Graphene-Coated Iron Nitride Streptavidin Magnetic Beads: Preparation and Application in SARS-CoV-2 Enrichment
Abstract
:1. Introduction
2. Materials and Methods
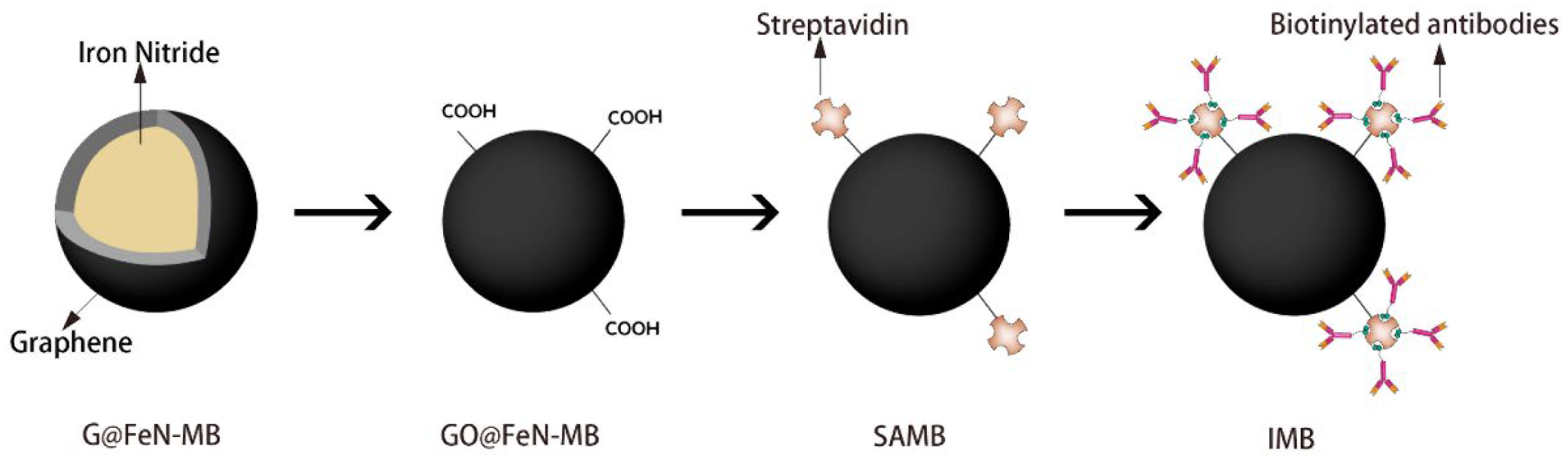
2.1. Surface Oxidation of G@FeN-MB
2.2. Preparation of IMB
2.3. Preparation of SARS-CoV-2 S-PS
2.4. IMB Capture SARS-CoV-2 S-PS Experiment
2.5. IMB Capture SARS-CoV-2 Pseudovirus Experiment
3. Results and Discussion
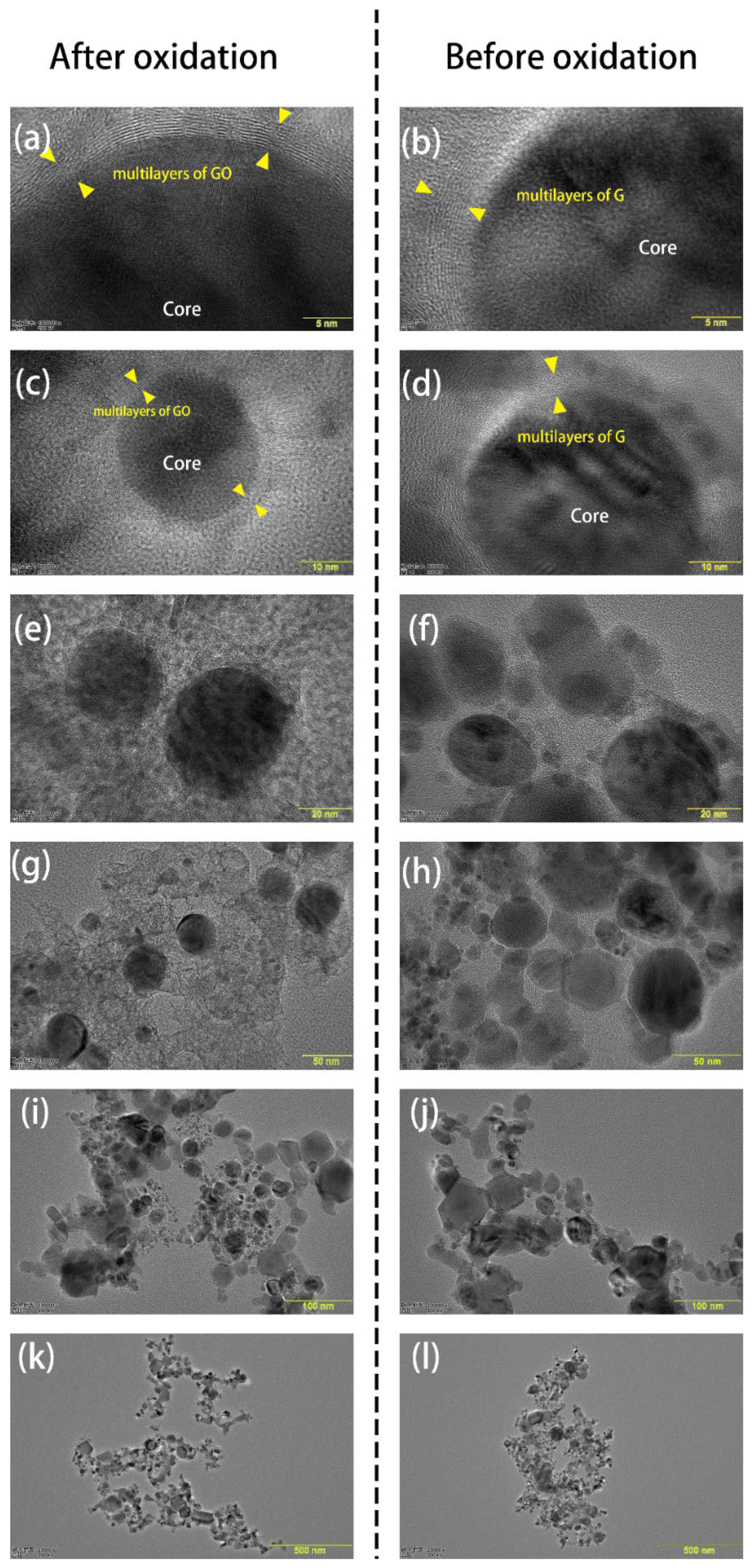
3.1. Characterization of GO@FeN-MB
3.2. Preparation of SAMBs
3.3. IMB Capture SARS-CoV-2 S-PS
3.4. IMB Captures SARS-CoV-2 Pseudoviruses
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Hui, D.S.; Azhar, E.I.; Madani, T.A.; Ntoumi, F.; Kock, R.; Dar, O.; Ippolito, G.; Mchugh, T.D.; Memish, Z.A.; Drosten, C.; et al. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health—The latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect. Dis. 2020, 91, 264–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Rathi, B. Coronavirus disease COVID-19: A new threat to public health. Curr. Top. Med. Chem. 2020, 20, 599–600. [Google Scholar] [CrossRef] [PubMed]
- National Health Commission. COVID-19 diagnosis and treatment plan (trial version 7). Infect. Dis. Inf. 2020, 33, 26. [Google Scholar]
- Wang, D.; Hu, B.; Hu, C.; Zhu, F.; Liu, X.; Zhang, J.; Wang, B.; Xiang, H.; Cheng, Z.; Xiong, Y.; et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus⁃infected pneumonia in Wuhan, China. JAMA 2020, 323, 1061–1069. [Google Scholar] [CrossRef]
