Abstract
Chrysanthemums represent the second most important cut flower after rose on the global commercial market. The phenomenal importance and global popularity of chrysanthemums have attracted breeders’ attention, resulting in the release of vast numbers of cultivars. Identifying these cultivars is crucial to protecting breeders’ intellectual property rights and improving the efficiency of breeding. Distinguishing chrysanthemum genotypes based on their morphological characteristics is challenging as they vary highly within this group, hence requiring the use of efficient molecular markers. In this study, we evaluated the genetic diversity of 57 spray-type chrysanthemum cultivars bearing white, ivory, and cream-colored flowers. A total of six loci were evaluated regarding their polymorphism efficiency across the tested cultivars. Allele numbers ranged from 2 to 6, with a mean of 3.5 alleles per locus. The average polymorphism information content (PIC) was 0.53 for six SSR markers. Cluster analysis of genetic relationships using the UPGMA method showed a genetic distance of 0.31 to 1.00, and the 57 white variants of chrysanthemum cultivars were characterized using the tested SSR markers. However, two sets of cultivars, namely, Pure Angel–Neba and Ladost–White wing, exhibited total genetic similarity and hence could not be discriminated. These results provide efficient SSR markers that can be used to identify chrysanthemum cultivars (and assess their genetic relationships) that cannot be discriminated based on phenotype.
1. Introduction
Chrysanthemums (Chrysanthemum morifolium), belonging to the family Asteraceae, are the second most popular ornamental crop after roses and account for the largest share of cut flower production across the global flower market [1]. The total revenue of the floral market was valued at 4.8 billion euros in 2019, and, despite the global pandemic caused by SARS-CoV-2, the annual trade of the flower market has since increased to 5.6 billion euros. Within this figure, the global cut flower turnover of chrysanthemums was estimated to account for 313 million euros (surpassed by roses, accounting for 634 million euros) in 2021 according to the Royal FloraHolland auction [2]. Their vibrant flower colors with a vast diversity in floral phenotype render chrysanthemums an excellent ornamental plant with high aesthetic value. They have been used as cut flowers, potted ornamental plants, a form of ground cover, and for landscaping [3]. Various chrysanthemum species contain substantial amounts of biologically active components and are used for dietary and medicinal purposes in Asia. Flower heads from Chrysanthemum morifolium and Chrysanthemum indicum possess highly nutritive and phytochemical components, which have been used as medicinal tea and for cosmetic purposes [4,5,6]. These components have been widely used to treat headaches, dizziness, and toxin-induced swelling. They have been reported to possess anti-bacterial, anti-oxidant, anti-inflammatory, and anti-arthritic properties [7,8]. Also, chrysanthemum leaves and stems contain major bioactive components similar to those of the flower heads including flavanoids, terpenoids, volatile oils, polysaccharides, and phenols [9,10,11]. These medicinal properties have been attributed to the presence of the main components of these parts, such as flavanoids. Among these flavanoids, linarin is reported to be the major flavone contributing to the medicinal properties of chrysanthemums [12,13]. In addition, flavanoids from chrysanthemum exhibited anti-tumor effects on MKN45 cells, showing the plant’s potential as an anti-tumor therapy for gastric cancer [4]. Chrysanthemum species generally possess ploidy levels ranging from diploid to decaploid [14]. However, cultivated chrysanthemums are usually hexaploids, but they also exhibit aneuploidy, constituting a stable conformation of 2n = 6x = 54. The allohexaploid background and complex genome of the cultivated chrysanthemum results in a remarkable diversity of floral colors, shapes, and architecture [15,16]. Genetic improvement and research regarding chrysanthemums are lacking due to the plant’s self-incompatibility and outbreeding nature.
The increasing annual demand for chrysanthemums globally compels breeders and researchers to produce cultivars with novel floral attributes. Substantial inbuilt genetic variations that can be easily manipulated via vegetative cultivation methods led to the development of a wide array of varieties, which resulted in the commercialization of hundreds of cultivars with diverse phenotypic variations [17]. Based on the adjustment in the required photoperiodism, the year-round breeding of these diverse cultivars has been improved to meet the increasing demand for these plants [18]. Although the morphological characteristics, including floral color, shape, and size, of each chrysanthemum variety are evaluated, a vast number of cultivars are commercialized with similar floral morphological features, making them difficult to discriminate solely on the basis of morphology. This is because these morphological characteristics can originate from distinct environments and are highly unstable. The commercialization of a vast number of chrysanthemum cultivars with similar morphological characteristics poses a major concern regarding the incorrect labelling of cultivars since there are numerous identical names among the local varieties. The substantial diversity of these varieties poses a major challenge with regard to identifying and classifying the cultivars on the market [19]. Apart from the floral color of the cultivars, floral shape also offers high aesthetic value. The ornamental floral part of a chrysanthemum called the capitulum is composed of a hermaphroditic central disc, which is either green or yellow, and male sterile marginal ray florets, which exhibit diverse colors specific to a given cultivar. Diverse combinations of the size of the petal, the number of ray florets, and floral organ fusion result in different flower types, like single, double, anemone, pompon, spoon, windmill, and pine needle [20]. The identification of cultivars with a similar flower color and shape is critical for chrysanthemum breeding, cultivar registration, trade, genetic improvement, and introduction into the market. The current method of discriminating cultivars based on phenotype is insufficient due to their enormous morphological diversity, rendering the overall process less informative and more time consuming. Also, phenotypic characteristics are multigenic, manifesting only during a particular growth phase, which is regulated by environmental factors, thus hindering the ability to assess them. Hence, the genetic characterization of chrysanthemums is required for the classification and accurate identification of cultivars in order to guarantee breeders’ intellectual property rights [21].
