Saccharomyces cerevisiae and Candida albicans Yeast Cells Labeled with Fe(III) Complexes as MRI Probes
Abstract
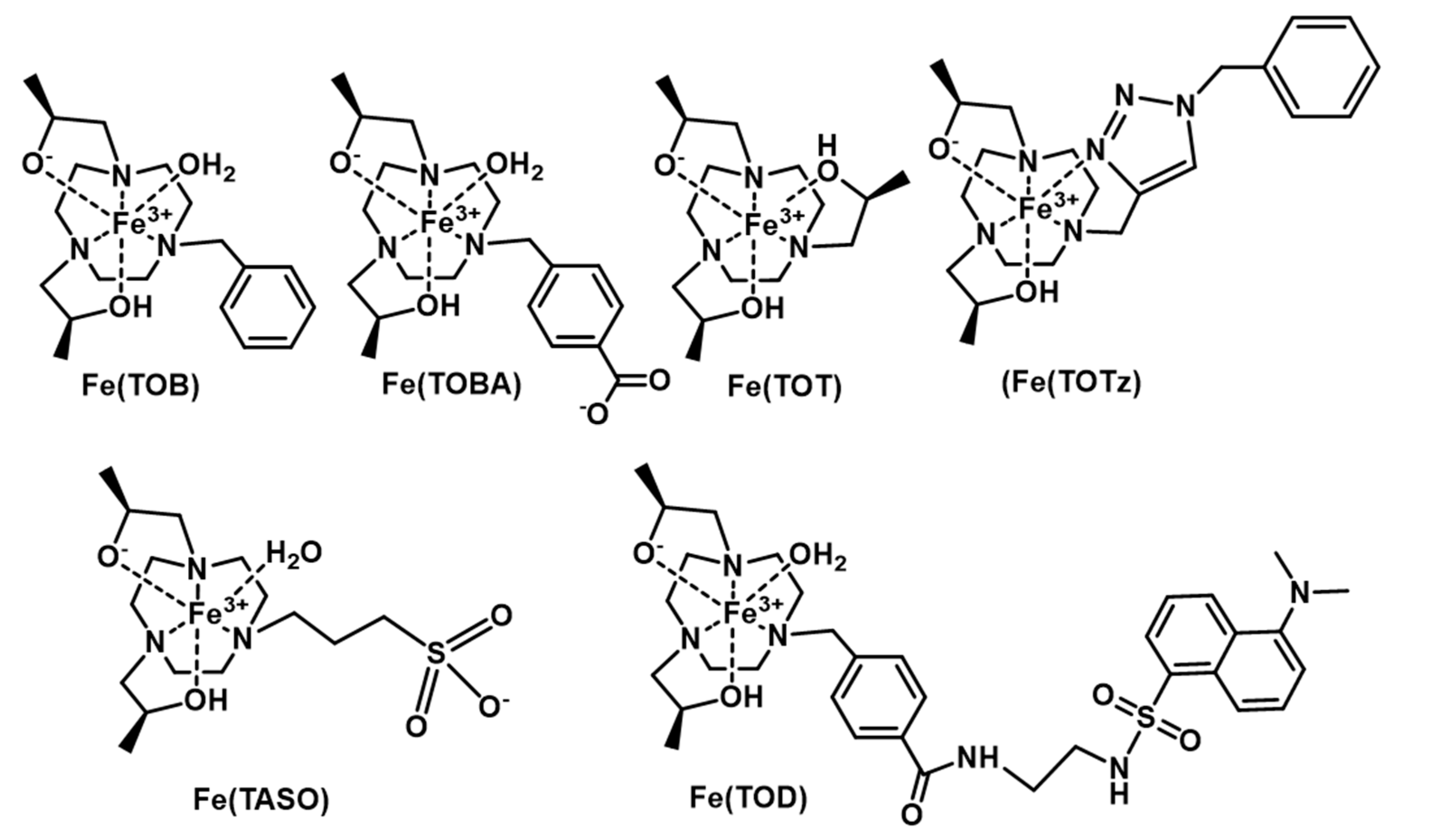
1. Introduction
2. Results
2.1. Yeast Cell Culture
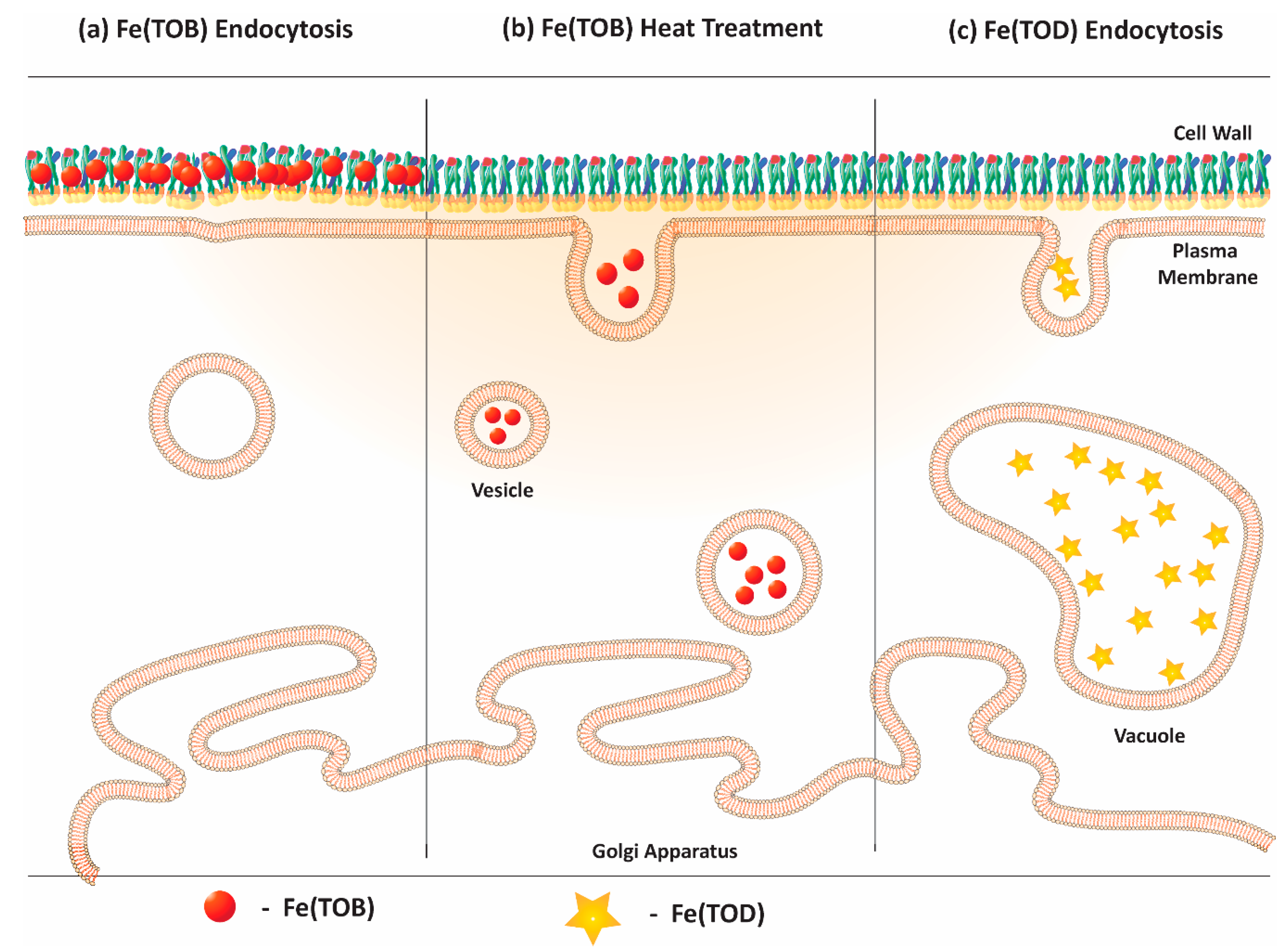
2.2. Labeling Yeast Cells
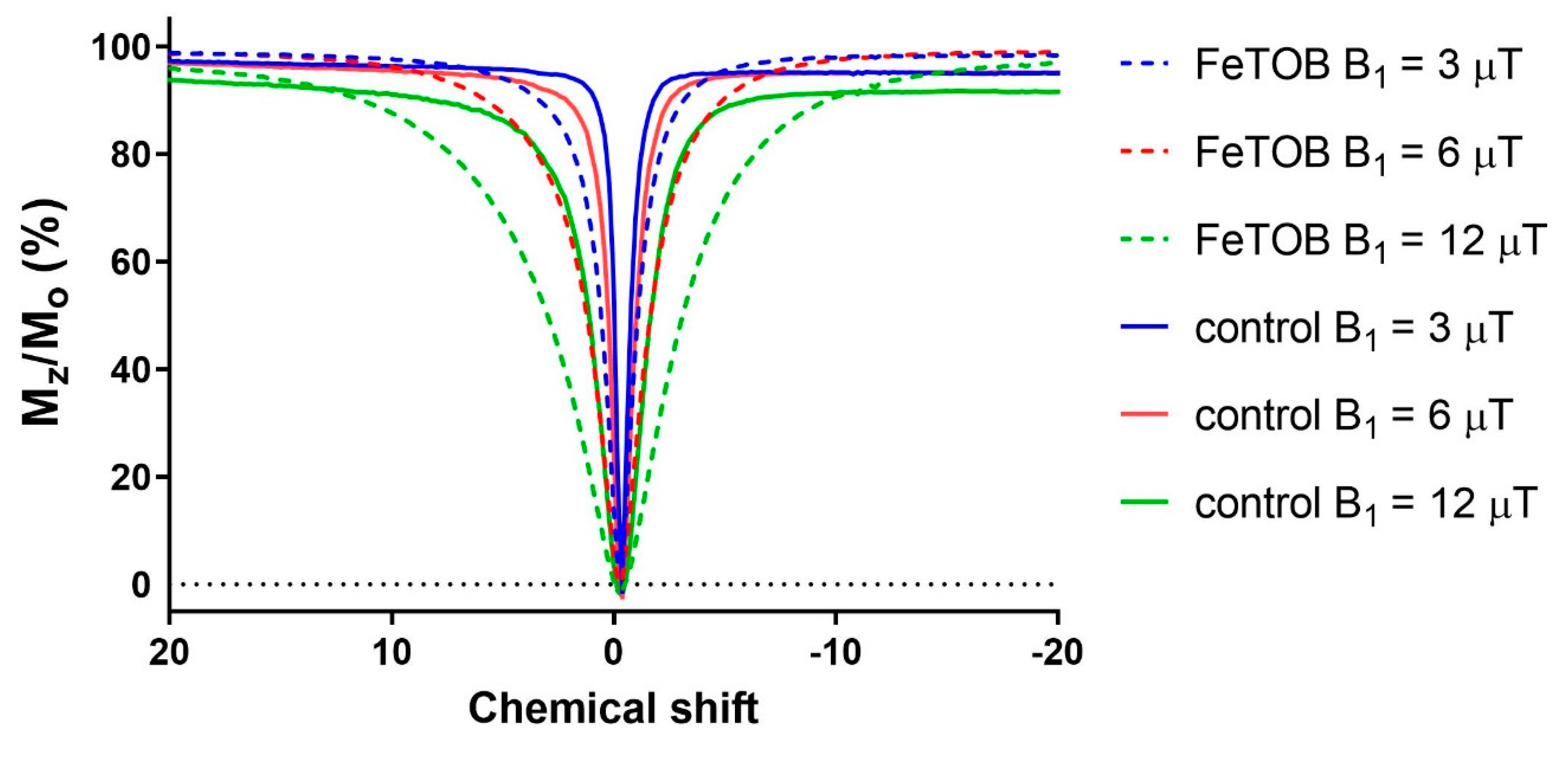
2.3. Z-Spectra Measurements
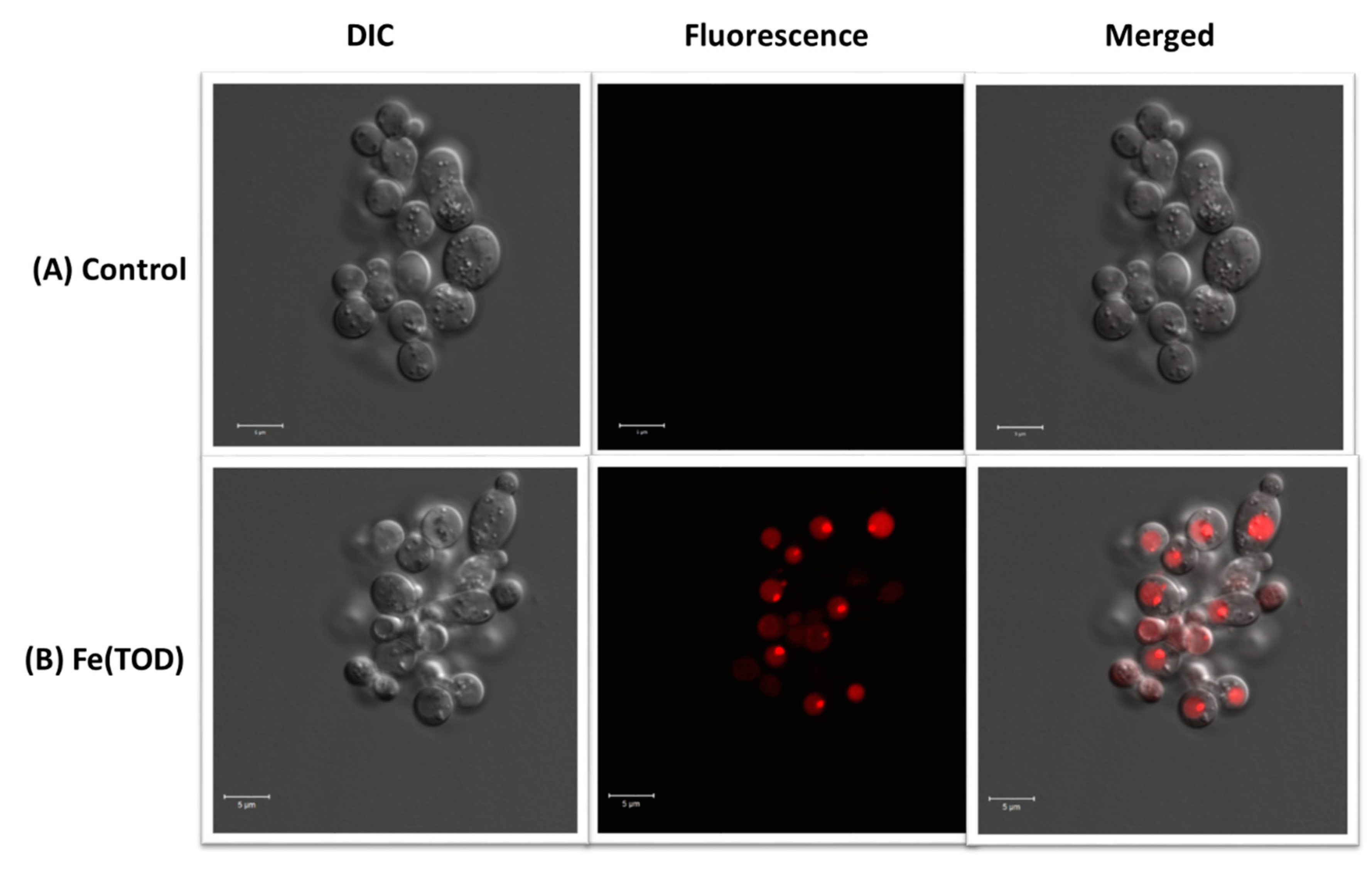
2.4. Fluorescence Microscopy on S. cerevisiae
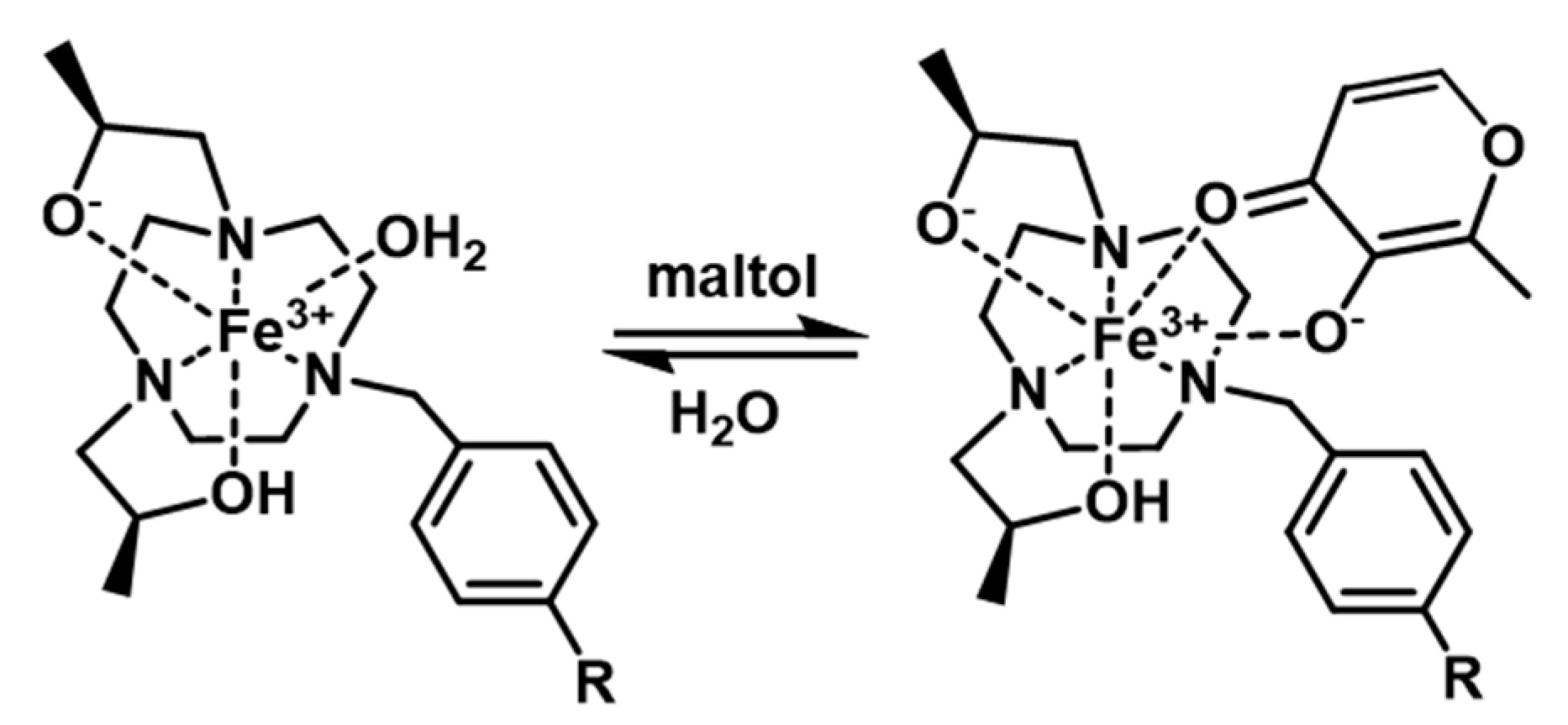
2.5. Sequestration of Fe(III) Complexes from Yeast with a Bidentate Chelator
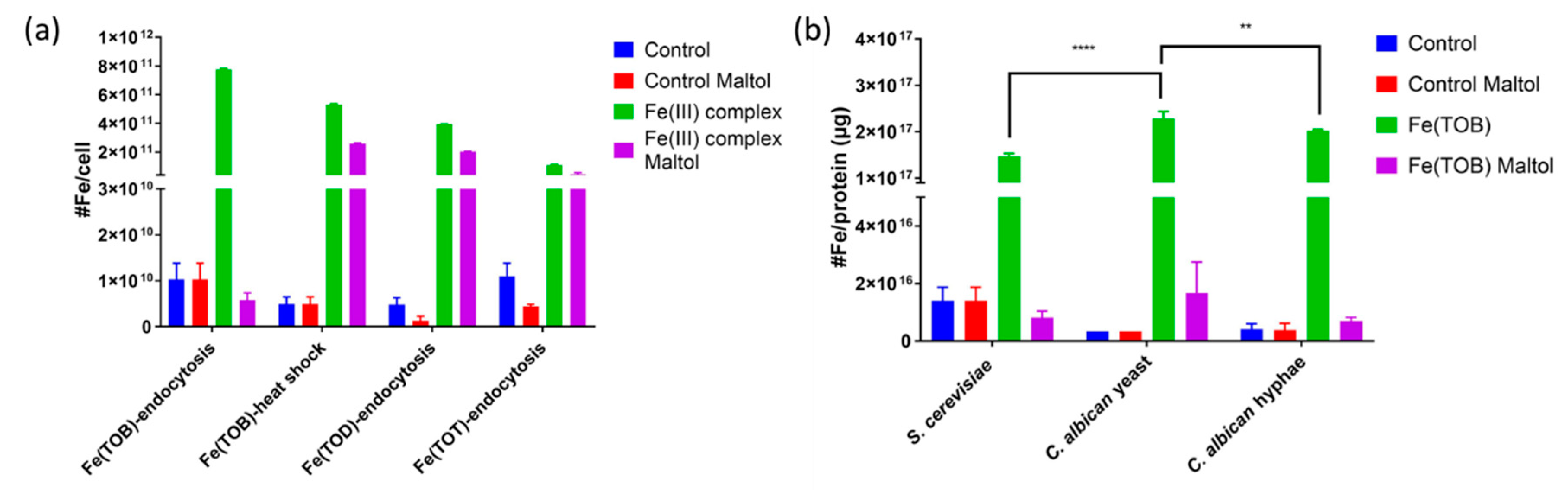
2.6. Determination of the Fe Content in the Yeast
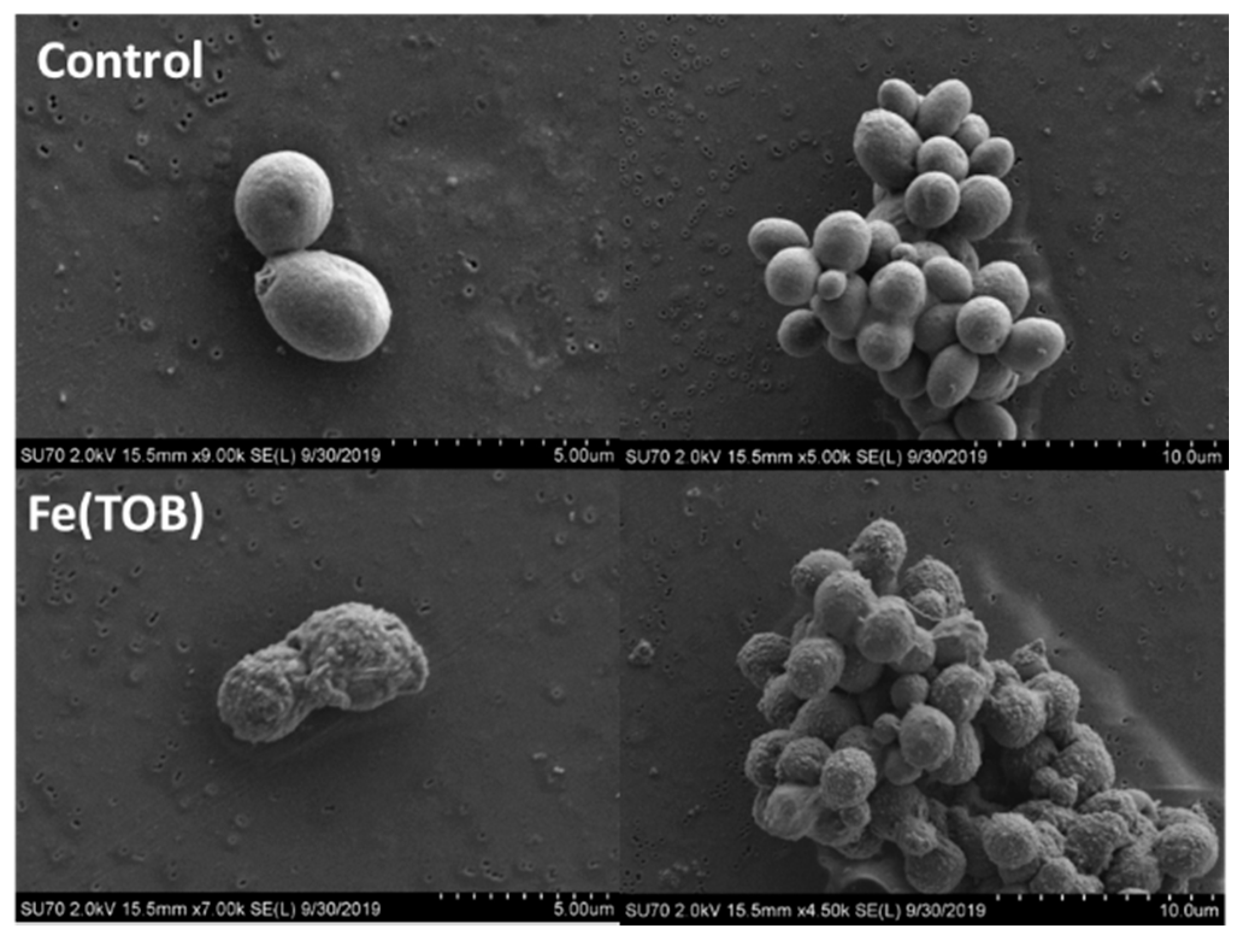
2.7. Scanning Electron Microscopy (SEM) Experiments
2.8. Optimizing the Matrix for T1 and T2 Water Proton Relaxation Measurements
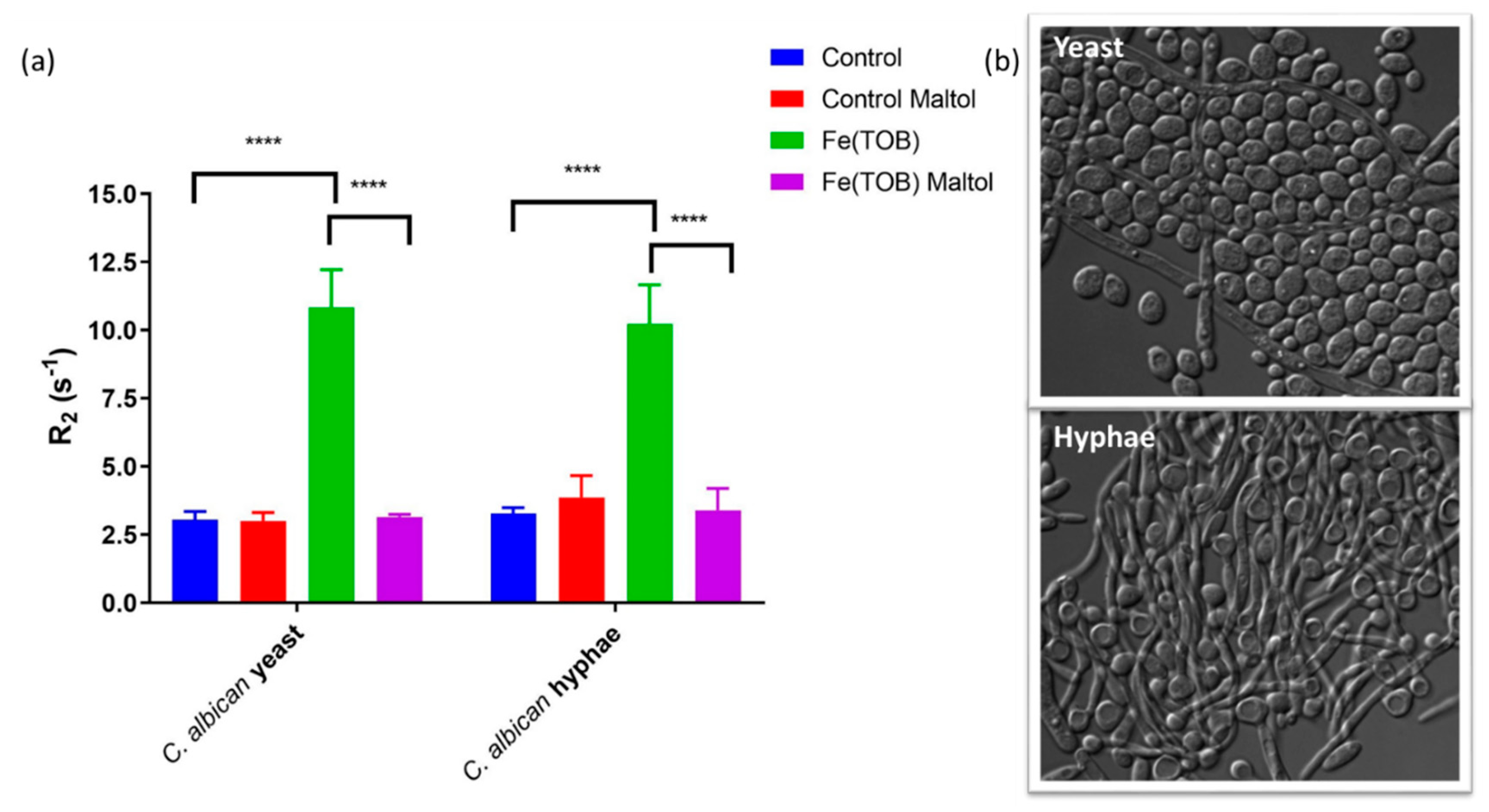
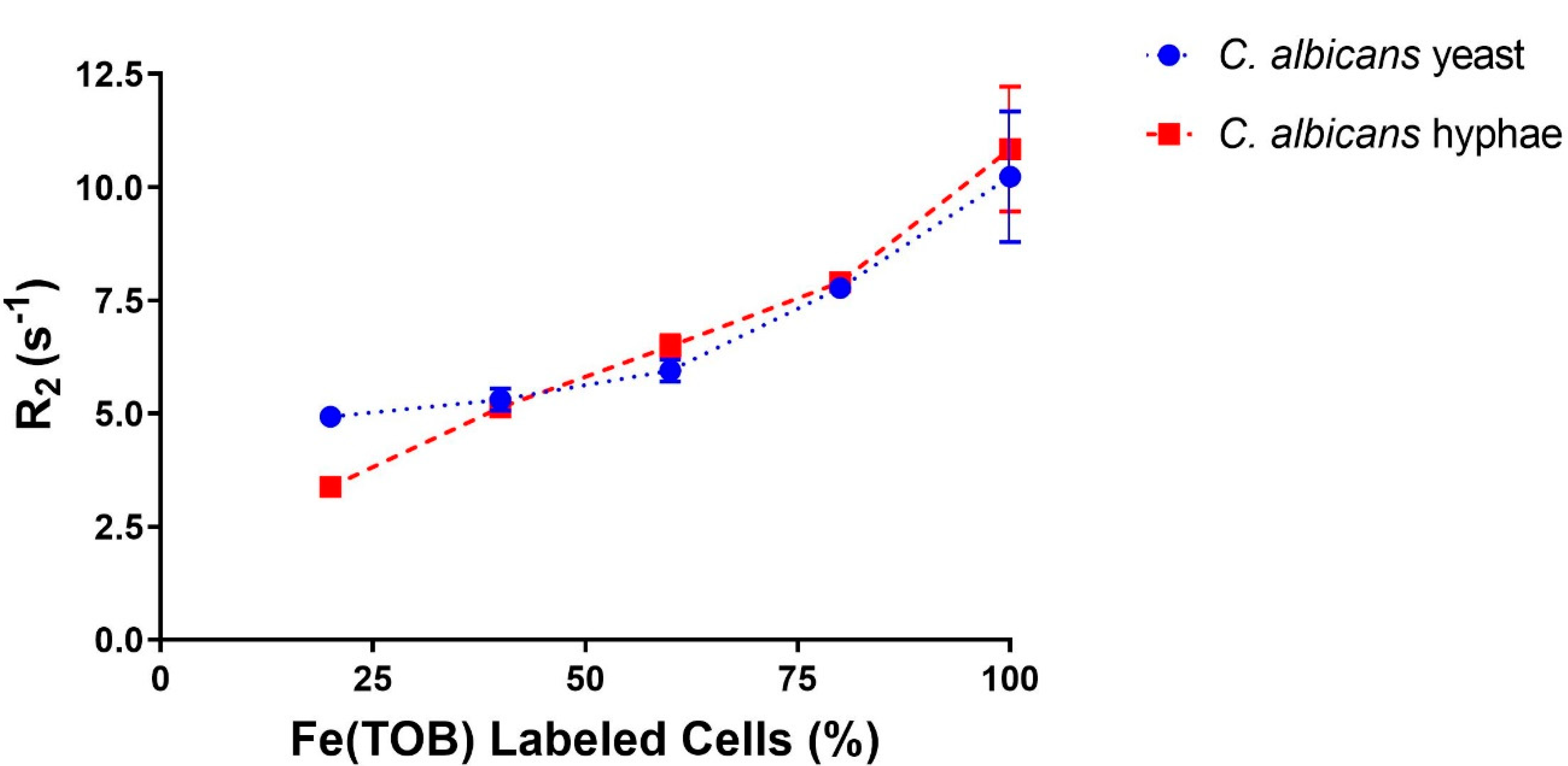
2.9. T1 and T2 Water Proton Relaxation Measurements of C. albicans Cells
2.10. Cell Viability
3. Discussion
4. Materials and Methods
4.1. Instrumentation
4.2. Materials
4.3. Cell Culture and Labeling
4.4. Fluorescence Microscopy
4.5. Pierce Protein Assay
4.6. Z-Spectra Measurements of the Fe(III)-Complex-Labeled Yeast
4.7. Maltol Removal of the Fe(TOB)-Labeled Yeast
4.8. Matrix Preparation
4.9. T1 and T2 Water Proton Relaxation Measurements of Yeast Cells and the Associated Data Analysis
4.10. Scanning Electron Microscopy on the Glucan Particles
4.11. Cell Viability Assay
4.12. Determination of Iron in the Yeast Cells
4.13. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Branski, L.K.; Gauglitz, G.G.; Herndon, D.N.; Jeschke, M.G. A review of gene and stem cell therapy in cutaneous wound healing. Burns 2009, 35, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Nicholls, F.J.; Ling, W.; Ferrauto, G.; Aime, S.; Modo, M. Simultaneous MR imaging for tissue engineering in a rat model of stroke. Sci. Rep. 2015, 5, 14597. [Google Scholar] [CrossRef] [PubMed]
- June, C.H.; O’Connor, R.S.; Kawalekar, O.U.; Ghassemi, S.; Milone, M.C. CAR T cell immunotherapy for human cancer. Science 2018, 359, 1361–1365. [Google Scholar] [CrossRef] [PubMed]
