TCP Transcription Factors in Pineapple: Genome-Wide Characterization and Expression Profile Analysis during Flower and Fruit Development
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Growth and Flower Induction
2.2. Identification of TCP Gene Family in Pineapple
2.3. Chromosomal Distribution and Evolutionary Analysis of the AcTCP Gene Family
2.4. Conserved Motifs and Gene Structure Analysis of AcTCP Gene Family
2.5. Gene Duplication and Syntenic Analysis of the Pineapple AcTCP Gene Family
2.6. Cis-Acting Regulatory Elements in the Promoter and Protein Interaction Prediction Analysis
2.7. RNA Isolation, qRT-PCR Analysis of AcTCPs in Different Issues, and the Stage of Development
2.8. Statistical Analysis
2.9. Yeast Two-Hybrid Analysis
3. Results
3.1. Identification and Classification of TCP Gene Family in Pineapple
3.2. Chromosomal Mapping of AcTCP Gene Family
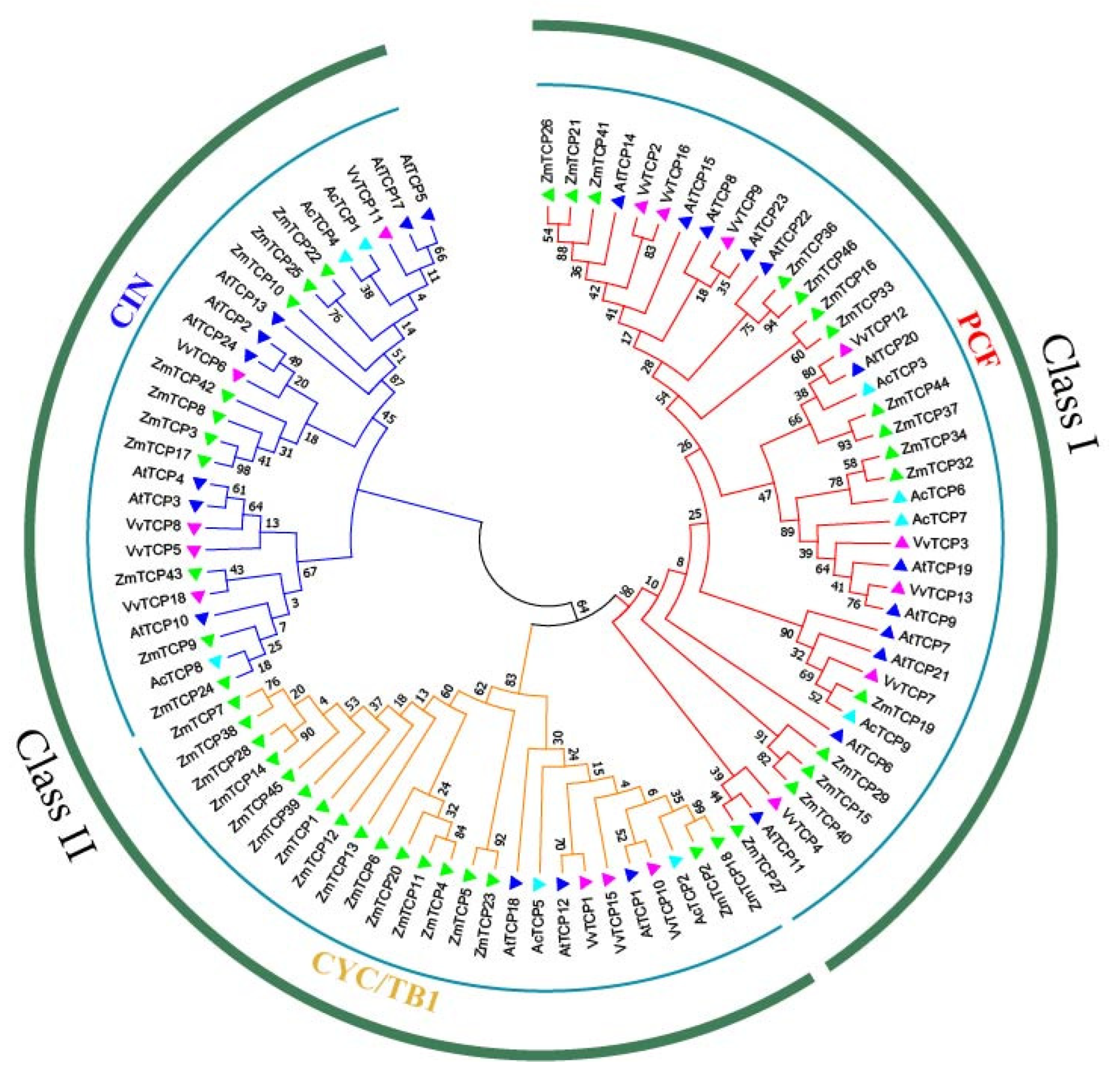
3.3. Phylogenetic Analysis
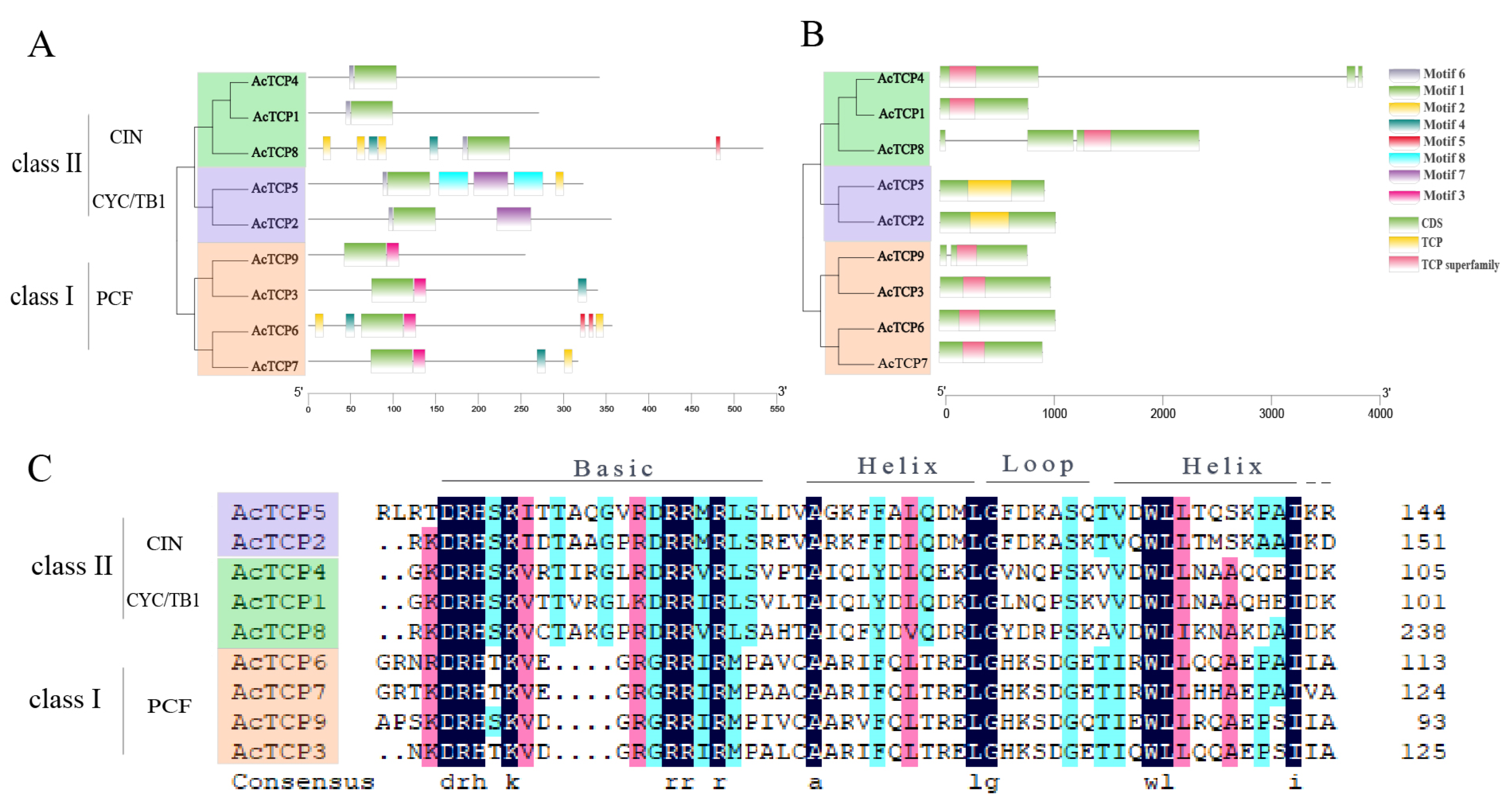
3.4. AcTCP Gene Structure, Motif Analysis, and Multiple-Sequence Alignment of AcTCP Protein Sequences
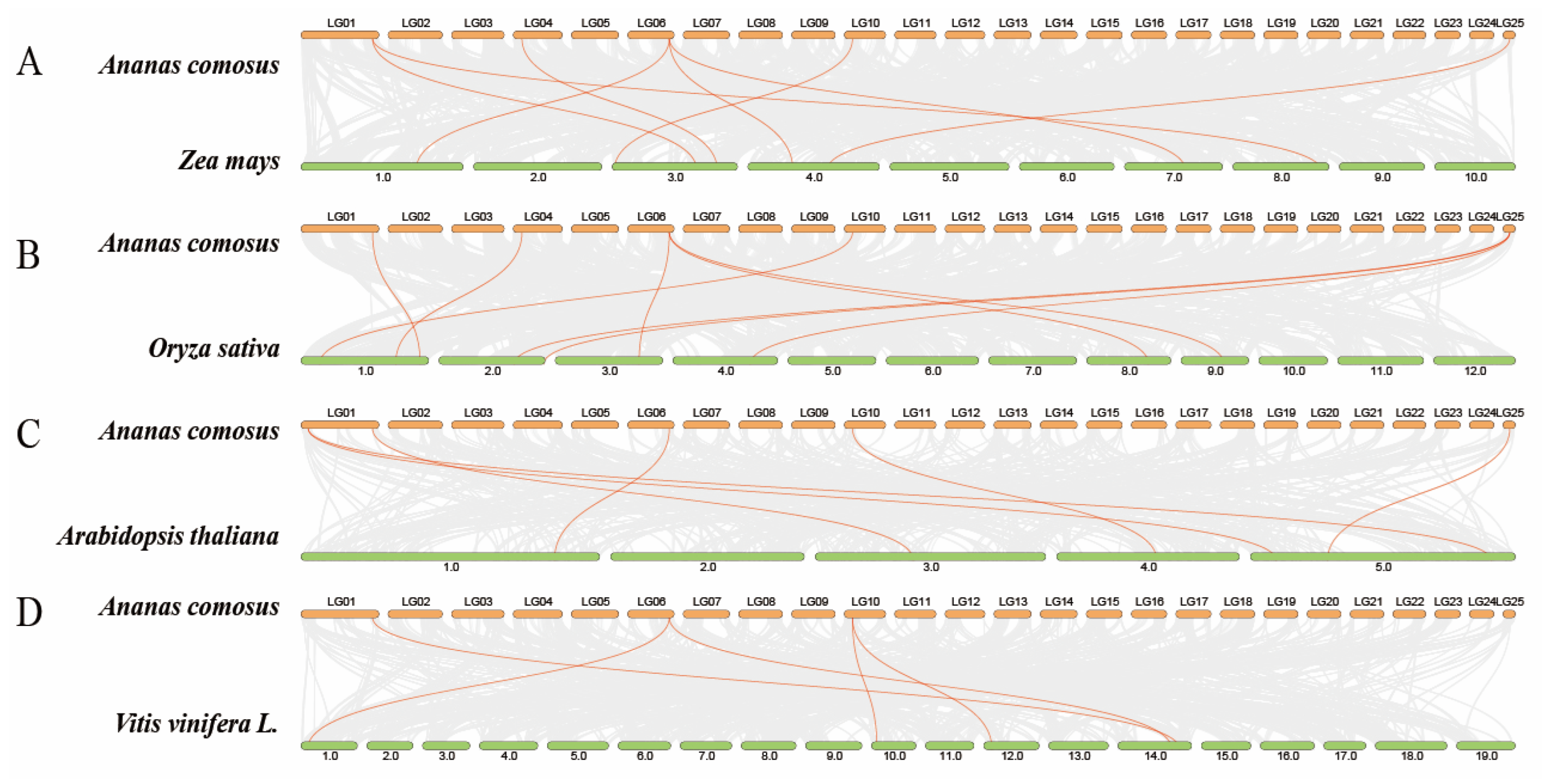
3.5. AcTCP Gene Covariance Analysis
3.6. Analysis of AcTCP Promoter cis-Acting Elements
3.7. Expression Profile of AcTCP Genes
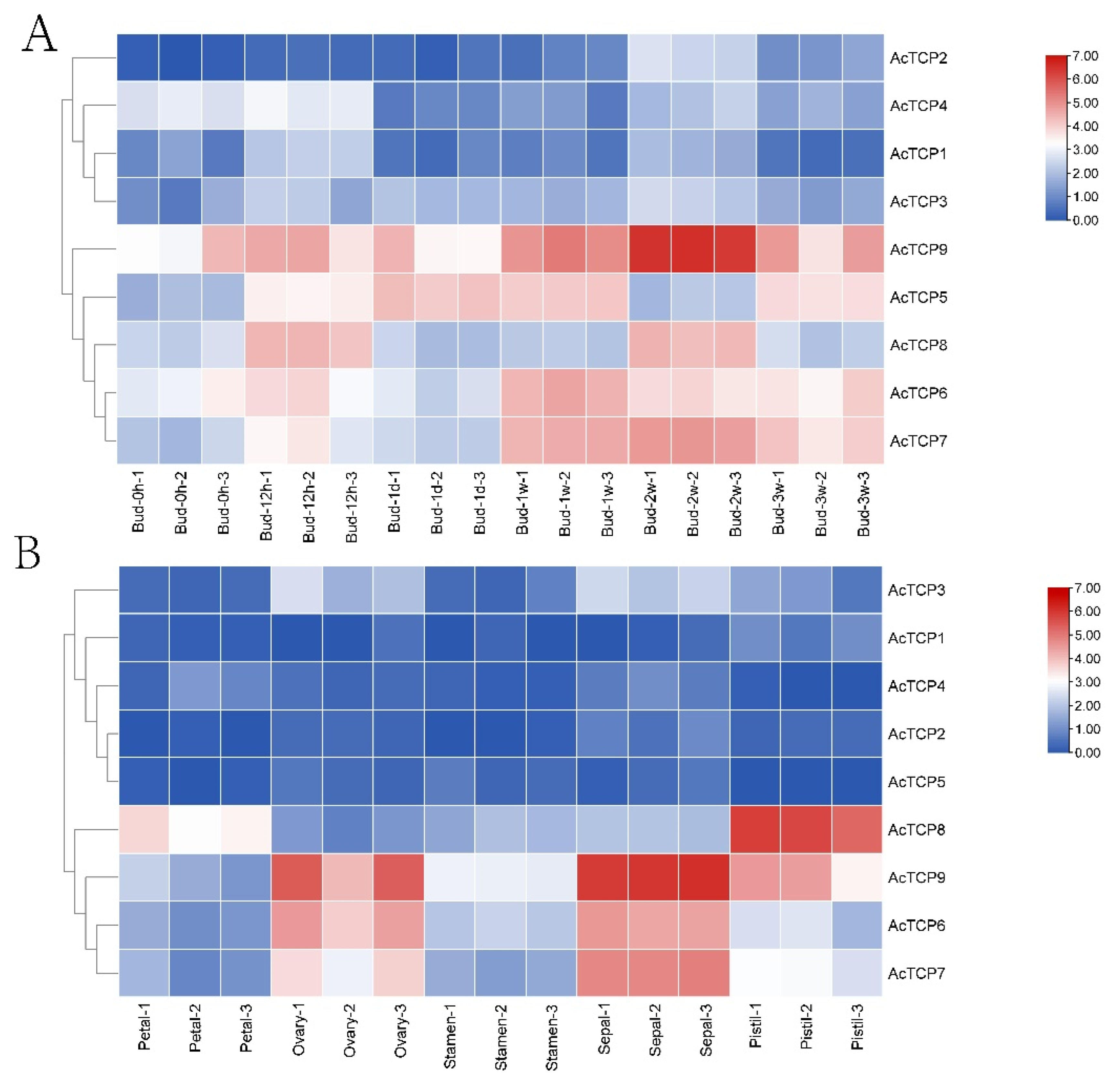
3.7.1. Expression Profile of AcTCP Genes during Flower Development
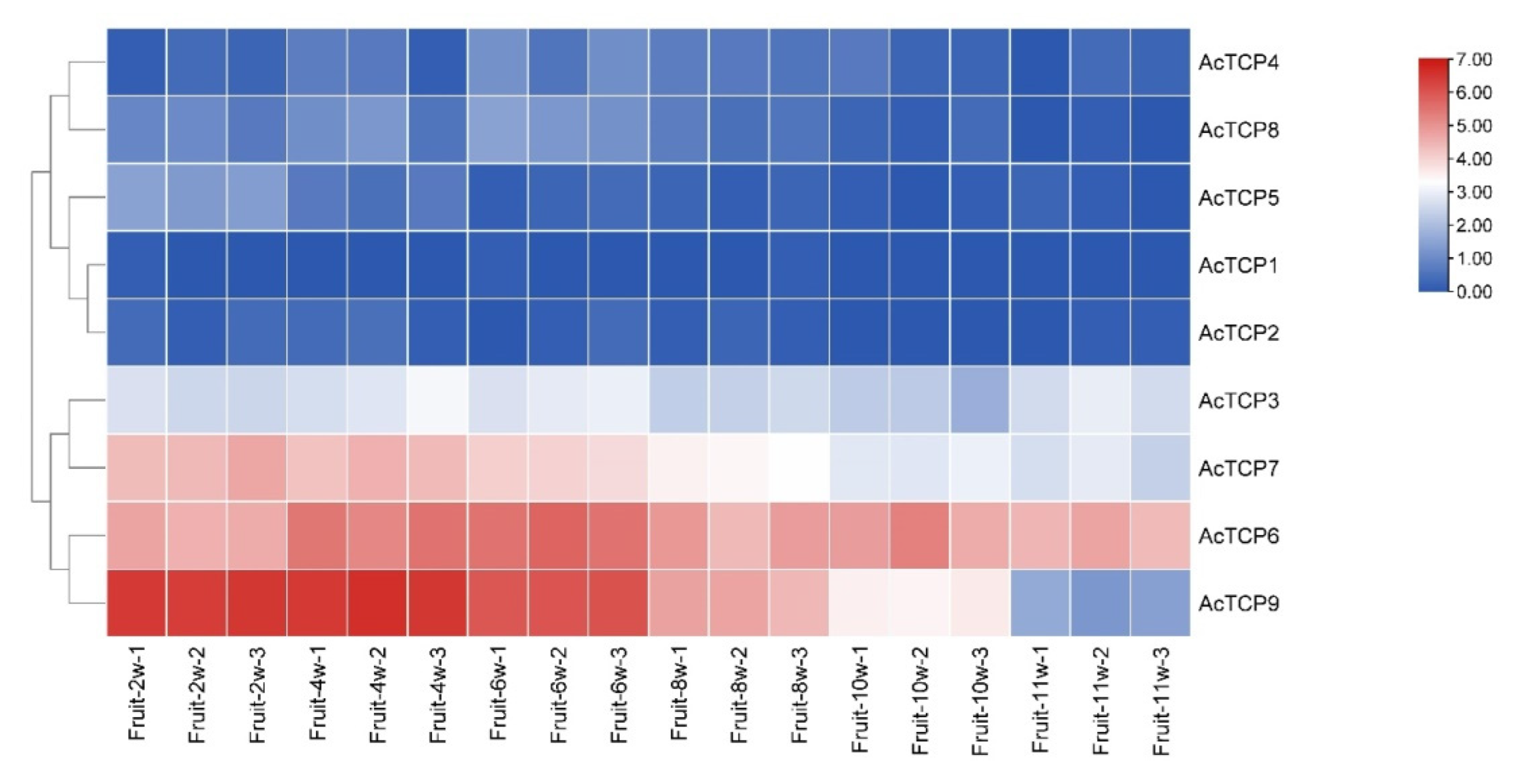
3.7.2. Expression Profile of AcTCPs Genes during Different Fruit Developmental Stages
3.8. qRT-PCR Assays of AcTCP Expression Patterns
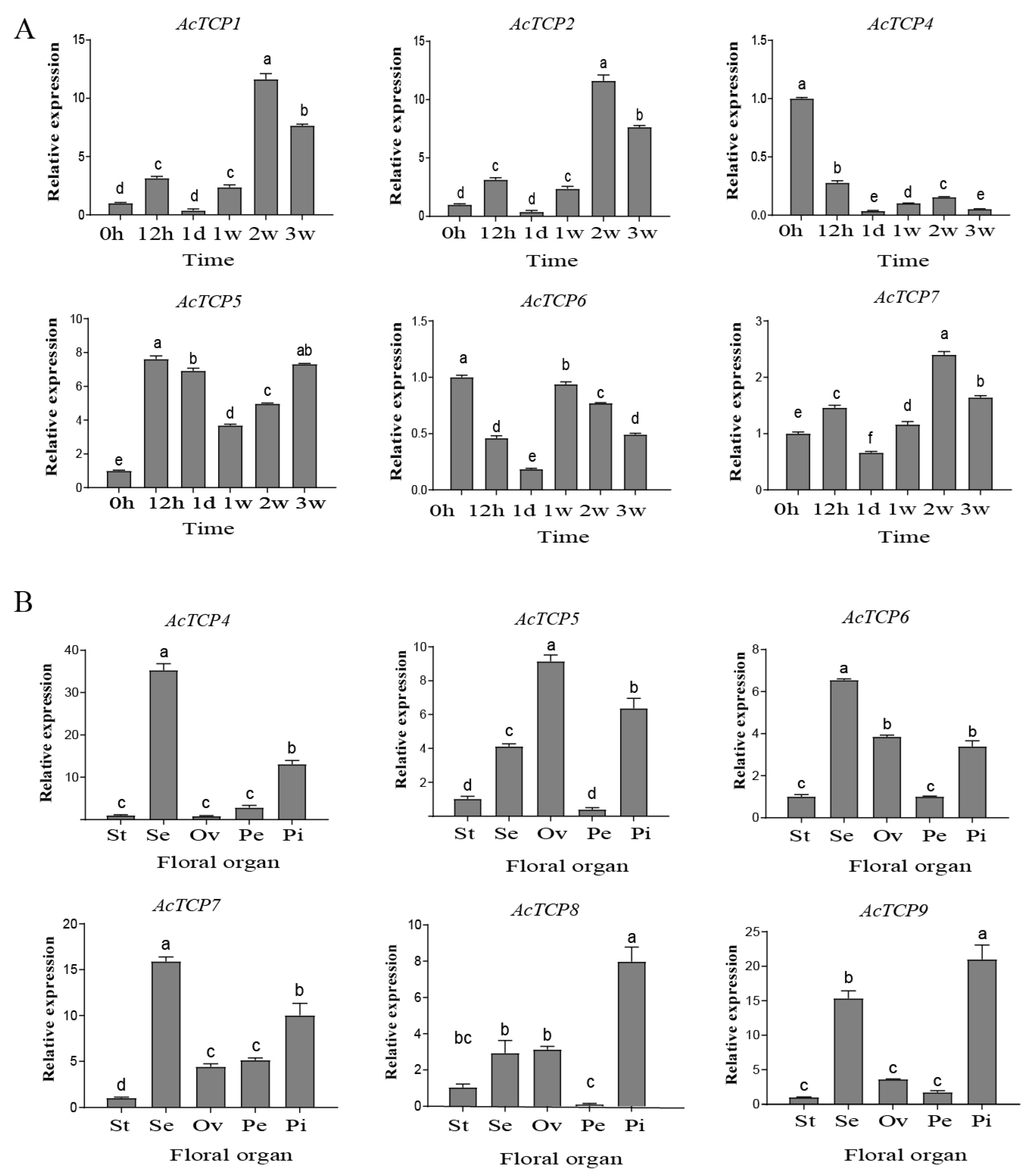
3.8.1. qRT-PCR Assays of AcTCP Expression Patterns during Flower Development
3.8.2. qRT-PCR Assays of AcTCP Expression Patterns during Fruit Development
