Genome-Wide Identification, Phylogenetic and Expression Analysis of the B-Box Gene Family in the Woodland Strawberry (Fragaria vesca)
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of BBX Gene Family Members in the Strawberry
2.2. Gene Structure and Motif Analysis of FveBBX Genes
2.3. Construction of FveBBX Genes Phylogenetic Tree
2.4. Cis-Element Prediction for FveBBX Gene Promoters
2.5. Transcriptome Data Analysis
2.6. Plant Materials and Photoperiodic Treatments
2.7. RNA Extraction, Quantitative Real-Time PCR Analysis
3. Results
3.1. Identification and Characterization of BBX Proteins in the Woodland Strawberry
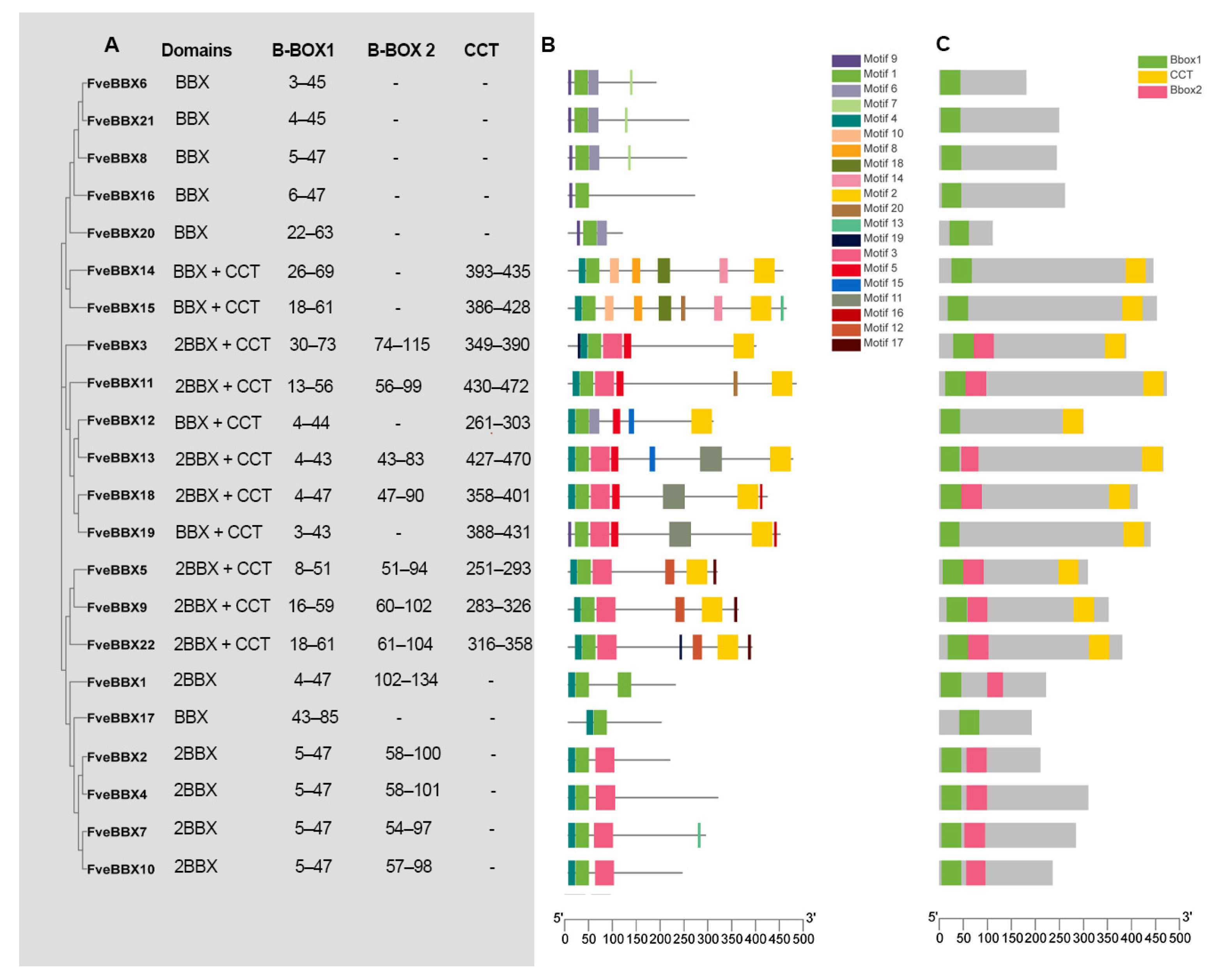
3.2. Conserved Domain and Motif Analysis of BBX Proteins in the Woodland Strawberry
3.3. Phylogeny and Gene Structure of the FveBBX Gene Family
3.4. Cis-Elements in the Promoters of FveBBX Genes
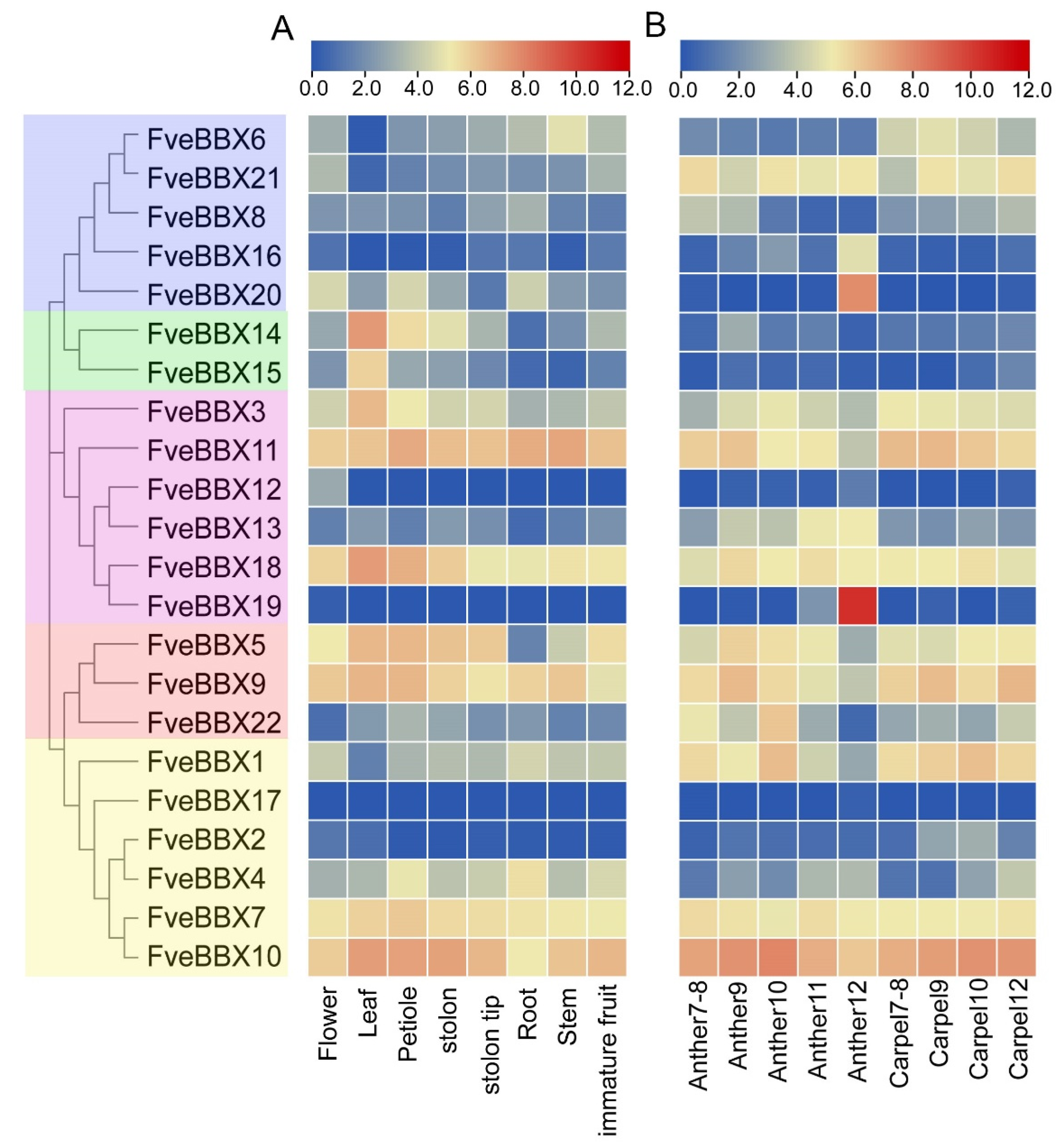
3.5. Expression Analysis of FveBBX Genes in Various Tissues and Organs
3.6. Expression Analysis of BBX Genes during the Development and Ripening of Woodland Strawberry Fruit
3.7. Expression Analysis of FveBBX Genes under Cold and Heat Stresses
3.8. Analysis of BBX Gene Expression in the Woodland Strawberry during Different Photoperiods
4. Discussion
4.1. Evolution of the BBX Gene Family in the Woodland Strawberry
4.2. Potential Role of FveBBX Gene in Strawberry Responses to Abiotic Stresses and Environmental Stimulus
4.3. The Potential Role of BBX Gene in Strawberry Growth and Development
4.4. The Potential Roles of Strawberry BBX Genes in the Development and Ripening of Fruit
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Shulaev, V.; Sargent, D.J.; Crowhurst, R.N.; Mockler, T.C.; Folkerts, O.; Delcher, A.L.; Jaiswal, P.; Mockaitis, K.; Liston, A.; Mane, S.P.; et al. The genome of woodland strawberry (Fragaria vesca). Nat. Genet. 2011, 43, 109–116. [Google Scholar] [CrossRef]
- Takatsuji, H. Zinc-finger transcription factors in plants. Cell. Mol. Life Sci. CMLS 1998, 54, 582–596. [Google Scholar] [CrossRef] [PubMed]
- Khanna, R.; Kronmiller, B.; Maszle, D.R.; Coupland, G.; Holm, M.; Mizuno, T.; Wu, S.H. The Arabidopsis B-box zinc finger family. Plant Cell 2009, 21, 3416–3420. [Google Scholar] [CrossRef]
- Meroni, G.; Diez-Roux, G. TRIM/RBCC, a novel class of ‘single protein RING finger’ E3 ubiquitin ligases. BioEssays 2005, 27, 1147–1157. [Google Scholar] [CrossRef] [PubMed]
- Gangappa, S.N.; Botto, J.F. The BBX family of plant transcription factors. Trends Plant Sci. 2014, 19, 460–470. [Google Scholar] [CrossRef] [PubMed]
