Effectiveness of a Novel Endophyte Bacillus velezensis Strain B1 in the Biocontrol of Pear Postharvest Ring Rot
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation of Antifungal Bacterial Strains
2.2. Screening of Antagonistic Bacteria against Botryosphaeria dothidea
2.3. Evaluating the Inhibitory Effect of Culture Filtrate of Strain B1
2.4. Inhibition Spectrum of Strain B1
2.5. Biocontrol of Pear Fruit Ring Rot by Strain B1
2.6. Genome Sequencing, Annotation and Phylogenetic Analysis
2.7. Genome Mining of Secondary Metabolites Biosynthetic Gene Clusters
2.8. Mass Spectrometry Analysis of Secondary Metabolites Produced by Strain B1
2.9. Statistical Analysis
3. Results
3.1. Isolation and Screening of Antagonistic Endophytes
3.2. Inhibitory Effect of Cell Suspension and Cell-Free Supernatants on Growth of B. dothidea
3.3. Antifungal Spectrum of Strain B1
3.4. Biocontrol Effect of Strain B1 on B. dothidea In Vivo
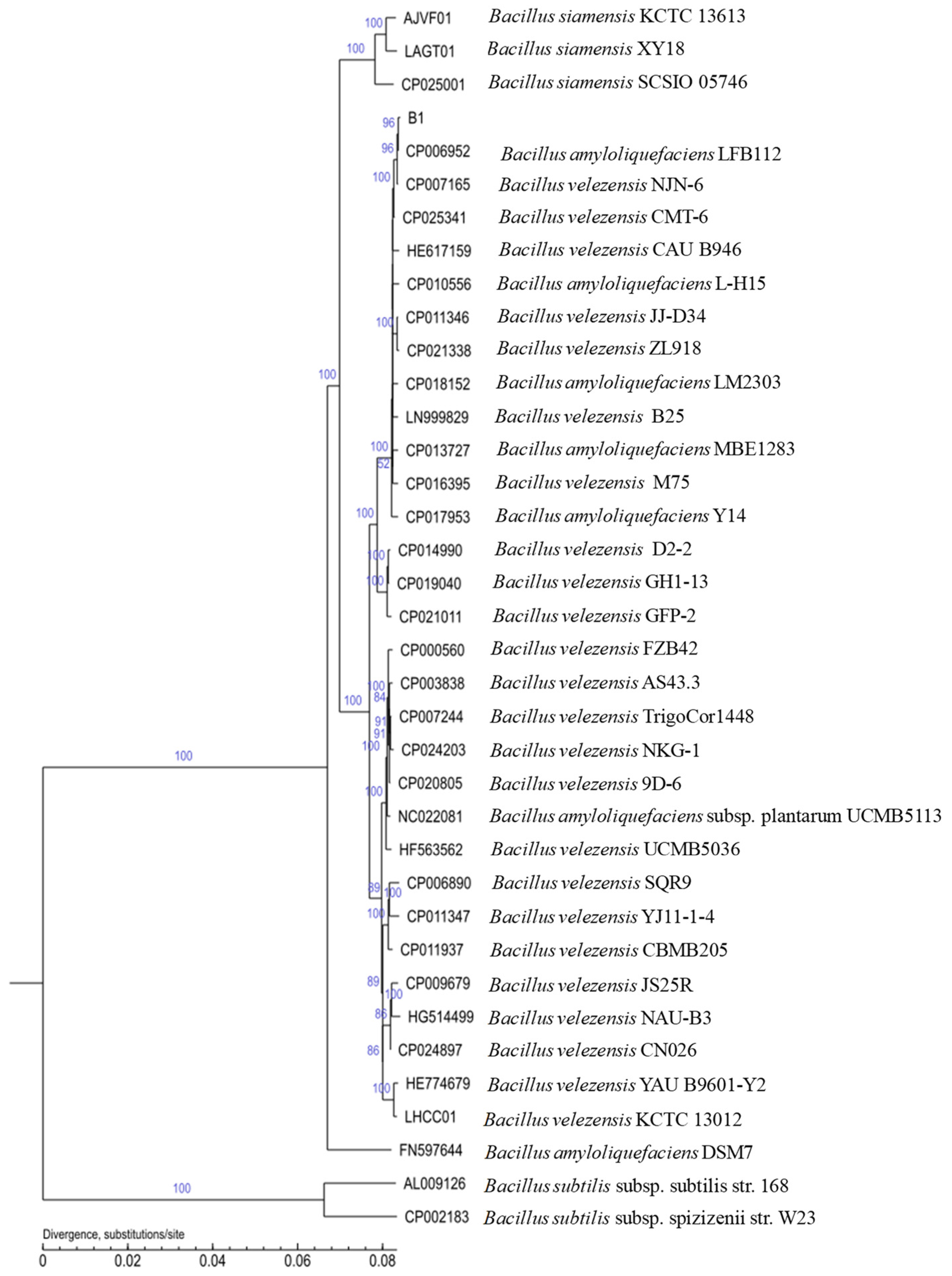
3.5. Genome General Characterization and Phylogenetic Analysis of Strain B1
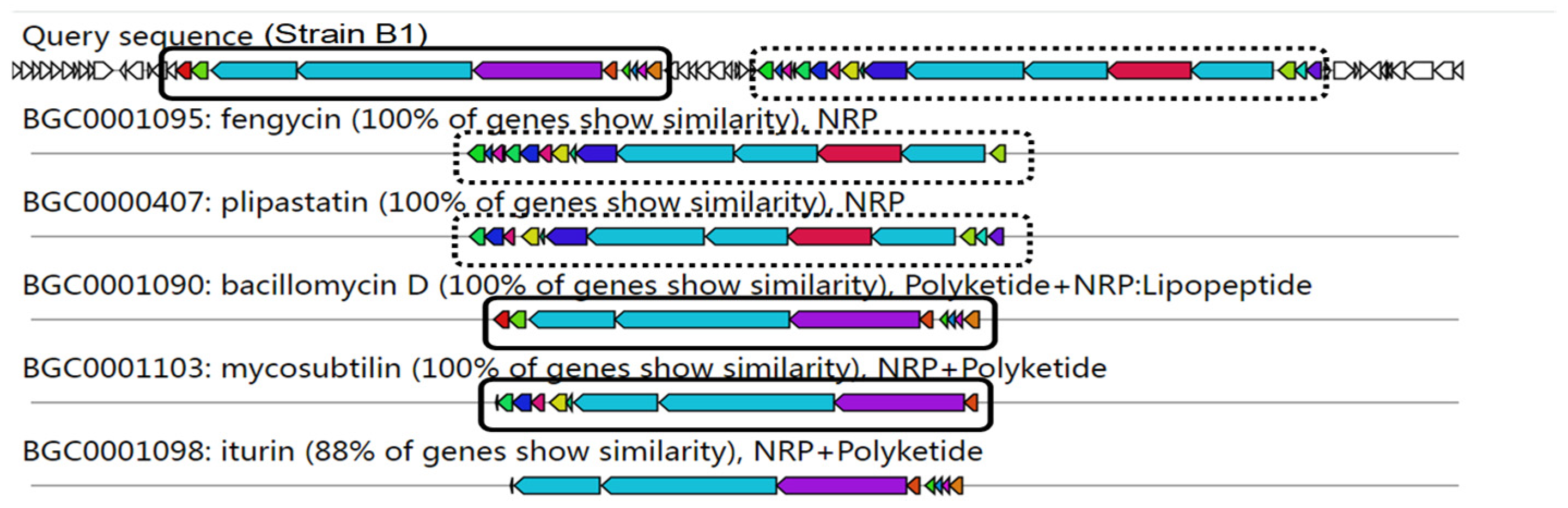
3.6. Genome Mining of Secondary Metabolites Biosynthetic Gene Clusters (BGCs)
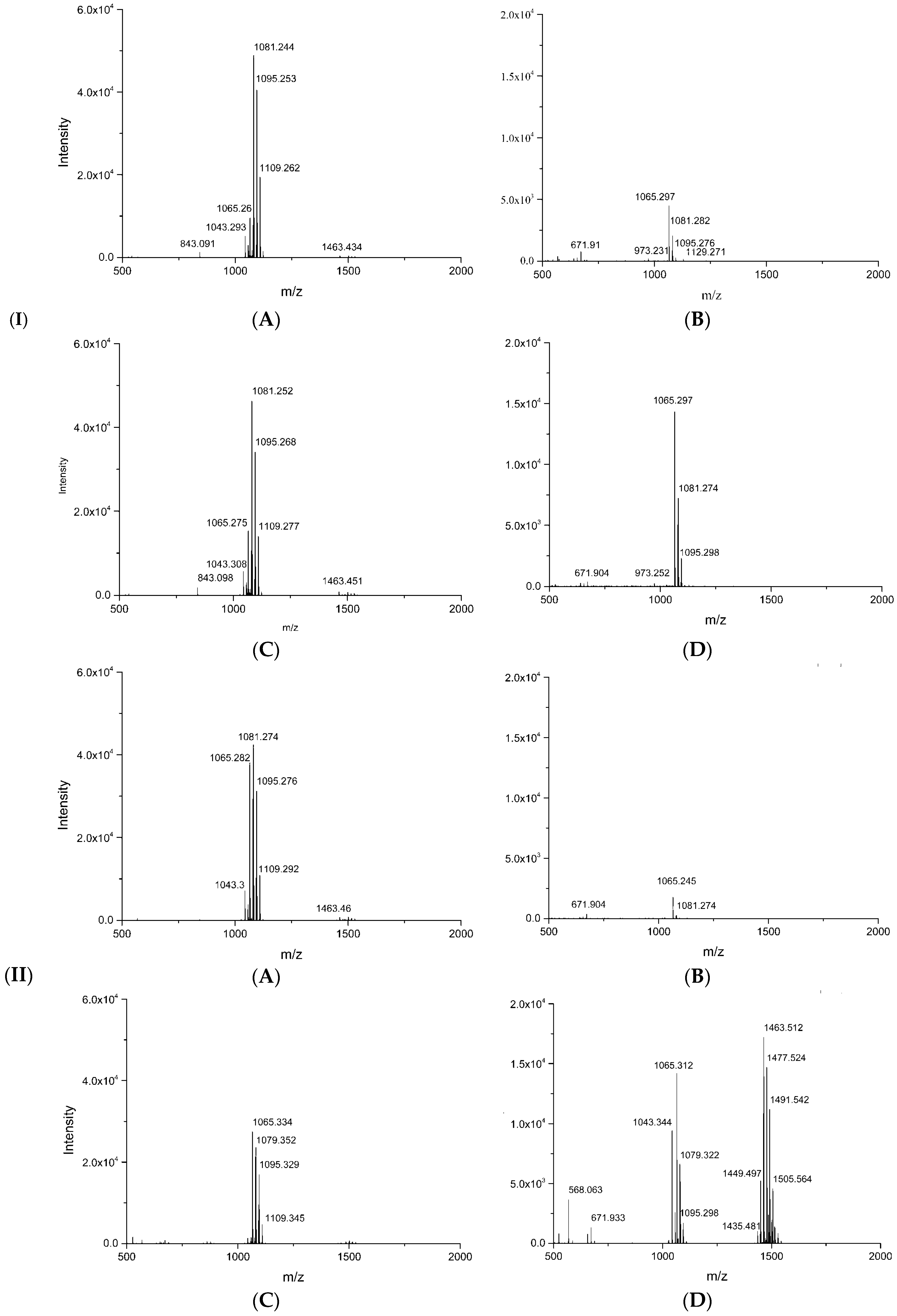
3.7. MALDI-TOF-MS Analysisof Secondary Metabolites
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Xu, B.; Zhang, H.; Chen, K.; Xu, Q.; Yao, Y.; Gao, H. Biocontrol of postharvest Rhizopus decay of peaches with Pichia caribbica. Curr. Microbiol. 2013, 67, 255–261. [Google Scholar] [CrossRef]
- Qu, H.; Zhao, L.Y.; Zhao, F.C.; Liu, Y.F.; Yang, Z.Y. Biocontrol of gray mold decay in pear by Bacillus amyloliquefaciens strain BA3 and its effect on postharvest quality parameters. Pol. J. Microbiol. 2016, 65, 171–176. [Google Scholar] [CrossRef]
- Marsberg, A.; Kemler, M.; Jami, F.; Nagel, J.H.; Postma-Smidt, A.; Naidoo, S.; Wingfield, M.J.; Crous, P.W.; Spatafora, J.W.; Hesse, C.N.; et al. Botryosphaeria dothidea: A latent pathogen of global importance to woody plant health. Mol. Plant Pathol. 2017, 18, 477–488. [Google Scholar] [CrossRef]
- Noguchi, H.; Hiruma, M.; Matsumoto, T.; Kano, R.; Tanaka, M.; Yaguchi, T.; Sonoda, K.; Ihn, H. Fungal melanonychia: Ungual phaeohyphomycosis caused by Botryosphaeria dothidea. Acta Derm. Venereol. 2017, 97, 765–766. [Google Scholar] [CrossRef]