3.9. Diversified and Conserved Protein Interaction Network of AcTCP Proteins
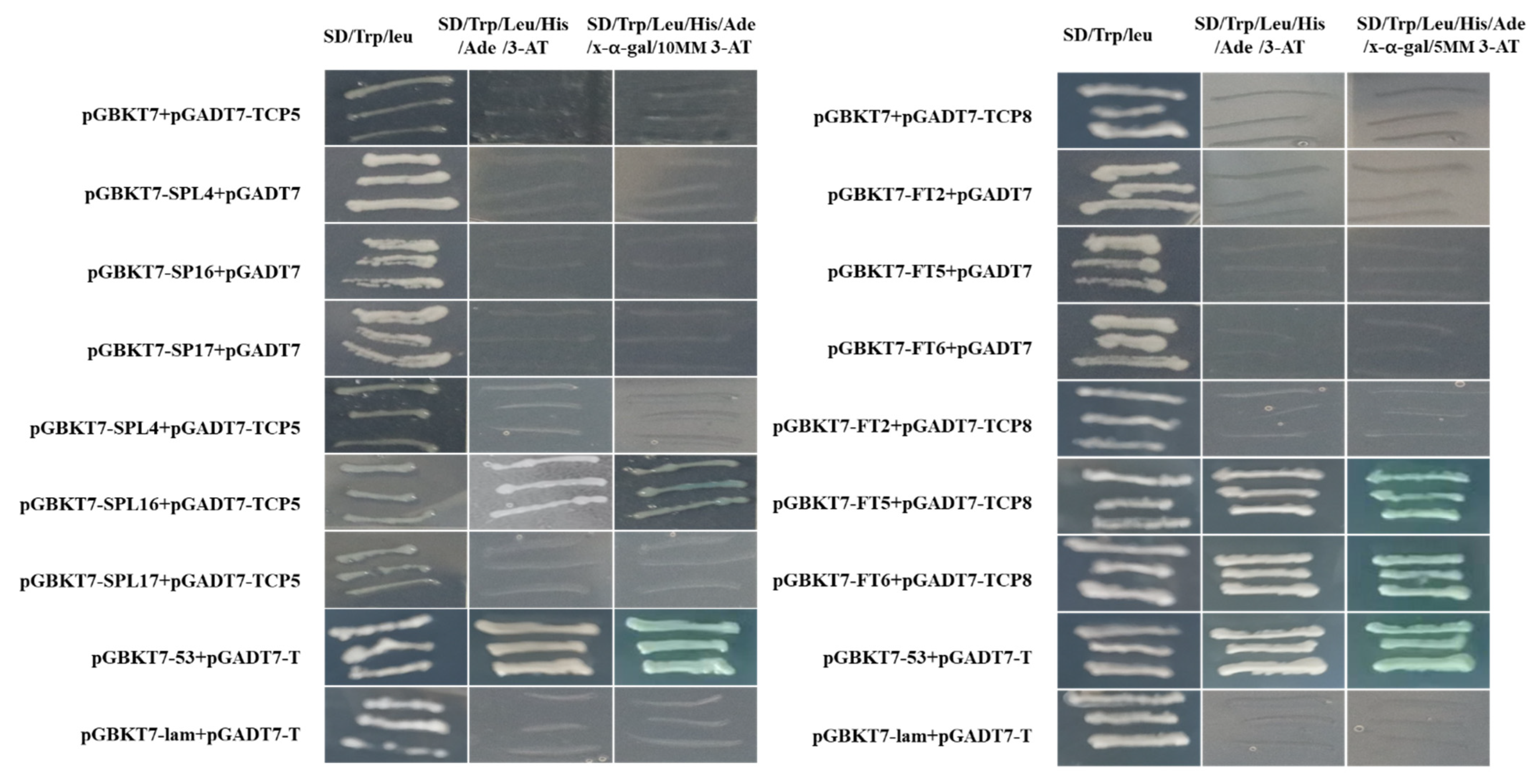
3.10. Y2H Validation of the Interaction of AcTCP with FT and SPL
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Doebley, J.; Stec, A.; Hubbard, L. The evolution of apical dominance in maize. Nature 1997, 386, 485–488. [Google Scholar] [CrossRef] [PubMed]
- Luo, D.; Carpenter, R.; Vincent, C.; Copsey, L.; Coen, E. Origin of floral asymmetry in Antirrhinum. Nature 1996, 383, 794–799. [Google Scholar] [CrossRef] [PubMed]
- Kosugi, S.; Ohashi, Y. PCF1 and PCF2 specifically bind to cis-elements in the rice proliferating cell nuclear antigen gene. Plant Cell 1997, 9, 1607–1619. [Google Scholar] [PubMed]
- Cubas, P.; Lauter, N.; Doebley, J.; Coen, E. The TCP domain: A motif found in proteins regulating plant growth and development. Plant J. 1999, 18, 215–222. [Google Scholar] [CrossRef]
- Martín-Trillo, M.; Cubas, P. TCP genes: A family snapshot ten years later. Trends Plant Sci. 2010, 15, 31–39. [Google Scholar] [CrossRef]
- Kosugi, S.; Ohashi, Y. DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J. 2002, 30, 337–348. [Google Scholar] [CrossRef]
- Yao, X.; Ma, H.; Wang, J.; Zhang, D. Genome-wide comparative analysis and expression pattern of TCP gene families in Arabidopsis thaliana and Oryza sativa. J. Integr. Plant Biol. 2007, 49, 885–897. [Google Scholar] [CrossRef]
- Chen, P.; Li, J.; Ye, X.; Tan, B.; Zheng, X.; Cheng, J.; Wang, W.; Wang, H.; Gu, L.; Feng, J. Genome-wide identification of Ziziphus jujuba TCP transcription factors and their expression in response to infection with jujube witches’ broom phytoplasma. Acta Physiol. Plant 2019, 41, 86. [Google Scholar] [CrossRef]