- Gendron, J.M.; Pruneda-Paz, J.L.; Doherty, C.J.; Gross, A.M.; Kang, S.E.; Kay, S.A. Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc. Natl. Acad. Sci. USA 2012, 109, 3167–3172. [Google Scholar] [CrossRef]
- Talar, U.; KiełbowiczMatuk, A. Beyond Arabidopsis: BBX regulators in crop plants. Int. J. Mol. Sci. 2021, 22, 2906. [Google Scholar] [CrossRef]
- Wei, H.R.; Wang, P.P.; Chen, J.Q.; Li, C.J.; Wang, Y.Z.; Yuan, Y.B.; Fang, J.G.; Leng, X.P. Genome-wide identification and analysis of B-BOX gene family in grapevine reveal its potential functions in berry development. BMC Plant Biol. 2020, 20, 72. [Google Scholar] [CrossRef]
- Liu, X.; Li, R.; Dai, Y.Q.; Chen, X.S.; Wang, X.Y. Genome-wide identification and expression analysis of the B-box gene family in the Apple (Malus domestica Borkh.) genome. Mol. Genet. Genom. 2018, 293, 303–315. [Google Scholar] [CrossRef]
- Zou, Z.Y.; Wang, R.H.; Wang, R.; Yang, S.L.; Yang, Y.J. Genome-wide identification, phylogenetic analysis, and expression profiling of the BBX family genes in pear. J. Hortic. Sci. Biotechnol. 2018, 93, 37–50. [Google Scholar] [CrossRef]
- Wang, Y.Y.; Zhai, Z.F.; Sun, Y.T.; Feng, C.; Peng, X.; Zhang, X.; Xiao, Y.Q.; Zhou, X.; Wang, W.L.; Jiao, J.L.; et al. Genome-wide identification of the B-BOX genes that respond to multiple ripening related signals in sweet cherry fruit. Int. J. Mol. Sci. 2021, 22, 1622. [Google Scholar] [CrossRef]
- Urszula, T.; Agnieszka, K.; Jagoda, C.; Tadeusz, R. Genome-wide survey of B-box proteins in potato (Solanum tuberosum)—Identification, characterization and expression patterns during diurnal cycle, etiolation and de-etiolation. PLoS ONE 2017, 12, e0177471. [Google Scholar]
- Bu, X.; Wang, X.J.; Yan, J.R.; Zhang, Y.; Zhou, S.Y.; Sun, X.; Yang, Y.X.; Ahammed, G.J.; Liu, Y.F.; Li, T.; et al. Genome-Wide Characterization of B-Box Gene Family and Its Roles in Responses to Light Quality and Cold Stress in Tomato. Front. Plant Sci. 2021, 12, 698525. [Google Scholar] [CrossRef]
- Huang, J.Y.; Zhao, X.B.; Weng, X.Y.; Wang, L.; Xie, W.B. The Rice B-Box Zinc Finger Gene Family: Genomic Identification, Characterization, Expression Profiling and Diurnal Analysis. PLoS ONE 2012, 7, e48242. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, S.B.; Shen, Y.; Chang, H.C.; Hou, Y.L.; Harris, A.; Ma, S.F.; McPartland, M.; Hymus, G.J.; Adam, L.; Marion, C.; et al. The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytol. 2010, 187, 57–66. [Google Scholar] [CrossRef]
- Steinbach, Y. The Arabidopsis thaliana CONSTANS-LIKE 4 (COL4)—A modulator of flowering time. Front. Plant Sci. 2019, 10, 651. [Google Scholar] [CrossRef] [PubMed]