- Pour, F.N.; Ferreira, V.; Félix, C.; Serôdio, J.; Alves, A.; Duarte, A.S.; Esteves, A.C. Effect of temperature on the phytotoxicity and cytotoxicity of Botryosphaeriaceae fungi. Fungal Biol. 2020, 124, 571–578. [Google Scholar] [CrossRef]
- Andrić, S.; Meyer, T.; Ongena, M. Bacillus responses to plant-associated fungal and bacterial communities. Front. Microbiol. 2020, 11, 1350. [Google Scholar] [CrossRef]
- Arnaouteli, S.; Bamford, N.C.; Stanley-Wall, N.R.; Kovács, Á.T. Bacillus subtilis biofilm formation and social interactions. Nat. Rev. Microbiol. 2021, 19, 600–614. [Google Scholar] [CrossRef]
- Erdogan, O.; Benlioglu, K. Biological control of Verticillium wilt on cotton by the use of fluorescent Pseudomonas spp. under field conditions. Biol. Control 2010, 53, 39–45. [Google Scholar] [CrossRef]
- Luo, J.; Ran, W.; Hu, J.; Yang, X.; Xu, Y.; Shen, Q. Application of bio-organic fertilizer significantly affected fungal diversity of soils. Soil Sci. Soc. Am. J. 2010, 74, 2039–2048. [Google Scholar] [CrossRef]
- Mizumoto, S.; Hirai, M.; Shoda, M. Enhanced iturin A production by Bacillus subtilis and its effect on suppression of the plant pathogen Rhizoctonia solani. Appl. Microbiol. Biotechnol. 2007, 75, 1267–1274. [Google Scholar] [CrossRef]
- Bhore, S.J.; Preveena, J.; Kandasamy, K.I. Isolation and identification of bacterial endophytes from pharmaceutical agarwood-producing Aquilaria species. Pharmacogn. Res. 2013, 5, 134–137. [Google Scholar] [CrossRef]
- Brader, G.; Stephane, C.; Birgit, M.; Friederike, T.; Angela, S. Metabolic potential of endophytic bacteria. Curr. Opin. Biotechnol. 2014, 27, 30–37. [Google Scholar] [CrossRef]
- Christina, A.; Christapher, V.; Bhore, S.J. Endophytic bacteria as a source of novel antibiotics: An overview. Pharmacogn. Rev. 2013, 7, 11–16. [Google Scholar]
- Li, C.; Cheng, P.; Zheng, L.; Li, Y.; Chen, Y.; Wen, S.; Yu, G. Comparative genomics analysis of two banana fusarium wilt biocontrol endophytes Bacillus subtilis R31 and TR21 provides insights into their differences on phytobeneficial trait. Genomics 2021, 1139, 900–909. [Google Scholar] [CrossRef]