Molecular markers offer researchers the ability to assess the genetic diversity within the germplasm and discriminate and identify cultivars. Genetic studies on chrysanthemums have employed a wide variety of markers [5,22,23]. However, simple sequence repeat (SSR) markers, which are widely used to study population genetics, are multi-allelic, abundant, co-dominant, and highly sensitive, enabling easy scoring in contrast to other markers that are dominant, non-reproducible, and laborious [23,24]. Among the spray- and standard-type chrysanthemums, the spray-type plants are genetically capable of producing a greater number of flowers per plant. The wide range of diverse colors and shapes of spray-type chrysanthemums are highly desired, as cut flowers fetch high market prices. A large number of spray cultivars of identical flower types and colors are commercialized annually. The application of SSR markers is efficient for the evaluation of genetic diversity and the discrimination of cultivars as SSR markers are polymorphic; additionally, they offer locus specificity, high reproducibility, and technical simplicity [25]. SSR markers have been previously used for the discrimination of cultivated chrysanthemum cultivars [26,27]; Shim et al. [28] used 14 SSR markers to assess the genetic characterization of 147 chrysanthemum cultivars. About 11 standard-type chrysanthemum cultivars were discriminated using two SSR markers [29]. Han et al. [30] developed the SSR database for chrysanthemum used to discriminate chrysanthemum cultivars. However, considering the vast number of chrysanthemum cultivars, the use of SSR markers to discriminate them has been relatively uncommon. In this study, we performed the genetic characterization of white-colored variants of different spray-type chrysanthemum cultivars in order to classify and discriminate them using SSR markers.
2. Materials and Methods
2.1. Plant Material
A total of 57 white, ivory, and cream-colored spray-type chrysanthemum cultivars were evaluated. Rooted plantlets were raised and maintained under natural greenhouse conditions at National Institute of Horticultural and Herbal Science (NIHHS), Korea. Plants were provided artificial light at an intensity of 100 μmol photons m−2 s−1 for 6 h on the initial 40 days, with a daily photoperiod of about 14 h to 16 h. Relative humidity and temperature of the green house was constantly maintained at 22 ± 5 °C and 70–75%, respectively. About ten plants from each variety were used for SSR analysis.
2.2. Genomic DNA Extraction
Fresh samples of leaves from the spray-type cultivar plants were collected and frozen in liquid nitrogen, and DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) was used to extract the genomic DNA. DNA quality and quantity were evaluated using Quick Drop (Molecular Devices, CA, USA). A total of 25 ng μL−1 of Diluted DNA was used for SSR analysis.
2.3. Analysis of SSR Markers via ABI Genetic Analyzer
A total of six primers were employed for the differentiation of 57 spray-type chrysanthemum varieties. Primer pairs with higher polymorphism from previous studies were used (20,22). List of primers’ details is presented in Table 1. PCR products were labelled using the M13 tailing method [31]. PCR analysis was performed by amplifying 25 ng of DNA in 20 μL PCR reaction volume containing 0.4 mM of dNTPs, Taq DNA polymerase (0.3U) coupled with 1× buffer, and 8 pmol (5′ FAM labelled) of primers. PCR was run by applying the following cycling conditions: 5 min of 94 °C; 35 cycles of 94 °C, 58 °C, and 72 °C for 60 s, 30 s, and 45 s, respectively; and a 30 min final extension at 72 °C. PCR product (2 μL) was initially analyzed via gel (2.5%) electrophoresis. All the primers produced clear and scorable amplification patterns. PCR analysis was performed at least thrice to determine reproducibility. For genetic analysis, 1 μL of each PCR product was mixed with 10 μL of Hi-Di formamide and 0.12 μL of GeneScan-500 ROX internal size standard per well; the resulting mixture was then analyzed using an ABI PRISM 3100XL Genetic Analyzer (Appplied Biosystems). GeneMapper (Version 3.7) software was used to read the fragment sizes of the PCR fragments of the corresponding SSR loci.

Table 1.
Details of six SSR primer pairs that were screened to discriminate the 57 spray-type white chrysanthemum cultivars.