- Leng, X.; Wei, H.; Xu, X.; Ghuge, S.A.; Jia, D.; Liu, G.; Wang, Y.; Yuan, Y. Genome-wide identification and transcript analysis of TCP transcription factors in grapevine. BMC Genom. 2019, 20, 786. [Google Scholar] [CrossRef]
- Wei, W.; Hu, Y.; Cui, M.Y.; Han, Y.T.; Gao, K.; Feng, J.Y. Identification and transcript analysis of the TCP transcription factors in the diploid woodland strawberry Fragaria vesca. Front. Plant Sci. 2016, 7, 1937. [Google Scholar] [CrossRef]
- Chai, W.; Jiang, P.; Huang, G.; Jiang, H.; Li, X. Identification and expression profiling analysis of TCP family genes involved in growth and development in maize. Physiol. Mol. Biol. Plants 2017, 23, 779–791. [Google Scholar] [CrossRef] [PubMed]
- Danisman, S. TCP Transcription factors at the interface between environmental challenges and the plant’s growth responses. Front. Plant Sci. 2016, 7, 1930. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zhang, H.; Mou, M.; Chen, Y.; Xiang, S.; Chen, L.; Yu, D. Arabidopsis Class II TCP Transcription Factors Integrate with the FT-FD Module to Control Flowering. Plant Physiol. 2019, 181, 97–111. [Google Scholar] [CrossRef] [PubMed]
- Sarvepalli, K.; Nath, U. Hyper-activation of the TCP4 transcription factor in Arabidopsis thaliana accelerates multiple aspects of plant maturation. Plant J. 2011, 67, 595–607. [Google Scholar] [CrossRef]
- Li, X.; Zhang, G.; Liang, Y.; Hu, L.; Zhu, B.; Qi, D.; Cui, S.; Zhao, H. TCP7 interacts with Nuclear Factor-Ys to promote flowering by directly regulating SOC1 in Arabidopsis. Plant J. 2021, 108, 1493–1506. [Google Scholar] [CrossRef]
- Huang, T.; Irish, V.F. Temporal control of plant organ growth by TCP transcription factors. Curr. Biol. 2015, 25, 1765–1770. [Google Scholar] [CrossRef]