- Ryan, R.P.; Germaine, K.; Franks, A.; Ryan, D.J.; Dowling, D.N. Bacterial endophytes: Recent developments and applications. FEMS Microbiol. Lett. 2008, 278, 1–9. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, R.H.; Cao, Y.H.; Chen, C.Y.; Bai, F.R.; Xu, T.J.; Zhao, R.; Zhang, X.; Zhao, J.R.; Cheng, C. Identification and antagonistic activity of endophytic bacterial strain Paenibacillus sp. 5 L8 isolated from the seeds of maize (Zea mays L., Jingke 968). Ann. Microbiol. 2016, 66, 653–660. [Google Scholar] [CrossRef]
- Chen, S.T.; Dai, J.; Song, X.W.; Jiang, X.P.; Zhao, Q.; Sun, C.B.; Chen, C.W.; Chen, N.F.; Han, B.X. Endophytic microbiota comparison of Dendrobium huoshanense root and stem in different growth years. Planta Med. 2020, 86, 967–975. [Google Scholar] [CrossRef]
- Yue, J.Y.; Yu, Y. Isolation and identification of endophytic fungi from Dendrobium huoshanense with their antibacterial and anti-inflammatory activities. Pak. J. Pharm. Sci. 2022, 35, 1143–1151. [Google Scholar]
- Zhu, Y.J.; Kong, Y.C.; Hong, Y.; Zhang, L.; Li, S.M.; Hou, S.R.; Chen, X.B.; Xie, T.; Hu, Y.; Wang, X.C. Huoshanmycins A–C, new polyketide dimers produced by endophytic Streptomyces sp. HS-3-L-1 from Dendrobium huoshanense. Front. Chem. 2022, 9, 807508. [Google Scholar] [CrossRef]
- Wu, L.Q.; Shang, H.Z.; Wang, Q.; Gu, H.K.; Liu, G.J.; Yang, S.L. Isolation and characterization of antagonistic endophytes from Dendrobium candidum Wall ex Lindl., and the biofertilizing potential of a novel Pseudomonas saponiphila strain. Appl. Soil Ecol. 2016, 105, 101–108. [Google Scholar] [CrossRef]
- Wu, L.Q.; Shang, H.Z.; Gu, H.K.; Zheng, J. Bacterial iturins mediate biocontrol activity of Bacillus sp. against postharvest pear fruitrotting fungi. J. Phytopathol. 2019, 167, 501–509. [Google Scholar] [CrossRef]
- Kurtz, S.; Phillippy, A.; Delcher, A.L.; Smoot, M.; Shumway, M.; Antonescu, C.; Salzberg, S.L. Versatile and open software for comparing large genomes. Genome Biol. 2004, 5, R12. [Google Scholar] [CrossRef] [PubMed]
- Harris, R.S. Improved Pairwise Alignment of Genomic DNA. Ph.D. Thesis, The Pennsylvania State University, University Park, PA, USA, 2007. [Google Scholar]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Vilella, A.J.; Severin, J.; Ureta-Vidal, A.; Heng, L.; Durbin, R.; Birney, E. EnsemblCompara Gene Trees: Complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 2009, 19, 327–335. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. antiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef]