- Ferrauto, G.; Di Gregorio, E.; Dastru’, W.; Lanzardo, S.; Aime, S. Gd-loaded-RBCs for the assessment of tumor vascular volume by contrast-enhanced-MRI. Biomaterials 2015, 58, 82–92. [Google Scholar] [CrossRef] [PubMed]
- Ferrauto, G.; Castelli, D.D.; Di Gregorio, E.; Langereis, S.; Burdinski, D.; Grüll, H.; Terreno, E.; Aime, S. Lanthanide-Loaded Erythrocytes As Highly Sensitive Chemical Exchange Saturation Transfer MRI Contrast Agents. J. Am. Chem. Soc. 2013, 136, 638–641. [Google Scholar] [CrossRef]
- Castelli, D.D.; Ferrauto, G.; Di Gregorio, E.; Terreno, E.; Aime, S. Sensitive MRI detection of internalizedT1contrast agents using magnetization transfer contrast. NMR Biomed. 2015, 28, 1663–1670. [Google Scholar] [CrossRef]
- Mota, F.; Ordonez, A.A.; Firth, G.; Ruiz-Bedoya, C.A.; Ma, M.T.; Jain, S.K. Radiotracer Development for Bacterial Imaging. J. Med. Chem. 2020, 63, 1964–1977. [Google Scholar] [CrossRef]
- Locke, L.W.; Shankaran, K.; Gong, L.; Stoodley, P.; Vozar, S.L.; Cole, S.L.; Tweedle, M.F.; Wozniak, D.J. Evaluation of peptide-based probes towards in vivo diagnostic imaging of bacterial biofilm-associated infections. ACS Infect. Dis. 2020. [Google Scholar] [CrossRef]
- Kullberg, B.-J.; Arendrup, M.C. Invasive Candidiasis. N. Engl. J. Med. 2015, 373, 1445–1456. [Google Scholar] [CrossRef]
- Pfaller, M.A.; Diekema, D.J. Epidemiology of Invasive Candidiasis: A Persistent Public Health Problem. Clin. Microbiol. Rev. 2007, 20, 133–163. [Google Scholar] [CrossRef]
- Vediyappan, G.; Dumontet, V.; Pélissier, F.; D’Enfert, C. Gymnemic Acids Inhibit Hyphal Growth and Virulence in Candida albicans. PLoS ONE 2013, 8, e74189. [Google Scholar] [CrossRef] [PubMed]
- Kato, H.; Yoshimura, Y.; Suido, Y.; Shimizu, H.; Ide, K.; Sugiyama, Y.; Matsuno, K.; Nakajima, H. Mortality and risk factor analysis for Candida blood stream infection: A multicenter study. J. Infect. Chemother. 2019, 25, 341–345. [Google Scholar] [CrossRef] [PubMed]