- Hassidim, M.; Harir, Y.; Yakir, E.; Kron, I.; Green, R.M. Over-expression of CONSTANS-LIKE 5 can induce flowering in short-day grown Arabidopsis. Planta 2009, 230, 481–491. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.F.; Wang, Z.Y. Overexpression of COL9, a CONSTANS-LIKE gene, delays flowering by reducing expression of CO and FT in Arabidopsis thaliana. Plant J. Cell Mol. Biol. 2005, 43, 758–768. [Google Scholar] [CrossRef]
- Natalia, O.H.; Trimborn, L.; Menje, M.; Henschel, M.; Robers, L.; Kaufholdt, D.; Hänsch, R.; Adrian, J.; Ponnu, J.; Hoecker, U. The transcription factor COL12 is a substrate of the COP1/SPA E3 ligase and regulates flowering time and plant architecture. Plant Physiol. 2018, 176, 1327–1340. [Google Scholar]
- Wang, C.Q.; Guthrie, C.; Sarmast, M.K.; Dehesh, K. BBX19 interacts with CONSTANS to repress FLOWERING LOCUS T transcription, defining a flowering time checkpoint in Arabidopsis. Plant Cell 2014, 26, 3589–3602. [Google Scholar] [CrossRef]
- Gangappa, S.N.; Crocco, C.D.; Johansson, H.; Datta, S.; Hettiarachchi, C.; Holm, M.; Botto, J.F. The Arabidopsis B-BOX protein BBX25 interacts with HY5, negatively regulating BBX22 expression to suppress seedling photomorphoge-nesis. Plant Cell 2013, 25, 1243–1257. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Lin, G.; Yin, C.M.; Fang, Y.D. B-box transcription factor 28 regulates flowering by interacting with constans. Sci. Rep. 2020, 10, 17789. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.; Jiang, Y.; Li, J.; Yan, T.T.; Fan, L.M.; Liang, J.S.; Chen, Z.J.; Xu, D.Q.; Deng, X.W. B-BOX DOMAIN PROTEIN28 Negatively Regulates Photomorph ogenesis by Repressing the Activity of Transcription Factor HY5 and Undergoes COP1-Mediated Degradation. Plant Cell 2018, 30, 2006–2019. [Google Scholar] [CrossRef]
- Song, Z.Q.; Yan, T.T.; Liu, J.J.; Bian, Y.T.; Heng, Y.Q.; Lin, F.; Jiang, Y.; Wang, D.X.; Xu, D.Q. BBX28/BBX29, HY5 and BBX30/31 form a feedback loop to fine-tune photomorphogenic development. Plant J. 2020, 104, 377–390. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.H.; Heng, Y.Q.; Wang, X.C.; Deng, X.W.; Xu, D.Q. A Positive Feedback Loop of BBX11-BBX21-HY5 Promotes Photomorphogenic Development in Arabidopsis. Plant Commun. 2020, 1, 100045. [Google Scholar] [CrossRef]
- Chang, C.S.; Li, Y.H.; Chen, L.T.; Chen, W.C.; Hsieh, W.P.; Shin, J.; Jane, W.N.; Chou, S.J.; Choi, G.; Hu, J.M.; et al. LZF1, a HY5-regulated transcriptional factor, functions in Arabidopsis de-etiolation. Plant J. 2008, 54, 205–219. [Google Scholar] [CrossRef]
- Zhang, X.Y.; Huai, J.L.; Shang, F.F.; Xu, G.; Tang, W.J.; Jing, Y.J.; Lin, R.C. A PIF1/PIF3-HY5-BBX23 Transcription Factor Cascade Affects Photomorphogenesis. Plant Physiol. 2017, 174, 2487–2500. [Google Scholar] [CrossRef]