- Borriss, R.; Chen, X.H.; Rueckert, C.; Blom, J.; Becker, A.; Baumgarth, B.; Fan, B.; Pukall, R.; Schumann, P.; Spröer, C.; et al. Relationship of Bacillus amyloliquefaciens clades associated with strains DSM7T and FZB42T: A proposal for Bacillus amyloliquefaciens subsp. amyloliquefaciens subsp. nov. and Bacillus amyloliquefaciens subsp. plantarum sub sp. nov. based on complete genome sequence comparisons. Int. J. Syst. Evol. Microbiol. 2011, 61, 1786–1801. [Google Scholar]
- Belbahri, L.; ChenariBouket, A.; Rekik, I.; Alenezi, F.N.; Vallat, A.; Luptakova, L.; Petrovova, E.; Oszako, T.; Cherrad, S.; Vacher, S.; et al. Comparative Genomics of Bacillus amyloliquefaciens strains reveals a core genome with traits for habitat adaptation and a secondary metabolites rich accessory genome. Front. Microbiol. 2017, 8, 1438. [Google Scholar] [CrossRef]
- Rooney, A.P.; Price, N.P.; Ehrhardt, C.; Swezey, J.L.; Bannan, J.D. Phylogeny and molecular taxonomy of the Bacillus subtilis species complex and description of Bacillus subtilis subsp. Inaquosorum subsp. nov. Int. J. Syst. Evol. Microbiol. 2009, 59, 2429–2436. [Google Scholar] [CrossRef]
- Dunlap, C.A.; Kim, S.J.; Kwon, S.W.; Rooney, A.P. Bacillus velezensis is not a later heterotypic synonym of Bacillus amyloliquefaciens; Bacillus methylotrophicus, Bacillus amyloliquefaciens subsp. plantarum and ‘Bacillus oryzicola’ are later heterotypic synonyms of Bacillus velezensis based on phylogenomics. Int. J. Syst. Evol. Microbiol. 2016, 66, 1212–1217. [Google Scholar]
- Rabbee, M.F.; Ali, M.S.; Choi, J.; Hwang, B.S.; Jeong, S.C.; Baek, K.H. Bacillus velezensis: A valuable member of bioactive molecules within plant microbiomes. Molecules 2019, 224, 1046. [Google Scholar] [CrossRef] [PubMed]
- Chun, B.H.; Kim, K.H.; Jeong, S.E.; Jeon, C.O. Genomic and metabolic features of the Bacillus amyloliquefaciens group–B. amyloliquefaciens, B. velezensis, and B. siamensis–revealed by pan-genome analysis. Food Microbiol. 2019, 77, 146–157. [Google Scholar] [PubMed]
- Fan, B.; Blom, J.; Klenk, H.P.; Borriss, R. Bacillus amyloliquefaciens, Bacillus velezensis, and Bacillus siamensis form an “operational group B. Amyloliquefaciens” within the B. Subtilis species complex. Front. Microbiol. 2017, 8, 3843. [Google Scholar] [CrossRef] [PubMed]