- Arendrup, M.C.; Andersen, J.S.; Holten, M.K.; Krarup, K.B.; Reiter, N.; Schierbeck, J.; Helleberg, M. Diagnostic Performance of T2Candida Among ICU Patients With Risk Factors for Invasive Candidiasis. Open Forum Infect. Dis. 2019, 6, ofz136. [Google Scholar] [CrossRef] [PubMed]
- Pappas, P.G.; Kauffman, C.A.; Andes, D.R.; Clancy, C.J.; Marr, K.A.; Ostrosky-Zeichner, L.; Reboli, A.C.; Schuster, M.G.; Vazquez, J.A.; Walsh, T.J.; et al. Clinical Practice Guideline for the Management of Candidiasis: 2016 Update by the Infectious Diseases Society of America. Clin. Infect. Dis. 2015, 62, e1–e50. [Google Scholar] [CrossRef]
- Saraswat, D.; Kumar, R.; Pande, T.; Edgerton, M.; Cullen, P.J. Signalling mucin Msb2 Regulates adaptation to thermal stress inCandida albicans. Mol. Microbiol. 2016, 100, 425–441. [Google Scholar] [CrossRef]
- Hosseinzadeh, A.; Urban, C.F. Novel Insight into Neutrophil Immune Responses by Dry Mass Determination of Candida albicans Morphotypes. PLoS ONE 2013, 8, e77993. [Google Scholar] [CrossRef]
- Berman, J.; Sudbery, P.E. Candida albicans: A molecular revolution built on lessons from budding yeast. Nat. Rev. Genet. 2002, 3, 918–931. [Google Scholar] [CrossRef]
- Noble, S.M.; French, S.; Kohn, L.A.; Chen, V.; Johnson, A.D. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat. Genet. 2010, 42, 590–598. [Google Scholar] [CrossRef]
- Orlowski, H.; McWilliams, S.; Mellnick, V.M.; Bhalla, S.; Lubner, M.G.; Pickhardt, P.J.; Menias, C.O. Imaging Spectrum of Invasive Fungal and Fungal-like Infections. Radiographics 2017, 37, 1119–1134. [Google Scholar] [CrossRef]
- Shih, R.Y.; Koeller, K.K. Bacterial, Fungal, and Parasitic Infections of the Central Nervous System: Radiologic-Pathologic Correlation and Historical Perspectives:From the Radiologic Pathology Archives. Radiographics 2015, 35, 1141–1169. [Google Scholar] [CrossRef]
- Sánchez–Portocarrero, J.; Pérez–Cecilia, E.; Corral, O.; Romero–Vivas, J.; Picazo, J.J. The central nervous system and infection by Candida species. Diagn. Microbiol. Infect. Dis. 2000, 37, 169–179. [Google Scholar] [CrossRef]