2.4. Data Analysis
Microsatellite allele data, including the total allele number, heterozygosity, genetic diversity, allele frequency, and polymorphism information content (PIC) were analyzed using Power Marker software v3.25 [32]. The SSR amplification bands were scored as follows: 1 for present and 0 for absent. Unweighted pair group method of arithmetic averages (UPGMA) was performed for the cluster analysis of genetic similarity, and DendroUPGMA software was used to obtain a dendrogram. Principle Component Analysis (PCA) was performed using PAST (v3.26) software with a bootstrap frequency of n = 500.
The number of subpopulations in the tested chrysanthemum germplasm was estimated by performing population structure analysis using STRUCTURE software (v2.3.4) (Stanford University, CA, USA) [33]. The model-based clustering carried out by STRUCTURE provided information about the origin and admixture of the alleles among the cultivars tested. The burn-in length and Markov Chain Monte Carlo (MCMC) iterations were 10,000 and 20,000, respectively [34]. The cultivars were tested for K = 1 to K = 10 using the ‘admixture model’. To quantify the variation in likelihood for each subpopulation (K), fifteen independent runs were evaluated for each fixed K. The appropriate K value was determined from LnP(K) value and the method based on the second order rate of change of likelihood (∆K) [33,34,35]. The true K value was determined according to the peak value of the ∆K plot. Calculations and graph construction were performed using STRUCTURE HARVESTER [36].
3. Results
3.1. Genetic Variation of SSR Markers
A total of 57 spray-type chrysanthemum cultivars with white, ivory, and cream floral colors were characterized using SSR markers. A list of the cultivars with different flower types is shown in Table 2. Among these, 38 cultivars bore white-colored flowers, 17 cultivars had ivory-colored flowers, and 2 cultivars possessed cream-colored flowers (Figure 1). A total of six loci were evaluated with respect to their efficiency of polymorphism across the selected 57 cultivars. All these SSR markers showed polymorphisms among the chrysanthemum cultivars (Figure 2). Among these, two markers individually amplified six and four alleles, followed by three alleles, each amplified by three markers, and one marker amplified two alleles. A total of about 804 scorable bands were obtained for 32 different alleles in the genetic variation analysis of six SSR markers. The average allele number was 3.5 per locus, with the range of allele numbers spanning from 2 (ChSSR_7) to 6 (ChSSR_16) alleles. The allele size of the markers ranged from 142 to 274 bp. The observed heterozygosity (HO) range was about 0.67 to 0.80, with a mean of 0.75. The major allele frequency (MAF) ranged from 0.34 to 0.65, with an average of 0.50 per locus (Table 3). The gene diversity [37] of the 57 chrysanthemum cultivars with respect to the tested SSR markers was about 0.52 to 0.76, with a mean of 0.57. The PIC values were not uniform among the markers, and the PIC values of the six SSR markers were in the range from a low of 0.33 (ChSSR_51) to a high of 0.73 (ChSSR_16), with a mean PIC of 0.53 (Table 3).

Table 2.
List of cultivars used in the study (arranged according to their flower type).
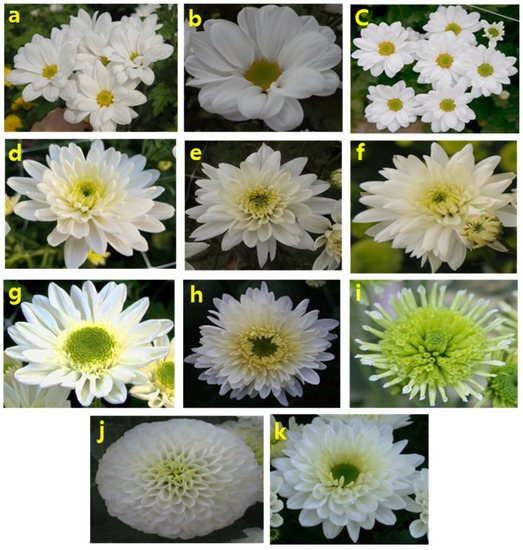
Figure 1.
Spray-type white-colored chrysanthemum cultivars used in this study. (a) White Wing; (b) Innocence; (c) Oh Blang (Single-type); (d) Arctic Queen; (e) Euro; (f) Euro Speedy (Double type) (g) Forest Aroma; (h) Inga; (i) Energy (Anemone type); (j) Snow Pompon (Pompon type); (k) Moon Festival (Semi-Double type).
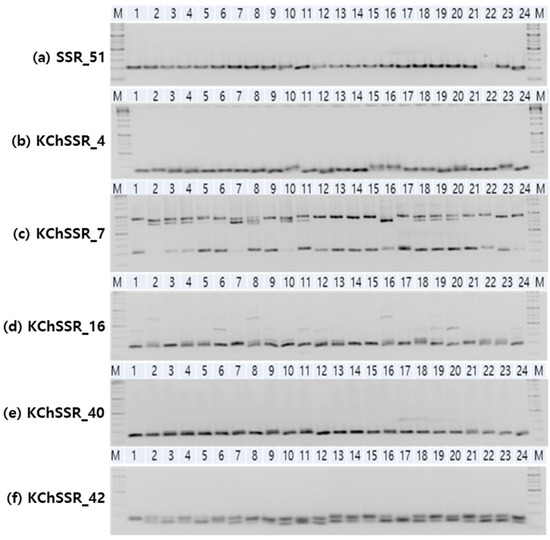
Figure 2.