- Palazzini, J.M.; Dunlap, C.A.; Bowman, M.J.; Chulze, S.N. Bacillus velezensis RC 218 as a biocontrol agent to reduce Fusarium head blight and deoxynivalenol accumulation: Genome sequencing and secondary metabolite cluster profiles. Microbiol. Res. 2016, 192, 30–36. [Google Scholar] [CrossRef]
- Koumoutsi, A.; Chen, X.H.; Henne, A.; Liesegang, H.; Hitzeroth, G.; Franke, P.L.; Vater, J.; Borriss, R. Structural and functional characterization of gene clusters directing nonribosomal synthesis of bioactive cyclic lipopeptides in Bacillus amyloliquefaciens Strain FZB42. J. Bacteriol. 2004, 186, 1084–1096. [Google Scholar] [CrossRef] [PubMed]
- Duitman, E.H.; Hamoen, L.W.; Rembold, M.; Venema, G.; Seitz, H.; Saenger, W.; Bernhard, F.; Reinhardt, R.; Schmidt, M.; Ulrich, C.; et al. The mycosubtilin synthetase of Bacillus subtilis ATCC 6633: A multifunctional hybrid between a peptide synthetase, an amino transferase, and a fatty acid synthase. Proc. Natl. Acad. Sci. USA 1999, 96, 13294–13299. [Google Scholar] [CrossRef] [PubMed]
- Vater, J.; Kablitz, B.; Wilde, C.; Franke, P.; Mehta, N.; Cameotra, S.S. Matrix-assisted laser desorption ionization–time of flight mass spectrometry of lipopeptide biosurfactants in whole cells and culture filtrates of Bacillus subtilis C-1 isolated from petroleum sludge. Appl. Environ. Microbiol. 2002, 68, 6210–6219. [Google Scholar]
- Shao, Y.; Yan, H.; Yin, T.; Sun, Z.; Xie, H.; Song, L.; Sun, K.; Li, W. New azaphilones from Penicillium variabile, a fungal endophyte from roots of Aconitum vilmorinianum. J. Antibiot. 2020, 73, 77–81. [Google Scholar] [CrossRef]
- Shiono, Y.; Yokoi, M.; Koseki, T.; Murayama, T.; Aburai, N.; Kimura, K. Allantopyrone A, a new α-pyrone metabolite with potent cytotoxicity from an endophytic fungus, Allantophomopsis lycopodina KS-97. J. Antibiot. 2010, 63, 251–253. [Google Scholar] [CrossRef]
- Yang, Z.; Wu, K.; Xu, Y.; Xia, X.; Wang, X.; Ge, M.; Shao, L. Three novel chromanones with biological activities from the endophytic fungus Phomopsis CGMCC No. 5416. J. Antibiot. 2020, 73, 194–199. [Google Scholar]
- Wicaksono, W.A.; Jones, E.E.; Casonato, S.; Monk, J.; Ridgway, H.J. Biological control of Pseudomonas syringae pv. Actinidiae (Psa), the causal agent of bacterial canker of kiwifruit, using endophytic bacteria recovered from a medicinal plant. Biol. Control 2018, 116, 103–112. [Google Scholar] [CrossRef]