- Zhang, P.; Lian, L.; Wang, F. Magnetic resonance imaging features of gelatinous pseudocysts in cryptococcal meningoencephalitis. Acta Neurol. Belg. 2018, 119, 265–267. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, I.D.; Lüttich, A.; Kurzai, O.; Hube, B.; Brock, M. In vivo imaging of disseminated murine Candida albicans infection reveals unexpected host sites of fungal persistence during antifungal therapy. J. Antimicrob. Chemother. 2014, 69, 2785–2796. [Google Scholar] [CrossRef] [PubMed]
- Asik, D.; Smolinski, R.; Abozeid, S.; Mitchell, T.B.; Turowski, S.G.; Spernyak, J.A.; Morrow, J.R. Modulating the Properties of Fe(III) Macrocyclic MRI Contrast Agents by Appending Sulfonate or Hydroxyl Groups. Molecules 2020, 25, 2291. [Google Scholar] [CrossRef]
- Snyder, E.M.; Asik, D.; Abozeid, S.M.; Burgio, A.; Bateman, G.; Turowski, S.G.; Spernyak, J.A.; Morrow, J.R. A Class of Fe III Macrocyclic Complexes with Alcohol Donor Groups as Effective T 1 MRI Contrast Agents. Angew. Chem. 2019, 132, 2435–2440. [Google Scholar] [CrossRef]
- Patel, A.; Asik, D.; Spernyak, J.A.; Cullen, P.J.; Morrow, J.R. MRI and fluorescence studies of Saccharomyces cerevisiae loaded with a bimodal Fe(III) T1 contrast agent. J. Inorg. Biochem. 2019, 201, 110832. [Google Scholar] [CrossRef]
- Patel, A.; Asik, D.; Snyder, E.M.; DiLillo, A.E.; Cullen, P.J.; Morrow, J.R. Binding and Release of FeIII Complexes from Glucan Particles for the Delivery of T 1 MRI Contrast Agents. ChemMedChem 2020, 15, 1050–1057. [Google Scholar] [CrossRef]
- Gimeno, C.J.; Ljungdahl, P.O.; Styles, C.A.; Fink, G.R. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: Regulation by starvation and RAS. Cell 1992, 68, 1077–1090. [Google Scholar] [CrossRef]
- Cullen, P.J.; Sabbagh, J.W.; Graham, E.; Irick, M.M.; Van Olden, E.K.; Neal, C.; Delrow, J.; Bardwell, L.; Sprague, J.G.F. A signaling mucin at the head of the Cdc42- and MAPK-dependent filamentous growth pathway in yeast. Genes Dev. 2004, 18, 1695–1708. [Google Scholar] [CrossRef]
- Patel, A.; Mohammed, A.S.; Cullen, P.J.; Morrow, J.R. Co(II) macrocyclic complexes with fluorescent tags as paraCEST and cellCEST agents. 2020. Unpublished work. [Google Scholar]