- Yuan, C.; Huang, D.; Yang, Y.; Sun, M.; Cheng, T.; Wang, J.; Pan, H.; Zhang, Q. CmCYC2-like transcription factors may interact with each other or bind to the promoter to regulate floral symmetry development in Chrysanthemum morifolium. Plant Mol. Biol. 2020, 103, 159–171. [Google Scholar] [CrossRef]
- Xie, Y.G.; Ma, Y.Y.; Bi, P.P.; Wei, W.; Liu, J.; Hu, Y.; Gou, Y.J.; Zhu, D.; Wen, Y.Q.; Feng, J.Y. Transcription factor FvTCP9 promotes strawberry fruit ripening by regulating the biosynthesis of abscisic acid and anthocyanins. Plant Physiol. Biochem. 2020, 146, 374–383. [Google Scholar] [CrossRef]
- Song, C.B.; Shan, W.; Yang, Y.Y.; Tan, X.L.; Fan, Z.Q.; Chen, J.Y.; Lu, W.J.; Kuang, J.F. Heterodimerization of MaTCP proteins modulates the transcription of MaXTH10/11 genes during banana fruit ripening. BBA-Gene Regul. Mech. 2018, 1861, 613–622. [Google Scholar] [CrossRef]
- Parapunova, V.; Busscher, M.; Busscher-Lange, J.; Lammers, M.; Karlova, R.; Bovy, A.G.; Angenent, G.C.; de Maagd, R.A. Identification, cloning and characterization of the tomato TCP transcription factor family. BMC Plant Biol. 2014, 14, 157. [Google Scholar] [CrossRef]
- Zheng, X.; Lan, J.; Yu, H.; Zhang, J.; Zhang, Y.; Qin, Y.; Su, X.D.; Qin, G. Arabidopsis transcription factor TCP4 represses chlorophyll biosynthesis to prevent petal greening. Plant Commun. 2022, 3, 100309. [Google Scholar] [CrossRef]
- Rubio-Somoza, I.; Zhou, C.M.; Confraria, A.; Martinho, C.; von Born, P.; Baena-Gonzalez, E.; Wang, J.W.; Weigel, D. Temporal control of leaf complexity by miRNA-regulated licensing of protein complexes. Curr. Biol. 2014, 24, 2714–2719. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.N.; Xu, D.B.; Yan, X.; Wu, Z.K.; Kayani, S.I.; Shen, Q.; Fu, X.Q.; Xie, L.H.; Hao, X.L.; Hassani, D.; et al. Jasmonate- and abscisic acid-activated AaGSW1-AaTCP15/AaORA transcriptional cascade promotes artemisinin biosynthesis in Artemisia annu. Plant Biotechnol. J. 2021, 19, 1412–1428. