.K.; Khurana, J.P. Genome-wide analysis, evolutionary expansion, and expression of early auxin-responsive SAUR gene family inrice (Oryza sativa). Genomics 2006, 88, 360–367. [Google Scholar] [CrossRef]
- Wang, S.; Bai, Y.; Shen, C.; Wu, Y.; Zhang, S.; Jiang, D.; Guilfoyle, T.J.; Chen, M.; Qi, Y. Auxin-related gene families in abiotic stress response in Sorghum bicolor. Funct. Integr. Genom. 2010, 10, 533–546. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Liu, S.; He, Y.; Guan, X.; Zhu, X.; Cheng, L.; Wang, J.; Lu, G. Genome-wide analysis of SAUR gene family in Solanaceae species. Gene 2012, 509, 38–50. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Kong, Y.; Wang, J. Research advances in auxin-responsive SAUR genes. Life Sci. 2014, 26, 407–413. [Google Scholar]
- Chen, Y.; Hao, X.; Cao, J. Small auxin upregulated RNA (SAUR) gene family in maize: Identification, evolution, and its phylogenetic comparison with Arabidopsis, rice, and sorghum. J. Integr. Plant Biol. 2014, 56, 133–150. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Shang, Q. Entification and expression analysis of cucumber SAUR gene family. Acta Hortic. Sin. 2019, 46, 1093–1111. [Google Scholar]
- Deng, G.; Huang, X.; Xie, L.; Tan, S.; Gbokie, T.; Bao, Y.; Xie, Z.; Yi, K. Identification and Expression of SAUR Genes in the CAM Plant Agave. Genes 2019, 10, 555. [Google Scholar] [CrossRef]
- Zhang, T.; Jiang, B.; Xie, R.; Xu, M.; Li, H.; Liu, K. Genome-wide identification and analysis of Medicago truncatula Small auxin upregulated RNA (SAUR) gene family uncover their roles in nodule formation. J. Plant Biochem. Biotechnol. 2020, 30, 126–137. [Google Scholar] [CrossRef]
- Jaillon, O.; Aury, J.M.; Noel, B.; Policriti, A. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 2007, 449, 463–467. [Google Scholar]
- Praveen, K.K.; Sunethra, D.; Nihal, D. SAUR53 regulates organ elongation and apical hook development in Arabidopsis. Plant Signal. Behav. 2018, 13, e1514896. [Google Scholar]
- Knauss, S.; Rohrmeier, T.; Lehle, L. The Auxin-induced Maize Gene ZmSAUR2 Encodes a Short-lived Nuclear Protein Expressed in Elongating Tissues. J. Biol. Chem. 2003, 278, 23936–23943. [Google Scholar] [CrossRef] [PubMed]
- Zhai, H.; Zhai, Y.; Tian, Y.; Zhang, Y.; Yang, L.; Wen, Z.; Chen, H. Genome-wide identification of peach SAUR gene family and characterization of PpSAUR5 gene. Acta Hortic. Sin. 2023, 50, 1–14. [Google Scholar]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bie, H.; Wang, H.; Wang, L.; Li, Y.; Fang, W.; Chen, C.; Wang, X.; Wu, J.; Cao, K. Mining Genes Related to Single Fruit Weight of Peach (Prunus persica) Based on WGCNA and GSEA. Horticulturae 2023, 9, 1335. https://doi.org/10.3390/horticulturae9121335
Bie H, Wang H, Wang L, Li Y, Fang W, Chen C, Wang X, Wu J, Cao K. Mining Genes Related to Single Fruit Weight of Peach (Prunus persica) Based on WGCNA and GSEA. Horticulturae. 2023; 9(12):1335. https://doi.org/10.3390/horticulturae9121335
Chicago/Turabian StyleBie, Hangling, Huimin Wang, Lirong Wang, Yong Li, Weichao Fang, Changwen Chen, Xinwei Wang, Jinlong Wu, and Ke Cao. 2023. "Mining Genes Related to Single Fruit Weight of Peach (Prunus persica) Based on WGCNA and GSEA" Horticulturae 9, no. 12: 1335. https://doi.org/10.3390/horticulturae9121335
APA StyleBie, H., Wang, H., Wang, L., Li, Y., Fang, W., Chen, C., Wang, X., Wu, J., & Cao, K. (2023). Mining Genes Related to Single Fruit Weight of Peach (Prunus persica) Based on WGCNA and GSEA. Horticulturae, 9(12), 1335. https://doi.org/10.3390/horticulturae9121335