Representative images of PCR amplicons of chrysanthemum varieties amplified by the six SSR markers run on agarose gel.

Table 3.
Summary of microsatellite allele data, including number of alleles, major allele frequency, observed heterozygosity, polymorphism information content, and gene diversity, revealed in 57 white-colored spray-type cultivars using six SSR markers.
3.2. Analysis of Genetic Relationships between the Cultivars
The genetic relationships of the 57 spray-type varieties of chrysanthemum were evaluated. The Jaccard’s similarity coefficient for all the tested SSR markers ranged from 0.31 to 1.00. An UPGMA-based dendrogram revealed the grouping of all 57 cultivars into three major clusters (Clusters I, II, and III) at a similarity coefficient of 0.41 (Figure 3). The seven cultivars that constituted Cluster I included three single-type white-flowered cultivars, three single-type ivory-flowered cultivars, and one anemone-type ivory-colored cultivar. Cluster III consists of an ivory single and a pompon cultivar, a white double cultivar, and two single-type cultivars, whereas Cluster II was a larger group consisting of about forty cultivars, with a higher number of white single cultivars [22], four anemone-type cultivars, four pompon-type cultivars, and one semi-double-type cultivar. Ivory-colored cultivars were represented by six single-type and four pompon-type cultivars. Both of the cream-colored cultivars tested were grouped in Cluster II. However, two cultivars, namely, Swan (a white anemone type) and Mannadu (an ivory single type), were divided separately, failing to form a group. A principal component analysis (PCA) plot was developed for the 57 cultivars using six SSR markers (Figure 4). The pattern of grouping in the PCA plot correlates with the cluster analysis derived via the UPGMA dendrogram and shows results that match those of the dendrogram.
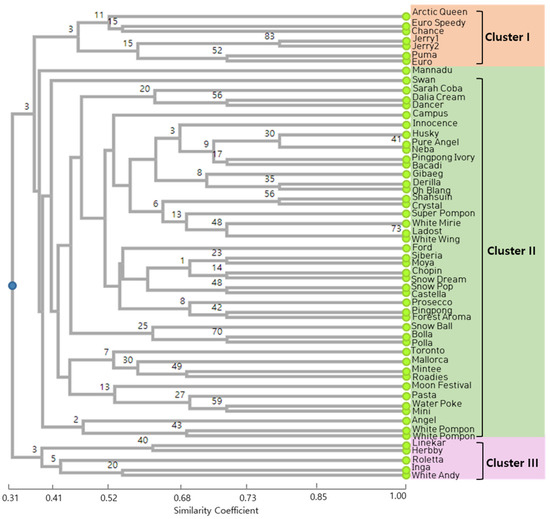
Figure 3.
Dendrogram depicting the classification of 57 spray-type white chrysanthemum cultivars constructed using UPGMA analysis based on six SSR markers. The clusters are marked on the right side of the dendrogram. The scale at the bottom is the Jaccard’s coefficient of similarity. Bootstrap values are given as nodal values along the branches.
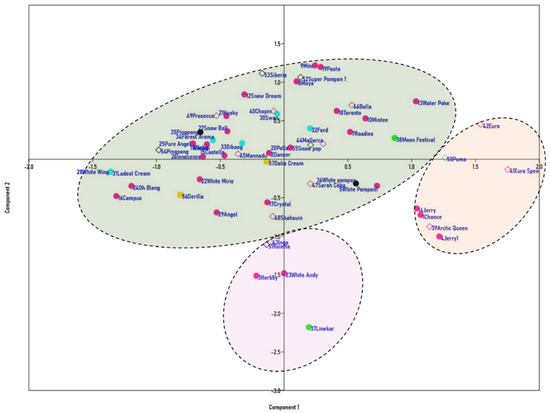
Figure 4.
Plot showing the principal component analysis (PCA), which was evaluated using PAST (v3.26) software with a bootstrap frequency of n = 500, conducted on 57 spray-type white chrysanthemum cultivars based on six SSR markers.
3.3. Population Structure Analysis
Relationships among the 57 white spray cultivars were tested via population structure, analysis which allows one to identify the presence of subpopulations and admixtures. The numbers of genetically distinct populations (K) and admixtures were estimated using the STRUCTURE program. The ∆K algorithm analysis showed that the mean ∆K values reached a maximum at K = 2 among the 15 runs. Hence, the most appropriate value was considered to be K = 2, which indicates two subpopulations (Figure 5). The mean value admixture plots from the 15 independent runs for K = 2 and K = 3 are shown in Figure 6a,b. At the optimal K = 2, all 57 cultivars were grouped into two subpopulations. Group 1 includes a total of 32 cultivars, and Group 2 consists of 25 cultivars. Among these cultivars, twenty-four single-, one semi-double-, one double-, three pompon-, and three anemone-type cultivars constituted Group 1, whereas Group 2 was made up of eighteen single-, four pompon-, and three anemone-type cultivars.