- Yuan, H.; Shi, B.; Wang, L.; Huang, T.; Zhou, Z.; Hou, H.; Tu, H. Isolation and characterization of Bacillus velezensis strain P2-1 for biocontrol of apple postharvest decay caused by Botryosphaeria dothidea. Front. Microbiol. 2022, 12, 808938. [Google Scholar]
- Azabou, M.C.; Gharbi, Y.; Medhioub, I.; Ennouri, K.; Barham, H.; Tounsi, S.; Triki, M.A. The endophytic strain Bacillus velezensis OEE1: An efficient biocontrol agent against Verticillium wilt of olive and a potential plant growth promoting bacteria. Biol. Control 2020, 142, 104168. [Google Scholar]
- Damasceno, C.L.; Duarte, E.A.A.; Santos, L.B.P.R.D.; Oliveira, T.A.S.D.; Jesus, F.N.D.; Oliveira, L.M.D.; Góes-Neto, A.; Soares, A.C.F. Postharvest biocontrol of anthracnose in bananas by endophytic and soil rhizosphere bacteria associated with sisal (agave sisalana) in brazil. Biol. Control 2019, 137, 104016. [Google Scholar]
- Mnif, I.; Ghribi, D. Review lipopeptides biosurfactants: Mean classes and new insights for industrial, biomedical, and environmental applications. Biopolymers 2015, 104, 129–147. [Google Scholar] [PubMed]
- Rautela, R.; Singh, A.K.; Shukla, A.; Cameotra, S.S. Lipopeptides from Bacillus strain AR2 inhibits biofilm formation by Candida albicans. Antonie Leeuwenhoek. 2014, 105, 809–821. [Google Scholar] [CrossRef]
- Li, B.; Li, Q.; Xu, Z.; Zhang, N.; Shen, Q.; Zhang, R. Responses of beneficial Bacillus amyloliquefaciens SQR9 to different soil borne fungal pathogens through the alteration of antifungal compounds production. Front. Microbiol. 2014, 5, 636. [Google Scholar] [CrossRef]
- Stein, T. Bacillus subtilis antibiotics: Structures, syntheses and specific functions. Mol. Microbiol. 2005, 56, 845–857. [Google Scholar]
- Thimon, L.; Peypoux, F.; Dana Maget, R.; Roux, B.; Michel, G. Interactions of bioactive lipopeptides, iturin A and surfactin from Bacillus subtilis. Biotechnol. Appl. Biochem. 1992, 16, 144–151. [Google Scholar]
- Cao, Y.; Pi, H.L.; Chandrangsu, P.; Li, Y.T.; Wang, Y.Q.; Zhou, H.; Xiong, H.Q.; Helmann, J.D.; Cai, Y.F. Antagonism of two plant-growth promoting Bacillus velezensis isolates against Ralstonia solanacearum and Fusarium oxysporum. Sci. Rep. 2018, 8, 4360. [Google Scholar]