- Kawai, S.; Hashimoto, W.; Murata, K. Transformation ofSaccharomyces cerevisiaeand other fungi. Bioeng. Bugs 2010, 1, 395–403. [Google Scholar] [CrossRef] [PubMed]
- Samira, M.; Abozeid, D.A.; Morrow, R.J. Liposomal Fe(III) complexes as T1, T2 and lipoCEST agents. 2020. Unpublished work. [Google Scholar]
- Mulas, G.; Ferrauto, G.; Dastru’, W.; Anedda, R.; Aime, S.; Terreno, E. Insights on the relaxation of liposomes encapsulating paramagnetic Ln-based complexes. Magn. Reson. Med. 2014, 74, 468–473. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.; Krohn, R.; Hermanson, G.; Mallia, A.; Gartner, F.; Provenzano, M.; Fujimoto, E.; Goeke, N.; Olson, B.; Klenk, D. Measurement of protein using bicinchoninic acid. Anal. Biochem. 1985, 150, 76–85. [Google Scholar] [CrossRef]
- Nguyen, T.H.; Fleet, G.H.; Rogers, P.L. Composition of the cell walls of several yeast species. Appl. Microbiol. Biotechnol. 1998, 50, 206–212. [Google Scholar] [CrossRef]
- Mitchell, M.D.; Kundel, H.L.; Axel, L.; Joseph, P.M. Agarose as a tissue equivalent phantom material for NMR imaging. Magn. Reson. Imaging 1986, 4, 263–266. [Google Scholar] [CrossRef]
- Hellerbach, A.; Schuster, V.; Jansen, A.; Sommer, J. MRI Phantoms—Are There Alternatives to Agar? PLoS ONE 2013, 8, e70343. [Google Scholar] [CrossRef]
- Young, S.; Wong, M.; Tabata, Y.; Mikos, A.G. Gelatin as a delivery vehicle for the controlled release of bioactive molecules. J. Control. Release 2005, 109, 256–274. [Google Scholar] [CrossRef]
- Komaiko, J.; McClements, D.J. Food-grade nanoemulsion filled hydrogels formed by spontaneous emulsification and gelation: Optical properties, rheology, and stability. Food Hydrocoll. 2015, 46, 67–75. [Google Scholar] [CrossRef]
- Dowling, M.B.; Lee, J.-H.; Raghavan, S.R. pH-Responsive Jello: Gelatin Gels Containing Fatty Acid Vesicles†. Langmuir 2009, 25, 8519–8525. [Google Scholar] [CrossRef]
- Ferrauto, G.; Di Gregorio, E.; Castelli, D.D.; Aime, S. CEST-MRI studies of cells loaded with lanthanide shift reagents. Magn. Reson. Med. 2018, 80, 1626–1637. [Google Scholar] [CrossRef] [PubMed]