- Tripathi, P.; Carvallo, M.; Hamilton, E.E.; Preuss, S.; Kay, S.A. Arabidopsis B-BOX32 interacts with CONSTAN S-LIKE3 to regulate flowering. Proc. Natl. Acad. Sci. USA 2017, 114, 172–177. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.G.; Zhang, Z.L.; Li, H.Y.; Zhao, X.Y.; Liu, X.M.; Ortiz, M.; Lin, C.; Liu, B. CONSTANS-LIKE 7 regulates branching and shade avoidance response in Arabidopsis. J. Exp. Bot. 2013, 64, 1017–1024. [Google Scholar] [CrossRef]
- Lyu, G.Z.; Li, D.B.; Li, S.S. Bioinformatics analysis of BBX family genes and its response to UV-B in Arabidopsis thaliana. Plant Signal. Behav. 2020, 15, 1782647. [Google Scholar] [CrossRef]
- Min, J.H.; Chung, J.S.; Lee, K.H.; Kim, C.S. The CONSTANS-like 4 transcription factor, AtCOL4, positively regulates abiotic stress tolerance through an abscisic acid-dependent manner in Arabidopsis. J. Integr. Plant Biol. 2015, 57, 313–324. [Google Scholar] [CrossRef]
- Wang, Q.M.; Tu, X.J.; Zhang, J.H.; Chen, X.B.; Rao, L.Q. Heat stress-induced BBX18 negatively regulates the thermotolerance in Arabidopsis. Mol. Biol. Rep. 2013, 40, 2679–2688. [Google Scholar] [CrossRef]
- Nagaoka, S.; Takano, T. Salt tolerance-related protein STO binds to a Myb transcription factor homologue and confers salt tolerance in Arabidopsis. J. Exp. Bot. 2003, 54, 2231–2237. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.F.; Wang, X.; Cao, Y.Z.; Zhang, H.Y.; Hua, R.; Liu, H.M.; Sui, S.Z. CpBBX19, a B-Box Transcription Factor Gene of Chimonanthus praecox, Improves Salt and Drought Tolerance in Arabidopsis. Genes 2021, 12, 1456. [Google Scholar] [CrossRef]
- Yang, Y.J.; Ma, C.; Xu, Y.J.; Wei, Q.; Imtiaz, M.; Lan, H.B.; Gao, S.; Cheng, L.; Wang, M.Y.; Fei, Z.J.; et al. A Zinc Finger Protein Regulates Flowering Time and Abiotic Stress Tolerance in Chrysanthemum by Modulating Gibberellin Biosynthesis. Plant Cell 2014, 26, 2038–2054. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Hannah, R.T.; Margaret, H.F.; He, Y.H.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.Y.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, 202–208. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Vande, P.Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Li, H.H.; Lu, X.Q.; Chen, L.Z.; Liu, J.; Wu, H. Identification and Expression Analysis of GRAS Transcription Factors to Elucidate Candidate Genes Related to Stolons, Fruit Ripening and Abiotic Stresses in Woodland Strawberry (Fragaria vesca). Int. J. Mol. Sci. 2019, 20, 4593. [Google Scholar] [CrossRef]