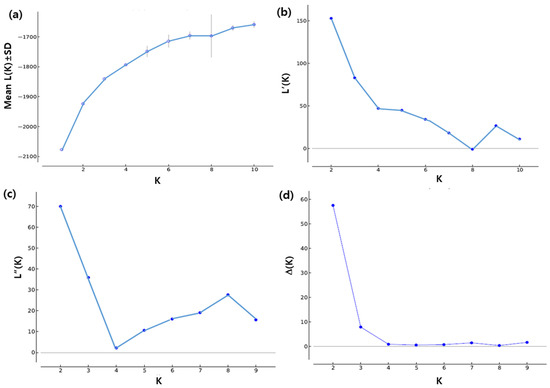
Figure 5.
Graphical representation of estimation of the best subpopulation numbers based on the appropriate K value. (a) Mean L (K) (±SD) over 15 runs for each K value; (b) rate of change of likelihood distribution; (c) absolute value of the second-order rate of change of the likelihood distribution; (d) mean of ∆K values of 15 independent runs, with K = 1 to K = 10 based on LnP(K) values. The mean of ∆K among the 15 runs reached a peak at K = 2.
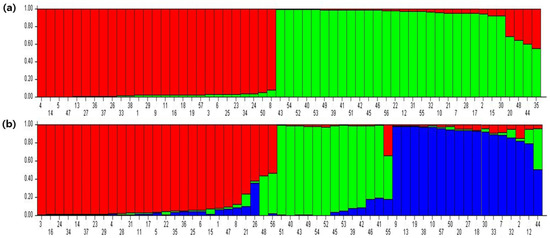
Figure 6.
Image representing the population structure for (a) K = 2 and (b) K = 3 for the 57 chrysanthemum cultivars tested. Each colored bar represents one test object according to the group to which it belongs.
4. Discussion
The chrysanthemum species, with its large, complex genome, exhibits hexaploid-based aneuploidy. More than 200,000 chrysanthemum cultivars are developed by breeding around the world and commercialized [38]. The discrimination of several hundreds of cultivars is highly difficult because the majority have similar morphological characteristics. Molecular markers constitute an efficient tool for the assessment of genetic characterization and the identification of a wide variety of cultivars. In this study, the genetic diversity of the 57 cultivars, including white, ivory, and cream-colored cultivars of single, double, pompon, and anemone flower types, was analyzed using six SSR markers. These six SSR markers generated 2 to 6 alleles with an average of 3.5 alleles per locus. The allele number per SSR locus is comparable with other results reporting an average of 2.7 alleles and 5.6 alleles per locus in 11 and 147 varieties of chrysanthemum tested with 7 and 14 SSR markers, respectively [29,30]. The degree of polymorphism (PIC) for the 57 white chrysanthemum cultivars with respect to six SSR markers was an average of 0.53, which is similar to that related in previous reports for chrysanthemum cultivars. However, higher PIC values of 0.9 and 0.88 were also observed for 32 and 88 chrysanthemum cultivars, respectively [39,40]. Since the PIC is used to evaluate the discriminatory ability of molecular markers and estimate the genetic diversity of genotypes, the variation in the PIC values among different chrysanthemum genotypes could be attributed to the genetic diversity of the genotypes and the variation in the tested number of SSR loci. The SSR markers used here enabled the characterization of the majority of chrysanthemum cultivars into moderately supported clusters and sub-clusters based on the UPGMA dendrogram, which revealed three clusters. Cluster I consisted of seven cultivars, Cluster II was a major group constituting forty-four cultivars, and Cluster III contained five cultivars. Group I included all single-type cultivars, except one anemone-type cultivar. Group II comprised five anemone, six pompon, and two semi-double varieties, while the remaining thirty-one varieties were single-type cultivars. Group III included one pompon-, one double-, and three single-type cultivars. However, Mannadu is a single-type cultivar that did not fall into any of these clusters and was separated individually. The SSR markers in the present study were observed to be effective in isolating the tested white-colored cultivars individually. However, Cluster I and Cluster II of the UPGMA dendrogram have a relatively lower degree of resolution, indicating weaker branch support between these clusters. Nevertheless, PCA analysis revealed the respective clusters clearly compared to the UPGMA dendrogram. This suggests that the tested number of loci were not sufficient to characterize all the cultivars; hence, an increased number of SSR markers might have to be employed to delimit the cultivars with stronger resolution. Also, four cultivars, namely, Pure Angel, Neba, Ladost, and White Wing, showed total genetic similarity. The relatively low genetic diversity of these cultivars suggests their development with a narrow genetic background. Two cultivars among the eleven standard types exhibited genetic similarity [25]. Analysis of genetic identity among the varieties is useful for the protection of breeders’ intellectual property rights. The standard chrysanthemum varieties Pingpong and White Runner have shown higher degrees of genetic similarity that preclude discrimination [30]. In contrast, Feng et al. [41] reported a genetic distance of 0.97, proving the higher genetic diversity of the tested cultivars and confirming the effectiveness of the SSR markers. In another study, the average genetic distance was about 0.67, which allowed for the differentiation of 97 chrysanthemum cultivars into small-, large-, and medium-flower varieties using 14 SSR markers. Although the small-flower cultivars were well differentiated, the large- and medium-flower varieties were not uniformly separated [19]. Population structure analysis using STRUCTURE program estimated that the appropriate K value of the tested 57 white variant cultivars was K = 2, grouping the tested genotypes into two subpopulations (Figure 6). It was observed that the two clusters shared a mixed population ancestry. The cultivars Moya, Oh Blang, and Puma from Group 1 and Polla, Pingpong, Mallorca, and Shahsuin from Group 2 exhibited a certain degree of admixing. Earlier reports showed different degrees of of admixture in chrysanthemum populations assessed using AFLP and RAPD markers [42,43]. Self-incompatibility, resource interchange, and the domestication history of highly heterozygous chrysanthemums constitute the possible attributes for these mixed populations [23,43].