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.J.; An, J.P.; Gao, N.; Wang, X.; Chen, X.X.; Wang, X.F.; Zhang, S.; You, C.X. MdTCP46 interacts with MdABI5 to negatively regulate ABA signalling and drought response in apple. Plant Cell Environ. 2022, 45, 3233–3248. [Google Scholar] [CrossRef] [PubMed]
- Nag, A.; King, S.; Jack, T. miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 22534–22539. [Google Scholar] [CrossRef]
- Cao, B.; Wang, H.; Bai, J.; Wang, X.; Li, X.; Zhang, Y.; Yang, S.; He, Y.; Yu, X. MiR319-regulated TCP3 modulates silique development associated with seed shattering in Brassicaceae. Cells 2022, 11, 3096. [Google Scholar] [CrossRef]
- Baulies, J.L.; Bresso, E.G.; Goldy, C.; Palatnik, J.F.; Schommer, C. Potent inhibition of TCP transcription factors by miR319 ensures proper root growth in Arabidopsis. Plant Mol. Biol. 2022, 108, 93–103. [Google Scholar] [CrossRef]
- Bresso, E.G.; Chorostecki, U.; Rodriguez, R.E.; Palatnik, J.F.; Schommer, C. Spatial control of gene expression by miR319-regulated TCP transcription factors in leaf development. Plant Physiol. 2018, 176, 1694–1708. [Google Scholar] [CrossRef]
- Mohd Ali, M.; Hashim, N.; Abd Aziz, S.; Lasekan, O. Pineapple (Ananas comosus): A comprehensive review of nutritional values, volatile compounds, health benefits, and potential food products. Food Res. Int. 2020, 137, 109675. [Google Scholar] [CrossRef]
- Xu, H.; Yu, Q.; Shi, Y.; Hua, X.; Tang, H.; Yang, L.; Ming, R.; Zhang, J. PGD: Pineapple genomics database. Hortic. Res. 2018, 5, 66. [Google Scholar] [CrossRef]