- Dorazio, S.J.; Olatunde, A.O.; Spernyak, J.A.; Morrow, J.R. CoCEST: Cobalt(II) amide-appended paraCEST MRI contrast agents. Chem. Commun. 2013, 49, 10025–10027. [Google Scholar] [CrossRef] [PubMed]
- Maaloum, M.; Pernodet, N.; Tinland, B. Agarose gel structure using atomic force microscopy: Gel concentration and ionic strength effects. Electrophoresis 1998, 19, 1606–1610. [Google Scholar] [CrossRef] [PubMed]
- Stellwagen, J.; Stellwagen, N.C. Internal Structure of the Agarose Gel Matrix. J. Phys. Chem. 1995, 99, 4247–4251. [Google Scholar] [CrossRef]
- Griess, G.; Guiseley, K.; Serwer, P. The relationship of agarose gel structure to the sieving of spheres during agarose gel electrophoresis. Biophys. J. 1993, 65, 138–148. [Google Scholar] [CrossRef]
- Hutchens, M.; Luker, G.D. Applications of bioluminescence imaging to the study of infectious diseases. Cell. Microbiol. 2007, 9, 2315–2322. [Google Scholar] [CrossRef]
- Andreu, N.; Zelmer, A.; Wiles, S. Noninvasive biophotonic imaging for studies of infectious disease. FEMS Microbiol. Rev. 2011, 35, 360–394. [Google Scholar] [CrossRef]
- Morad, H.O.J.; Wild, A.-M.; Wiehr, S.; Davies, G.; Maurer, A.; Pichler, B.J.; Thornton, C.R. Pre-clinical Imaging of Invasive Candidiasis Using ImmunoPET/MR. Front. Microbiol. 2018, 9, 1996. [Google Scholar] [CrossRef]
- Bleeker-Rovers, C.; Warris, A.; Drenth, J.; Corstens, F.; Oyen, W.J.; Kullberg, B.-J.; Bleeker-Rovers, C.P. Diagnosis of Candida lung abscesses by 18F-fluorodeoxyglucose positron emission tomography. Clin. Microbiol. Infect. 2005, 11, 493–495. [Google Scholar] [CrossRef]
- Lesage, G.; Bussey, H. Cell Wall Assembly in Saccharomyces cerevisiae. Microbiol. Mol. Boil. Rev. 2006, 70, 317–343. [Google Scholar] [CrossRef]
- Dörr, T.; Moynihan, P.J.; Mayer, C. Editorial: Bacterial Cell Wall Structure and Dynamics. Front. Microbiol. 2019, 10, 2051. [Google Scholar] [CrossRef] [PubMed]
- Romaniuk, J.A.H.; Cegelski, L. Bacterial cell wall composition and the influence of antibiotics by cell-wall and whole-cell NMR. Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 20150024. [Google Scholar] [CrossRef] [PubMed]