5. Conclusions
In conclusion, this study provides an evaluation of the genetic relationships and a characterization of the closely related white-colored variants of chrysanthemum cultivars. The availability of enormous number of chrysanthemum cultivars with similar morphological characteristics on the market and the existence of numerous synonyms for the domestic varieties leads to the incorrect labelling of the respective cultivars. This poses a major problem with respect to the correct identification of such cultivars. Although phenotypic screening offers a means for the direct discrimination of genotypes, it requires a specific growth stage to be visible, which is also prone to environment-dependent changes. Also, it is highly difficult to assess the degree of variation if the phenotype is similar; such is the case of the white-colored flower analyzed in the present study. Hence, cultivar identification using molecular markers, such as the SSR markers used in the present study, offers a practical alternative for the accurate differentiation of closely related genotypes. Therefore, the results of this study serve as a testament to the efficacy of SSR markers with respect to the genetic characterization of 57 white-colored variants of chrysanthemums using six SSR marker sets. These SSR markers could also be used in the certification of protected varieties and for the pedigree analysis of white-colored chrysanthemum varieties. The study provides the basic data required to increase the utilization of SSR markers in genetic analysis and breed discrimination and to broaden the selection of genetic resources through chrysanthemum breeding. Although these cultivars have been well characterized, further studies employing a greater number of SSR markers would be essential to discriminate the cultivars according to their flower types.
Author Contributions
Conceptualization, J.-A.J.; methodology, M.M.; investigation, M.M.; software, M.M.; validation and formal analysis, M.M.; resources, H.-Y.S. and S.-H.L.; writing—original draft preparation, M.M.; writing—review and editing, M.M. and J.-A.J.; funding acquisition, J.-A.J. All authors have read and agreed to the published version of the manuscript.
Funding
This study was financially supported by the National Institute of Horticultural and Herbal Science, Rural Development Administration, Republic of Korea under the project grant PJ01098202.
Data Availability Statement
All datasets generated and analyzed in the current study are available from the corresponding author upon request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Teixeira da Silva, J.A.; Shinoyama, H.; Aida, R.; Matsushita, Y.; Raj, S.K.; Chen, F. Chrysanthemum biotechnology: Quo vadis. Crit. Rev. Plant Sci. 2013, 32, 21–52. [Google Scholar] [CrossRef]
- Royal Flora Holland in Facts and Figures. Annual Report. 2020. Available online: https://www.royalfloraholland.com/en (accessed on 4 July 2023).