- Poole, R.L. The TAIR database. Methods Mol. Biol. 2007, 406, 179–212. [Google Scholar] [PubMed]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Chen, Y.; Lu, C.; Huang, J.K. Prediction of protein subcellular localization. Proteins 2006, 64, 643–651. [Google Scholar] [CrossRef] [PubMed]
- Chao, J.; Li, Z.; Sun, Y.; Aluko, O.O.; Wu, X.; Wang, Q.; Liu, G. MG2C: A user-friendly online tool for drawing genetic maps. Mol. Hortic. 2021, 1, 16. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar] [PubMed]
- Mukesh, J.; Akhilesh, K.T.; Jitendra, P.K. Genome-wide analysis, evolutionary expansion, and expression of early auxin-responsive SAUR gene family in rice (Oryza sativa). Genomics 2006, 88, 360–371. [Google Scholar]
- Ferreira e Silva, G.F.; Silva, E.M.; Azevedo, M.d.a.S.; Guivin, M.A.; Ramiro, D.A.; Figueiredo, C.R.; Carrer, H.; Peres, L.E.; Nogueira, F.T. microRNA156-targeted SPL/SBP box transcription factors regulate tomato ovary and fruit development. Plant J. 2014, 78, 604–618. [Google Scholar] [CrossRef]
- Kojima, S.; Takahashi, Y.; Kobayashi, Y.; Monna, L.; Sasaki, T.; Araki, T.; Yano, M. Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell 2002, 43, 1096–1105. [Google Scholar] [CrossRef]
- Zheng, L.; Zhou, X.J.; Ma, Y.T.; Guo, M.X. Genome-wide identification and characterization of TCP family genes associated with flower and fruit development in Fragaria vesca. Pak. J. Bot. 2019, 51, 513–519. [Google Scholar] [CrossRef]
- Zhang, S.; Zhou, Q.; Chen, F.; Wu, L.; Liu, B.; Li, F.; Zhang, J.; Bao, M.; Liu, G. Genome-wide identification, characterization and expression analysis of TCP transcription factors in Petunia. Int. J. Mol. Sci. 2020, 21, 6594. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Sun, P.; Jia, F.; Lu, L.; Li, Y.; Zhang, S.; Huang, J. Genome-wide analysis of TCP transcription factor gene family in Malus domestica. J. Genet. 