- Tee, Y.K.; Donahue, M.J.; Harston, G.W.J.; Payne, S.J.; Chappell, M. Quantification of amide proton transfer effect pre- and post-gadolinium contrast agent administration. J. Magn. Reson. Imaging 2013, 40, 832–838. [Google Scholar] [CrossRef] [PubMed]
- Kurki, T.; Niemi, P.T.; Lundbom, N. Gadolinium-enhanced magnetization transfer contrast imaging of intracranial tumors. J. Magn. Reson. Imaging 1992, 2, 401–406. [Google Scholar] [CrossRef] [PubMed]
- Mukaremera, L.; Lee, K.K.; Mora-Montes, H.M.; Gow, N.A.R. Candida albicans Yeast, Pseudohyphal, and Hyphal Morphogenesis Differentially Affects Immune Recognition. Front. Immunol. 2017, 8, 629. [Google Scholar] [CrossRef]
- Creger, P.E.; Blankenship, J.R. Analysis of gene expression in filamentous cells of Candida albicans grown on agar plates. J. Boil. Methods 2018, 5, e84. [Google Scholar] [CrossRef]
- Terreno, E.; Crich, S.G.; Belfiore, S.; Biancone, L.; Cabella, C.; Esposito, G.; Manazza, A.D.; Aime, S. Effect of the intracellular localization of a Gd-based imaging probe on the relaxation enhancement of water protons. Magn. Reson. Med. 2006, 55, 491–497. [Google Scholar] [CrossRef]
- Li, F.; Svarovsky, M.J.; Karlsson, A.J.; Wagner, J.P.; Marchillo, K.; Oshel, P.; Andes, D.; Palecek, S.P. Eap1p, an Adhesin That Mediates Candida albicans Biofilm Formation In Vitro and In Vivo. Eukaryot. Cell 2007, 6, 931–939. [Google Scholar] [CrossRef]
- Liu, Y.; Filler, S.G. Candida albicans Als3, a Multifunctional Adhesin and Invasin. Eukaryot. Cell 2010, 10, 168–173. [Google Scholar] [CrossRef]
- McCall, A.D.; Pathirana, R.U.; Prabhakar, A.; Cullen, P.J.; Edgerton, M. Candida albicans biofilm development is governed by cooperative attachment and adhesion maintenance proteins. NPJ Biofilms Microbiomes 2019, 5, 1–12. [Google Scholar] [CrossRef]