- Gonzalez-Jaramillo, L.M.; Aranda, F.J.; Teruel, J.A.; Villegas-Escobar, V.; Ortiz, A. Antimycotic activity of fengycin C biosurfactant and its interaction with phosphatidylcholine model membranes. Colloids Surf. B Biointerfaces 2017, 156, 114–122. [Google Scholar] [PubMed]
- Romero, D.; De, V.A.; Rakotoaly, R.H.; Dufour, S.E.; Veening, J.W.; Arrebola, E.; Cazorla, F.M.; Kuipers, O.P.; Paquot, M.; Perez-Garcia, A. The iturin and fengycin families of lipopeptides are key factors in antagonism of Bacillus subtilis toward Podosphaerafusca. Mol. Plant Microbe Interact. 2007, 20, 430–440. [Google Scholar] [CrossRef] [PubMed]
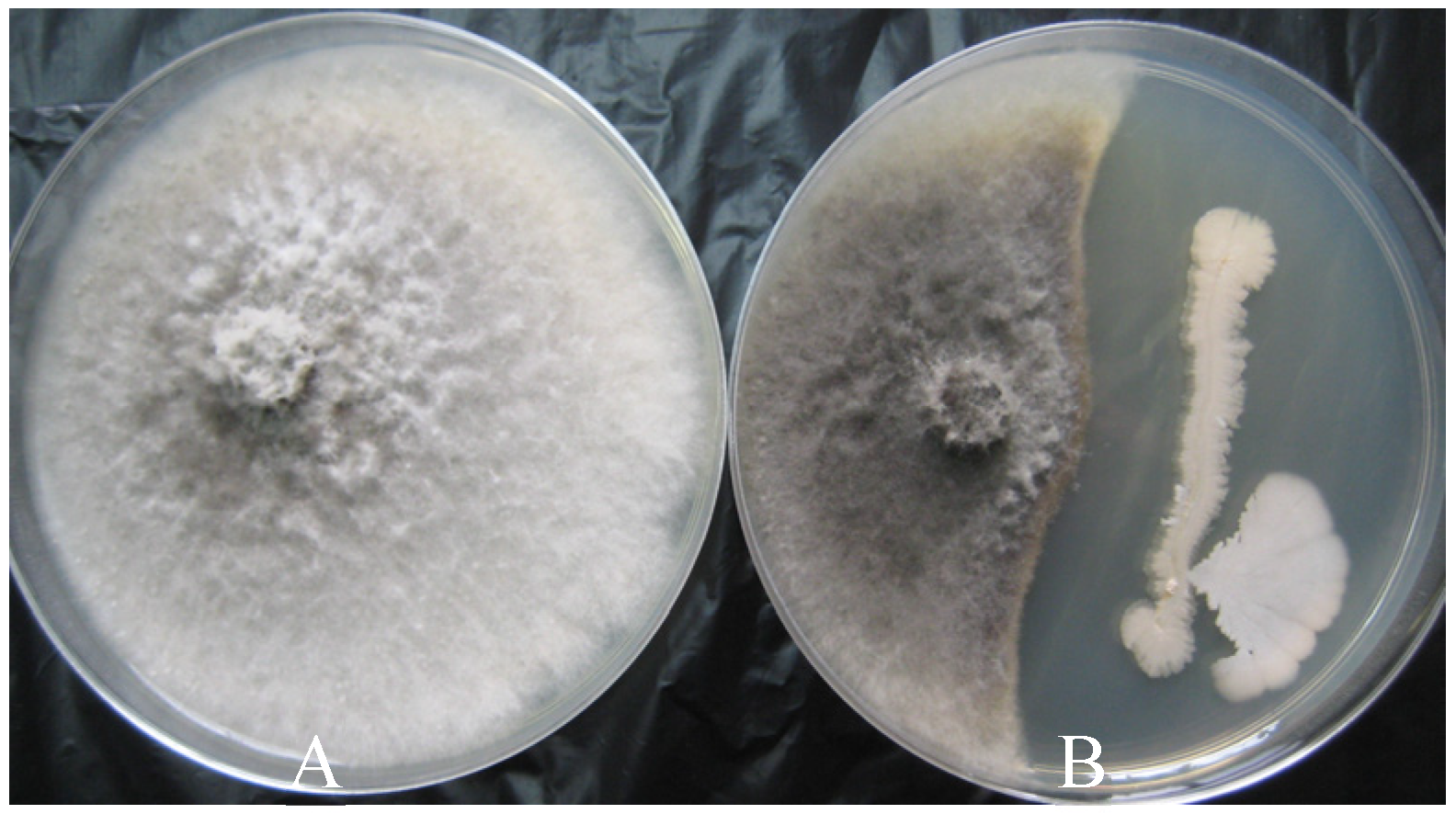

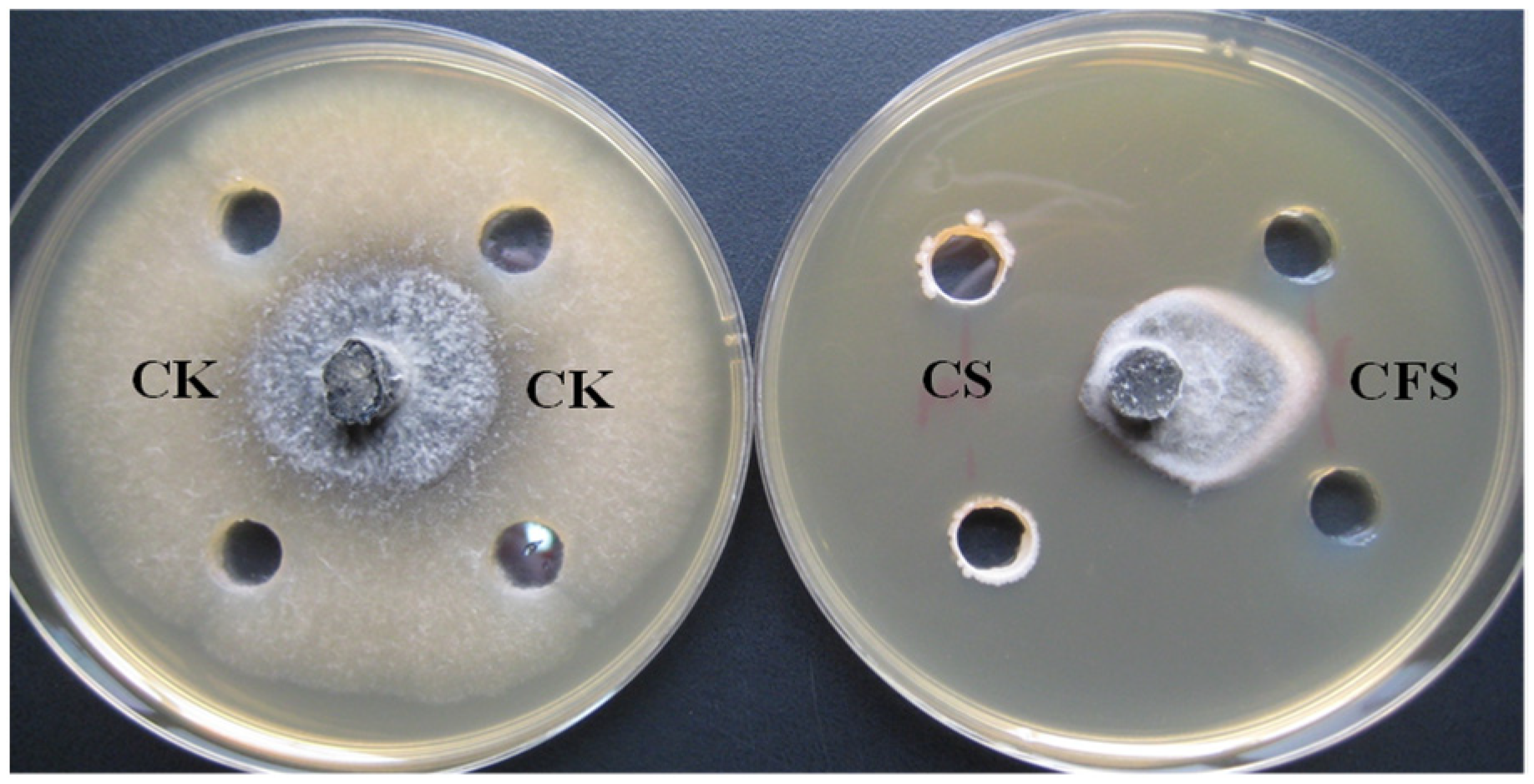

| Treatment | Inhibition Zones (cm) * |
|---|---|
| CS | 1.30 ±0.07 a |
| CFS | 0.52 ± 0.06 b |
| Pathogens | Inhibition Zone/cm |
|---|---|
| Alternaria alstroemeriae | 1.5 ± 0.14 |
| Verticillium dahliae | 2.4 ± 0.28 |
| Rhizoctonia solani | 1.2 ± 0.13 |
| Alternaria brassicae | 1.3 ± 0.12 |
| Botrytis cinerea | 1.2 ± 0.21 |
| Fusarium graminaerum | 1.3 ± 0.22 |
| Fusarium oxysporum | 0.4 ± 0.02 |
| Pyricularia oryzae | 2.0 ± 0.23 |
| Treatment | 3d | 5d | 7d | |||
|---|---|---|---|---|---|---|
| Rot Diameter (cm) | Inhibition Rate (%) | Rot Diameter (cm) | Inhibition Rate (%) | Rot Diameter (cm) | Inhibition Rate (%) | |
| CK | 1.3 ± 0.3 a | 2.81 ± 0.7 a | 4.31 ± 1.1 a | |||
| B1 | 0 b | 100 | 0 b | 100 | 1.3 ± 0.6 b | 73.4 ± 18.6 |
| Mass Peak (m/z) | Family | Assignment |
|---|---|---|
| 1044.3, 1060.3 | Surfactin | C14Surfactin [M + Na, K]+ |
| 1058.3, 1074.3 | C15Surfactin [M + Na, K]+ | |
| 1043.3, 1065.3 | Iturin | C14Iturin A/C14Mycosubtilin [M + H, Na]+ |
| 1057.3, 1079.3 | C15Iturin A/C15Mycosubtilin [M + H, Na]+ | |
| 1081.3 | C14Iturin A/C14Mycosubtilin [M + K]+/C16Bacillomycin D [M + Na]+ | |
| 1095.3 | C15Iturin A/C15Mycosubtilin [M + K]+/C17Bacillomycin D [M + Na]+ | |
| 1109.3 | C16Iturin A/C16Mycosubtilin [M + K]+ | |
| 1435.5 | Fengycin | Ala-6-C14 Fengycin [M + H]+ |
| 1449.5 | Ala-6-C15 Fengycin [M + H]+ | |
| 1463.5, 1485.5, 1501.5 | Ala-6-C16 Fengycin [M + H, Na, K]+ | |
| 1477.5, 1515.5 | Ala-6-C17 Fengycin [M +H, K]+ | |
| 1491.5, 1529.5 | Val-6-C16 Fengycin [M + H, K]+ | |
| 1505.5 | Val-6-C17 Fengycin [M + H]+ |
| Treatment | Mass Peak (m/z) | Assignment | Intensity |
|---|---|---|---|
| Control | 1065.3 | C14Iturin A/C14Mycosubtilin [M + Na]+ | 1745 |