- Anderson, N.O. Chrysanthemum. In Flower Breeding and Genetics; Springer: Dordrecht, The Netherlands, 2007; pp. 389–437. [Google Scholar]
- Liu, Y.H.; Mou, X.; Zhou, D.Y.; Zhou, D.Y.; Shou, C.M. Extraction of flavonoids from Chrysanthemum morifolium and antitumor activity in vitro. Exp. Ther. Med. 2018, 15, 1203–1210. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, A.; da Silva, J.A.T. Molecular systematics in Chrysanthemum × grandiflorum (Ramat) Kitamura. Sci. Hortic. 2006, 109, 379–384. [Google Scholar] [CrossRef]
- Lin, L.; Harnly, J.M. Identification of the phenolic components of chrysanthemum flower (Chrysanthemum morifolium Ramat). Food Chem. 2010, 120, 319–326. [Google Scholar] [CrossRef]
- Li, Y.; Yang, P.; Luo, Y.; Gao, B.; Sun, J.; Lu, W.; Liu, J.; Chen, P.; Zhang, Y.; Yu, L.L. Chemical compositions of chrysanthemum teas and their anti-inflammatory and antioxidant properties. Food Chem. 2019, 286, 8–16. [Google Scholar] [CrossRef]
- Wu, J.Y.; Chen, Y.J.; Fu, X.Q.; Li, J.K.; Chou, J.Y.; Yin, C.L.; Bai, J.X.; Wu, Y.; Wang, X.Q.; Li, A.S.; et al. Chrysoeriol suppresses hyperproliferation of rheumatoid arthritis fibroblast-like synoviocytes and inhibits JAK2/STAT3 signaling. BMC Complement. Med. Ther. 2022, 22, 73. [Google Scholar] [CrossRef]
- Chen, S.; Liu, J.; Dong, G.; Zhang, X.; Liu, Y.; Sun, W. Flavonoids and caffeoylquinic acids in Chrysanthemum morifolium Ramat flowers: A potentially rich source of bioactive compounds. Food Chem. 2020, 344, 128733. [Google Scholar] [CrossRef]
- Liang, F.; Hu, C.; He, Z.; Pan, Y. An arabinogalactan from flowers of Chrysanthemum morifolium: Structural and bioactivity studies. Carbohydr. Res. 2014, 387, 37–41. [Google Scholar] [CrossRef]
- Kuang, C.; Lv, D.; Shen, G.; Li, S.; Luo, Q.; Zhang, Z. Chrysanthemum morifolium Chemical composition and antimicrobial activities of volatile oil extracted from Ramat. J. Food Sci. Technol. 2018, 55, 2786–2794. [Google Scholar] [CrossRef]
- Mottaghipisheh, J.; Taghrir, H.; Boveiri Dehsheikh, A.; Zomorodian, K.; Irajie, C.; Mahmoodi Sourestani, M.; Iraji, A. Linarin, a Glycosylated Flavonoid, with Potential Therapeutic Attributes: A Comprehensive Review. Pharmaceuticals 2021, 14, 1104. [Google Scholar] [CrossRef]
- Qi, W.; Chen, Y.; Sun, S.; Xu, X.; Zhan, J.; Yan, Z.; Shang, P.; Pan, X.; Liu, H. Inhibiting TLR4 signaling by linarin for preventing inflammatory response in osteoarthritis. Aging 2021, 13, 5369–5382. [Google Scholar] [CrossRef] [PubMed]
- El-Twab, M.H.A.; Kondo, K. Visualization of genomic relationships in allotetraploid hybrids between Chrysanthemum lavandulifolium × C. chanetii by fluorescence in situ hybridization. Chromosome Bot. 2008, 3, 19–25. [Google Scholar] [CrossRef]
- Dowrick, G.J. The chromosomes of Chrysanthemum, II: Garden varieties. Heredity 1953, 7, 59–72. [Google Scholar] [CrossRef]
- Roxas, N.J.; Tashiro, Y.; Miyazaki, S.; Isshiki, S.; Takeshita, A. Meiosis and pollen fertility in Higo chrysanthemum (Dendranthema × grnadiflorum (Ramat.) Kitam. J. Jpn. Soc. Hortic. Sci. 1995, 64, 161–168. [Google Scholar] [CrossRef]
- Shudo, A.; Goeku, S.; Tarora, K.; Gima, N.; Urasaki, N.; Tokunaga, T.; Ureshino, K.; Miyagi, E.; Sekizuka, S.; Adaniya, S. Identification of chrysanthemum (Chrysanthemum morifolium) cultivars using flower pigment related genes. Trop. Agr. Dev. 2011, 55, 166–169. [Google Scholar]
- Shiroto, C.S.; Peres, N.V.; Sabbag, O.J. Economic viability of potted chrysanthemums production in Atibaia, Sao Paulo state. Ornam. Hortic. 2016, 22, 130–137. [Google Scholar] [CrossRef]
- Olejnik, A.; Parkitna, K.; Bartosz, K.; Florczak, S.; Matkowski, J.; Nowosad, K. Assessment of the genetic diversity of Chrysanthemum cultivars using SSR markers. Agronomy 2021, 11, 2318. [Google Scholar] [CrossRef]
- Su, J.; Jiang, J.; Zhang, F.; Liu, Y.; Ding, L.; Chen, S.; Chen, F. Current achievements and future prospects in the genetic breeding of chrysanthemum: A review. Hortic. Res. 2019, 6, 109. [Google Scholar] [CrossRef]
- Korir, N.H.; Han, J.; Shangguan, L.; Wang, C.; Kayesh, E.; Zhang, Y.; Fang, J. Plant variety and cultivar identification: Advances and prospects. Crit. Rev. Biotechnol. 2013, 33, 111–125. [Google Scholar] [CrossRef]