2014, 93, 733–746. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Shi, T.; Ni, X.; Xu, Y.; Qu, S.; Gao, Z. The role of miR319a and its target gene TCP4 in the regulation of pistil development in Prunus mume. Genome 2018, 61, 43–48. [Google Scholar] [CrossRef]
- Iqbal, S.; Pan, Z.; Wu, X.; Shi, T.; Ni, X.; Bai, Y.; Gao, J.; Khalil-Ur-Rehman, M.; Gao, Z. Genome-wide analysis of PmTCP4 transcription factor binding sites by ChIP-Seq during pistil abortion in Japanese apricot. Plant Genome 2020, 13, e20052. [Google Scholar] [CrossRef]
- Mo, W.; Zhang, J.; Zhang, L.; Yang, Z.; Yang, L.; Yao, N.; Xiao, Y.; Li, T.; Li, Y.; Zhang, G.; et al. Arabidopsis cryptochrome 2 forms photobodies with TCP22 under blue light and regulates the circadian clock. Nat. Commun. 2022, 13, 2631. [Google Scholar] [CrossRef]
- Liu, S.; Yin, X.; Feng, T.; Kang, Z.; Zhang, X.; Dong, J.; Liang, Z. Genome-wide identification and expression analysis of the TCP genes in Senna tora reveal the regulatory mechanism of their response to MeJA. Ind. Crops Prod. 2022, 187, 115511. [Google Scholar] [CrossRef]
- Jung, J.H.; Lee, H.J.; Ryu, J.Y.; Park, C.M. SPL3/4/5 integrate developmental aging and photoperiodic signals into the FT-FD module in Arabidopsis flowering. Mol. Plant 2016, 9, 1647–1659. [Google Scholar] [CrossRef] [PubMed]




| Gene Name | Gene ID | Maximum ORF Length (bp) | Amino Acid Length (aa) | MW (kDa) | Theoretical pI | Aliphatic Index | GRAVY | Predicted Subcellular Location |
|---|---|---|---|---|---|---|---|---|
| AcTCP1 | Aco012417.1 | 807 | 269 | 30.21 | 7.89 | 75.43 | −0.607 | Nuclear |
| AcTCP2 | Aco024489.1 | 1062 | 354 | 39.43 | 9.13 | 69.77 | −0.503 | Nuclear |
| AcTCP3 | Aco006659.1 | 1014 | 338 | 34.37 | 6.59 | 62.07 | −0.51 | Nuclear |