| 1079.3 | C15 Iturin A/C15Mycosubtilin [M + Na]+ | 237 | |
| 1081.3 | C14Iturin A/C14Mycosubtilin [M + K]+/C16Bacillomycin D [M + Na]+ | 285 | |
| Dual culture | 1044.3 | C14Surfactin [M + Na]+ | 3976 |
| 1058.3 | C15Surfactin [M + Na]+ | 893 | |
| 1043.3 | C14Iturin A/C14Mycosubtilin [M + H]+ | 9431 | |
| 1057.3 | C15 Iturin A/C15Mycosubtilin [M + H]+ | 2561 | |
| 1065.3 | C14IturinA/C14Mycosubtilin [M + Na]+ | 14,196 | |
| 1079.3 | C15 Iturin A/C15Mycosubtilin [M + Na]+ | 6615 | |
| 1081.3 | C14Iturin A/C14Mycosubtilin [M + K]+/C16Bacillomycin D [M + Na]+ | 5150 | |
| 1095.3 | C15 Iturin A/C15Mycosubtilin [M + K]+/C17Bacillomycin D [M + Na]+ | 1714 | |
| 1435.5 | Ala-6-C14 Fengycin [M + H]+ | 1020 | |
| 1449.5 | Ala-6-C15 Fengycin [M + H]+ | 5251 | |
| 1463.5 | Ala-6-C16 Fengycin [M + H]+ | 17,214 | |
| 1477.5 | Ala-6-C17 Fengycin [M + H]+ | 14,696 | |
| 1485.5 | Ala-6-C16 Fengycin [M + Na]+ | 2368 | |
| 1491.5 | Val-6-C16 Fengycin [M + H]+ | 11,158 | |
| 1501.5 | Ala-6-C16 Fengycin [M + K]+ | 1943 | |
| 1505.5 | Val -6-C17 Fengycin [M + H]+ | 4584 | |
| 1515.5 | Ala-6-C17 Fengycin [M + K]+ | 1288 | |
| 1529.5 | Val-6-C16 Fengycin [M + K]+ | 825 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, S.; Song, M.; Qi, L.; Liu, G.; Zhou, S.; Qiao, Y.; Wang, P.; Gu, H.; Wu, L. Effectiveness of a Novel Endophyte Bacillus velezensis Strain B1 in the Biocontrol of Pear Postharvest Ring Rot. Horticulturae 2023, 9, 1049. https://doi.org/10.3390/horticulturae9091049
Yang S, Song M, Qi L, Liu G, Zhou S, Qiao Y, Wang P, Gu H, Wu L. Effectiveness of a Novel Endophyte Bacillus velezensis Strain B1 in the Biocontrol of Pear Postharvest Ring Rot. Horticulturae. 2023; 9(9):1049. https://doi.org/10.3390/horticulturae9091049
Chicago/Turabian StyleYang, Suling, Meifang Song, Limei Qi, Guijun Liu, Sijing Zhou, Yuchen Qiao, Ping Wang, Haike Gu, and Liqin Wu. 2023. "Effectiveness of a Novel Endophyte Bacillus velezensis Strain B1 in the Biocontrol of Pear Postharvest Ring Rot" Horticulturae 9, no. 9: 1049. https://doi.org/10.3390/horticulturae9091049
APA StyleYang, S., Song, M., Qi, L., Liu, G., Zhou, S., Qiao, Y., Wang, P., Gu, H., & Wu, L. (2023). Effectiveness of a Novel Endophyte Bacillus velezensis Strain B1 in the Biocontrol of Pear Postharvest Ring Rot. Horticulturae, 9(9), 1049. https://doi.org/10.3390/horticulturae9091049