- Li, Y.P.; Pi, M.T.; Gao, Q.; Liu, Z.C.; Kang, C.Y. Author Correction: Updated annotation of the wild strawberry Fragaria vesca V4 genome. Hortic. Res. 2019, 6, 90. [Google Scholar] [CrossRef] [PubMed]
- Gu, T.T.; Jia, S.F.; Huang, X.R.; Wang, L.; Fu, W.M.; Huo, G.T.; Gan, L.J.; Ding, J.; Li, Y. Transcriptome and hormone analyses provide insights into hormonal regulation in strawberry ripening. Planta 2019, 250, 145–162. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.Y.; Yu, F.H.; Guo, Q.; Wang, Y.; Zhang, Z.H.; Liu, Y.X. Genome-Wide Identification, Characterization, and Expression Profile Analysis of CONSTANS-like Genes in Woodland Strawberry (Fragaria vesca). Front. Plant Sci. 2022, 13, 931721. [Google Scholar] [CrossRef]
- Yu, D.Q.; Tang, H.R.; Zhang, Y.; Zhen, D.; Yu, H.W.; Chen, Q. Comparison and Improvement of Different Methods of RNA Isolation from Strawberry (Fragria × ananassa). J. Agric. Sci. 2012, 4, 51–56. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data using Real-Time Quantitative PCR. Methods 2002, 25, 402–408. [Google Scholar] [CrossRef]
- Abdullah, S.; Fan, S.; Jia, P.; Li, G.F.; Izhar, M.; Li, Y.M.; Rahat, S.; Dong, F.; Zuo, X.Y.; Li, K.; et al. Genome Identification of B-BOX Gene Family Members in Seven Rosaceae Species and Their Expression Analysis in Response to Flower Induction in Malus domestica. Molecules 2018, 23, 1763. [Google Scholar]
- Cao, Y.P.; Meng, D.D.D.; Han, Y.H.; Chen, T.Z.; Jiao, C.Y.; Chen, Y.; Jin, Q.; Cai, Y.P. Comparative analysis of B-BOX genes and their expression pattern analysis under various treatments in Dendrobium officinale. BMC Plant Biol. 2019, 19, 245. [Google Scholar] [CrossRef] [PubMed]
- Soitamo, A.; Piippo, M.; Allahverdiyeva, Y.; Battchikova, N.; Aro, E.M. Light has a specific role in modulating Arabidopsis gene expression at low temperature. BMC Plant Biol. 2008, 8, 13. [Google Scholar] [CrossRef]
- Riboni, M.; Robustelli, T.A.; Galbiati, M.; Tonelli, C.; Conti, L. ABA-dependent control of GIGANTEA signaling enables drought escape via up-regulation of FLOWERING LOCUS T in Arabidopsis thaliana. J. Exp. Bot. 2016, 67, 6309–6322. [Google Scholar] [CrossRef]
- Liu, J.H.; Shen, J.Q.; Xu, Y.; Li, X.H.; Xiao, J.H.; Xiong, L.Z. Ghd2, a CONSTANS-like gene, confers drought sensitivity through regulation of senescence in rice. J. Exp. Bot. 2016, 67, 5785–5798. [Google Scholar] [CrossRef]
- Song, Z.Q.; Bian, Y.T.; Liu, J.J.; Sun, Y.T.; Xu, D.Q. B-box proteins: Pivotal players in light-mediated development in plants. J. Integr. Plant Biol. 2020, 62, 1293–1309. [Google Scholar] [CrossRef]