- Rupert, C.B.; Heltzel, J.M.H.; Taylor, D.J.; Rusché, L. Sporadic Gene Loss After Duplication Is Associated with Functional Divergence of Sirtuin Deacetylases AmongCandidaYeast Species. G3 Genes Genomes Genet. 2016, 6, 3297–3305. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gillum, A.M.; Tsay, E.Y.H.; Kirsch, D.R. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Genet. Genom. 1984, 198, 179–182. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Kohler, J.; Fink, G. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 1994, 266, 1723–1726. [Google Scholar] [CrossRef] [PubMed]
- Rines, D.R.; Thomann, D.; Dorn, J.F.; Goodwin, P.; Sorger, P.K. Live Cell Imaging of Yeast. Cold Spring Harb. Protoc. 2011, 2011, 1026–1041. [Google Scholar] [CrossRef]
- Farhat, Y. Protocol for Cell-Seeded Collagen Gels. Available online: http://protocol-place.com/cell-culture/cell-seeded-collagen-gel-protocol/ (accessed on 2 October 2019).
- McIntosh, L.P. CPMG. In Encyclopedia of Biophysics; Springer Science and Business Media LLC: Berlin, Germany, 2013; p. 386. [Google Scholar]
- Chow, J.; Notaro, M.; Prabhakar, A.; Free, S.J.; Cullen, P.J. Impact of Fungal MAPK Pathway Targets on the Cell Wall. J. Fungi 2018, 4, 93. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Patel, A.; Asik, D.; Snyder, E.M.; Spernyak, J.A.; Cullen, P.J.; Morrow, J.R. Saccharomyces cerevisiae and Candida albicans Yeast Cells Labeled with Fe(III) Complexes as MRI Probes. Magnetochemistry 2020, 6, 41. https://doi.org/10.3390/magnetochemistry6030041
Patel A, Asik D, Snyder EM, Spernyak JA, Cullen PJ, Morrow JR. Saccharomyces cerevisiae and Candida albicans Yeast Cells Labeled with Fe(III) Complexes as MRI Probes. Magnetochemistry. 2020; 6(3):41. https://doi.org/10.3390/magnetochemistry6030041
Chicago/Turabian StylePatel, Akanksha, Didar Asik, Eric M. Snyder, Joseph A. Spernyak, Paul J. Cullen, and Janet R. Morrow. 2020. "Saccharomyces cerevisiae and Candida albicans Yeast Cells Labeled with Fe(III) Complexes as MRI Probes" Magnetochemistry 6, no. 3: 41. https://doi.org/10.3390/magnetochemistry6030041
APA StylePatel, A., Asik, D., Snyder, E. M., Spernyak, J. A., Cullen, P. J., & Morrow, J. R. (2020). Saccharomyces cerevisiae and Candida albicans Yeast Cells Labeled with Fe(III) Complexes as MRI Probes. Magnetochemistry, 6(3), 41. https://doi.org/10.3390/magnetochemistry6030041