| AcTCP4 | Aco002292.1 | 1020 | 340 | 37.89 | 5.89 | 71.38 | −0.694 | Nuclear |
| AcTCP5 | Aco003020.1 | 963 | 321 | 35.66 | 6.27 | 60.59 | −0.795 | Nuclear |
| AcTCP6 | Aco021664.1 | 1065 | 355 | 37.42 | 5.47 | 63.94 | −0.646 | Nuclear |
| AcTCP7 | Aco015741.1 | 945 | 315 | 32.55 | 6.17 | 65.05 | −0.499 | Nuclear |
| AcTCP8 | Aco010666.1 | 1596 | 532 | 57.70 | 5.35 | 52.44 | −0.848 | Nuclear |
| AcTCP9 | Aco010326.1 | 759 | 253 | 25.54 | 10.35 | 67.83 | −0.255 | Nuclear |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Ouyang, Y.; Pan, X.; Zhang, X.; Zhao, L.; Wang, C.; Xu, R.; Zhang, H.; Wei, Y. TCP Transcription Factors in Pineapple: Genome-Wide Characterization and Expression Profile Analysis during Flower and Fruit Development. Horticulturae 2023, 9, 799. https://doi.org/10.3390/horticulturae9070799
Li Z, Ouyang Y, Pan X, Zhang X, Zhao L, Wang C, Xu R, Zhang H, Wei Y. TCP Transcription Factors in Pineapple: Genome-Wide Characterization and Expression Profile Analysis during Flower and Fruit Development. Horticulturae. 2023; 9(7):799. https://doi.org/10.3390/horticulturae9070799
Chicago/Turabian StyleLi, Ziqiong, Yanwei Ouyang, Xiaolu Pan, Xiaohan Zhang, Lei Zhao, Can Wang, Rui Xu, Hongna Zhang, and Yongzan Wei. 2023. "TCP Transcription Factors in Pineapple: Genome-Wide Characterization and Expression Profile Analysis during Flower and Fruit Development" Horticulturae 9, no. 7: 799. https://doi.org/10.3390/horticulturae9070799
APA StyleLi, Z., Ouyang, Y., Pan, X., Zhang, X., Zhao, L., Wang, C., Xu, R., Zhang, H., & Wei, Y. (2023). TCP Transcription Factors in Pineapple: Genome-Wide Characterization and Expression Profile Analysis during Flower and Fruit Development. Horticulturae, 9(7), 799. https://doi.org/10.3390/horticulturae9070799