- Bai, S.L.; Tao, R.Y.; Tang, Y.X.; Yin, L.; Ma, Y.J.; Ni, J.B.; Yan, X.H.; Yang, Q.S.; Wu, Z.Y.; Zeng, Y.L.; et al. BBX16, a B-box protein, positively regulates light-induced anthocyanin accumulation by activating MYB10 in red pear. Plant Biotechnol. J. 2019, 17, 1985–1997. [Google Scholar] [CrossRef]
- Cao, Y.P.; Han, Y.H.; Meng, D.D.; Li, D.H.; Jiao, C.Y.; Jin, Q.; Lin, Y.; Cai, Y.P. B-BOX genes: Genome-wide identification, evolution and their contribution to pollen growth in pear (Pyrus bretschneideri Rehd.). BMC Plant Biol. 2017, 17, 156. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.Q.; Li, J.G.; Gangappa, S.N.; Hettiarachchi, C.; Lin, F.; Andersson, M.X.; Jiang, Y.; Deng, X.W.; Holm, M. Convergence of light and ABA signaling on the ABI5 promoter. PLoS Genet. 2014, 10, e1004197. [Google Scholar] [CrossRef]
- Jim, G. Molecular biology of fruit maturation and ripening. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 725–749. [Google Scholar]
- Cappellini, F.; Marinelli, A.; Toccaceli, M.; Tonelli, C.; Petroni, K. Anthocyanins: From Mechanisms of Regulation in Plants to Health Benefits in Foods. Front. Plant Sci. 2021, 12, 748049. [Google Scholar] [CrossRef]
- Krga, I.; Milenkovic, D. Anthocyanins: From sources and bioavailability to cardiovascular-Health benefits and molecular mechanisms of action. J. Agric. Food Chem. 2019, 67, 1771–1783. [Google Scholar] [CrossRef] [PubMed]
- Datta, S.; Hettiarachchi, C.; Johansson, H.; Holm, M. SALT TOLERANCE HOMOLOG2, a B-box protein in Arabidopsis that activates transcription and positively regulates light-mediated development. Plant Cell 2007, 19, 3242–3255. [Google Scholar] [CrossRef]
- Bai, S.L.; Saito, T.; Honda, C.; Hatsuyama, Y.; Ito, A.; Moriguchi, T. An apple B-box protein, MdCOL11, is involved in UV-B- and temperature-induced anthocyanin biosynthesis. Planta 2014, 240, 1051–1062. [Google Scholar] [CrossRef]
- An, J.P.; Wang, X.F.; Espley, R.V.; Lin, W.K.; Bi, S.Q.; You, C.X.; Hao, Y.J. An Apple B-Box Protein MdBBX37 Modulates Anthocyanin Biosynthesis and Hypocotyl Elongation Synergistically with MdMYBs and MdHY5. Plant Cell Physiol. 2020, 61, 130–143. [Google Scholar] [CrossRef] [PubMed]
- Bai, S.L.; Tao, R.Y.; Yin, L.; Ni, J.B.; Yang, Q.S.; Yan, X.H.; Yang, F.; Guo, X.P.; Li, H.X.; Teng, Y.W. Two B-box proteins, PpBBX18 and PpBBX21, antagonistically regulate anthocyanin biosynthesis via competitive association with Pyrus pyrifolia ELONGATED HYPOCOTYL 5 in the peel of pear fruit. Plant J. 2019, 100, 1208–1223. [Google Scholar] [CrossRef]