- Zhang, F.; Chen, S.M.; Chen, F.D.; Fang, W.M.; Li, F.T. A preliminary genetic linkage map of chrysanthemum (Chrysanthemum morifolium) cultivars using RAPD, ISSR and AFLP markers. Sci. Hortic. 2010, 125, 422–428. [Google Scholar] [CrossRef]
- Zhang, F.; Chen, S.; Chen, F.; Fang, W.; Deng, Y.; Chang, Q.; Liu, P. Genetic analysis and associated SRAP markers for flowering traits of chrysanthemum (Chrysanthemum morifolium). Euphytica 2011, 177, 15–24. [Google Scholar] [CrossRef]
- Powell, W.; Machray, G.C.; Provan, J. Polymorphism revealed by simple sequence repeats. Trends Plant Sci. 1996, 1, 215–222. [Google Scholar] [CrossRef]
- Kalia, R.K.; Rai, M.K.; Kalia, S.; Singh, R.; Dhawan, A.K. Microsatellite markers: An overview of the recent progress in plants. Euphytica 2010, 177, 309–334. [Google Scholar] [CrossRef]
- Zhang, Y.; Dai, S.L.; Hong, Y.; Song, X.B. Application of genomic SSR locus polymorphisms on the identification and classification of Chrysanthemum cultivars in China. PLoS ONE 2014, 9, 104856. [Google Scholar] [CrossRef] [PubMed]
- Jo, K.M.; Jo, Y.H.; Chu, H.S.; Lian, S.; Cho, W.K. Development of EST-derived SSR markers using next-generation sequencing to reveal the genetic diversity of 50 chrysanthemum cultivars. Biochem. Syst. Ecol. 2015, 60, 37–45. [Google Scholar] [CrossRef]
- Shim, E.J.; Heo, E.J.; Yoon, M.K.; Soh, E.H.; Hong, J.H. Construction of SSR marker database of Chrysanthemum varieties collected in Korea. Korean J. Breed. Sci. 2015, 47, 366–375. [Google Scholar] [CrossRef]
- Mekapogu, M.; Kwon, O.K.; Hyun, D.Y.; Lee, K.J.; Ahn, M.S.; Park, J.T.; Jung, J.A. Identification of standard type cultivars in chrysanthemum (Dendranthema grandiflorum) using SSR markers. Hortic. Environ. Biotech. 2020, 61, 153–161. [Google Scholar] [CrossRef]
- Han, J.Y.; Kim, J.B.; Lee, J.H.; Hong, C.P.; Park, H.S.; Kim, T.S. Development of EST-SSR markers for cultivar determination and genetic diversity studies of commercial chrysanthemums in the Korean floral market. Korean J. Breed. Sci. 2019, 51, 201–208. [Google Scholar] [CrossRef]
- Schuelke, M. An economic method for the fluorescent labeling of PCR fragments. Nat. Biotechnol. 2000, 18, 233–234. [Google Scholar] [CrossRef]
- Liu, K.; Muse, S.V. Power Marker: An integrated analysis environment for genetic marker analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Rosenberg, N.A.; Burke, T.K.; Feldmann, M.W.; Freidlin, P.J.; Mam, G. Empirical evaluation of genetic clustering methods using multi-locus genotypes from 20 chicken breeds. Genetics 2001, 159, 699–713. [Google Scholar] [CrossRef]
- Earl, D.A.; vonHoldt, B.M. Structure Harvester: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Nei, M. Analysis of gene diversity in subdivided populations. Proc. Natl. Acad. Sci. USA 1973, 70, 3321–3323. [Google Scholar] [CrossRef]
- Wang, F.; Zhang, F.J.; Chen, F.D.; Fang, W.M.; Teng, N.J. Identification of Chrysanthemum (Chrysanthemum morifolium) self-incompatibility. Sci. World J. 2014, 2014, 625658. [Google Scholar] [CrossRef]
- Feng, S.G.; He, R.F.; Jiang, M.Y.; Lu, J.J.; Shen, X.X.; Liu, J.J.; Wang, Z.A.; Wang, H.Z. Genetic diversity and relationships of medicinal Chrysanthemum morifolium revealed by start codon targeted (SCoT) markers. Sci. Hortic. 2016, 201, 118–123. [Google Scholar] [CrossRef]
- Luo, C.; Chen, D.; Cheng, X.; Liu, H.; Li, H.; Li, Y.; Huang, C. SSR analysis of genetic relationship and classification in Chrysanthemum germplasm collection. Hortic. Plant J. 2018, 4, 73–82. [Google Scholar] [CrossRef]
- Feng, S.; He, R.; Li, J.; Jiang, M.; Shen, X.; Jiang, Y.; Wang, Z.; Wang, H. Development of SSR markers and assessment of genetic diversity in medicinal Chrysanthemum morifolium cultivars. Front. Genet. 2016, 7, 113. [Google Scholar] [CrossRef] [PubMed]
- Roein, Z.; Asil, M.H.; Sabouri, A.; Dadras, A.R. Genetic structure of Chrysanthemum genotypes from Iran assessed by AFLP markers and phenotypic traits. Plant Syst. Evol. 2014, 300, 493–503. [Google Scholar] [CrossRef]
- Kumar, S.; Kumar, M.; Yadav, H.K.; Sharma, S.; Kumar, S. Genetic diversity and population structure analysis of Chrysanthemum (Dendranthema grandiflora Tzvelev) germplasm based on RAPD markers. J. Environ. Biol. 2017, 38, 457–464. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).