- Chen, J.; Chen, J.Y.; Wang, J.N.; Kuang, J.F.; Shan, W.; Lu, W.J. Molecular characterization and expression profiles of MaCOL1, a CONSTANS -like gene in banana fruit. Gene 2012, 496, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Lira, B.S.; Oliveira, M.J.; Shiose, L.; Wu, R.T.; Rosado, D.; Lupi Alessandra, C.D.; Freschi, L.; Rossi, M. Light and ripening-regulated BBX protein-encoding genes in Solanum lycopersicum. Sci. Rep. 2020, 10, 19235. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.Q.; Ye, Y.T.; Wang, Y.P.; Jiang, L.Y.; Yue, M.L.; Tang, L.; Jin, M.S.X.; Zhang, Y.T.; Lin, Y.X.; Tang, H.R. B-Box Transcription Factor FaBBX22 Promotes Light-Induced Anthocyanin Accumulation in Strawberry (Fragaria × ananassa). Int. J. Mol. Sci. 2022, 23, 7757. [Google Scholar] [CrossRef] [PubMed]




| Protein Name | Gene ID | CDS | Protein/AA | pI | MW | Introns | Chr Srart | Chr End | Chrom |
|---|---|---|---|---|---|---|---|---|---|
| FveBBX1 | FvH4_1g12110.t3 | 675 | 225 | 6.24 | 24,772.05 | 3 | 6615641 | 6618432 | Fvb1 |
| FveBBX2 | FvH4_2g10070.t1 | 639 | 213 | 6.17 | 23,958.62 | 1 | 8897464 | 8899003 | Fvb2 |
| FveBBX3 | FvH4_2g24910.t1 | 1182 | 394 | 5.74 | 43,576.53 | 3 | 20252750 | 20254655 | Fvb2 |
| FveBBX4 | FvH4_2g38990.t1 | 942 | 314 | 6.62 | 34,463.57 | 2 | 28109150 | 28111588 | Fvb2 |
| FveBBX5 | FvH4_2g41420.t2 | 939 | 313 | 6.33 | 34,506.73 | 1 | 29305073 | 29306617 | Fvb2 |
| FveBBX6 | FvH4_3g03640.t1 | 552 | 184 | 4.37 | 20,264.61 | 1 | 2045154 | 2047400 | Fvb3 |
| FveBBX7 | FvH4_3g17750.t1 | 864 | 288 | 5.12 | 30,812.60 | 2 | 11299422 | 11301298 | Fvb3 |
| FveBBX8 | FvH4_3g21230.t1 | 744 | 248 | 4.30 | 27,112.50 | 1 | 14224305 | 14225858 | Fvb3 |
| FveBBX9 | FvH4_4g08980.t1 | 1071 | 357 | 5.63 | 38,993.97 | 1 | 10078076 | 10079592 | Fvb4 |
| FveBBX10 | FvH4_4g10930.t1 | 717 | 239 | 4.71 | 26,293.62 | 3 | 14596852 | 14598977 | Fvb4 |
| FveBBX11 | FvH4_4g23090.t1 | 1437 | 479 | 5.66 | 51,772.28 | 3 | 25667711 | 25670086 | Fvb4 |
| FveBBX12 | FvH4_4g26540.t1 | 912 | 304 | 6.25 | 33,281.15 | 2 | 27896068 | 27898487 | Fvb4 |
| FveBBX13 | FvH4_4g26550.t1 | 1416 | 472 | 5.62 | 52,959.81 | 4 | 27899275 | 27901406 | Fvb4 |
| FveBBX14 | FvH4_4g27390.t1 | 1353 | 451 | 5.56 | 50,333.90 | 1 | 28409309 | 28411538 | Fvb4 |
| FveBBX15 | FvH4_5g12150.t1 | 1374 | 458 | 5.56 | 51,231.43 | 1 | 6852249 | 6854527 | Fvb5 |
| FveBBX16 | FvH4_6g37140.t1 | 795 | 265 | 8.99 | 28,638.63 | 0 | 29197326 | 29198120 | Fvb6 |
| FveBBX17 | FvH4_6g37790.t1 | 585 | 195 | 8.30 | 20,963.21 | 2 | 29750190 | 29757327 | Fvb6 |
| FveBBX18 | FvH4_6g40380.t1 | 1254 | 418 | 5.45 | 45,246.46 | 5 | 31931413 | 31935765 | Fvb6 |
| FveBBX19 | FvH4_6g43570.t1 | 1335 | 445 | 5.43 | 48,826.65 | 4 | 33682838 | 33686104 | Fvb6 |
| FveBBX20 | FvH4_6g43580.t1 | 339 | 113 | 8.75 | 12,618.81 | 0 | 33686668 | 33687006 | Fvb6 |
| FveBBX21 | FvH4_6g44270.t1 | 759 | 253 | 4.71 | 27,901.25 | 2 | 34125442 | 34128499 | Fvb6 |
| FveBBX22 | FvH4_6g45860.t1 | 1158 | 386 | 5.35 | 42,345.92 | 1 | 35108088 | 35110508 | Fvb6 |
| Gene Name | Hormone Responsive | Light Responsive | Stress Responsive | Other Responsive | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ABA | GA | IAA | MeJA | SA | Anaerobic | Drought | Low-Temperature | Defense | |||
| FveBBX1 | + | + | + | + | + | + | + | + | |||
| FveBBX2 | + | + | + | + | + | ||||||
| FveBBX3 | + | + | + | + | + | + | + | + | + | ||
| FveBBX4 | + | + | + | + | + | + | |||||
| FveBBX5 | + | + | + | + | + | + | |||||
| FveBBX6 | + | + | + | + | + | + | + | + | |||
| FveBBX7 | + | + | + | + | + | + | + | ||||
| FveBBX8 | + | + | + | + | + | + | + | + | + | ||
| FveBBX9 | + | + | + | + | + | + | + | ||||
| FveBBX10 | + | + | + | + | + | + | |||||
| FveBBX11 | + | + | + | + | + | + | + | ||||
| FveBBX12 | + | + | + | + | + | + | |||||
| FveBBX13 | + | + | + | + | + | + | + | + | + | ||
| FveBBX14 | + | + | + | + | + | + | + | + | + | ||
| FveBBX15 | + | + | + | + | + | + | |||||
| FveBBX16 | + | + | + | + | + | + | + | ||||
| FveBBX17 | + | + | + | + | + | + | + | + | |||
| FveBBX18 | + | + | + | + | + | + | + | + | |||
| FveBBX19 | + | + | + | + | + | + | + | + | |||
| FveBBX20 | + | + | + | + | + | + | + | ||||
| FveBBX21 | + | + | + | + | + | + | + | + | |||
| FveBBX22 | + | + | + | + | + | + | + | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, D.; Wang, H.; Feng, X.; Ma, Y.; Huang, Y.; Wang, Y.; Ding, J.; Chen, H.; Wu, H. Genome-Wide Identification, Phylogenetic and Expression Analysis of the B-Box Gene Family in the Woodland Strawberry (Fragaria vesca). Horticulturae 2023, 9, 842. https://doi.org/10.3390/horticulturae9070842
Xu D, Wang H, Feng X, Ma Y, Huang Y, Wang Y, Ding J, Chen H, Wu H. Genome-Wide Identification, Phylogenetic and Expression Analysis of the B-Box Gene Family in the Woodland Strawberry (Fragaria vesca). Horticulturae. 2023; 9(7):842. https://doi.org/10.3390/horticulturae9070842
Chicago/Turabian StyleXu, Dong, Hongkun Wang, Xiaotian Feng, Yuqing Ma, Yirui Huang, Yushan Wang, Jing Ding, Hong Chen, and Han Wu. 2023. "Genome-Wide Identification, Phylogenetic and Expression Analysis of the B-Box Gene Family in the Woodland Strawberry (Fragaria vesca)" Horticulturae 9, no. 7: 842. https://doi.org/10.3390/horticulturae9070842
APA StyleXu, D., Wang, H., Feng, X., Ma, Y., Huang, Y., Wang, Y., Ding, J., Chen, H., & Wu, H. (2023). Genome-Wide Identification, Phylogenetic and Expression Analysis of the B-Box Gene Family in the Woodland Strawberry (Fragaria vesca). Horticulturae, 9(7), 842. https://doi.org/10.3390/horticulturae9070842