- Tali, S.H.S.; LeBlanc, J.J.; Sadiq, Z.; Oyewunmi, O.D.; Camargo, C.; Nikpour, B.; Armanfard, N.; Sagan, S.M.; Jahanshahi-Anbuhi, S. Tools and Techniques for Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2)/COVID-19 Detection. Clin. Microbiol. Rev. 2021, 34, e00228-20. [Google Scholar] [CrossRef]
- Eileen, M. Validation of laboratory-developed molecular assays for infectious diseases. Clin. Microbiol. Rev. 2010, 23, 550–576. [Google Scholar]
- Weiyin, G.; Hong, Z.; Yang, L. False negative analysis and countermeasures in nucleic acid detection of novel coronavirus pneumonia. Int. J. Lab. Med. 2020, 41, 641–643. [Google Scholar]
- Naoya, K.; Naotaka, K. Analytical techniques for the determination of biologically active quinones in biological and environmental samples. J. Pharm. Biomed. Anal. 2014, 87, 261–270. [Google Scholar]
- Saito, Y.; Ueta, I.; Ogawa, M.; Hayashida, M.; Jinno, K. Miniaturized sample preparation needle: A versatile design for the rapid analysis of smoking-related compounds in hair and air samples. J. Pharm. Biomed. Anal. 2007, 44, 1–7. [Google Scholar] [CrossRef]
- Huang, Z.; Cui, X.; Xie, Q.Y.; Liu, D.F.; Lai, W.H. Short communication: A novel method using immunomagnetic separation with a fluorescent nanobeads lateral flow assay for the rapid detection of low-concentration Escherichia coliO157:H7 in raw milk. J. Dairy Sci. 2016, 99, 9581–9585. [Google Scholar] [CrossRef]
- Tekin, C.; Gijs, M. Ultrasensitive protein detection: A case for microfluidic magnetic bead-based assays. Lab. Chip 2013, 13, 4711–4759. [Google Scholar] [CrossRef] [PubMed]
- Hickstein, B.; Peuker, U.A. Characterization of protein capacity of nanocation exchanger particles as filling material for functional magnetic beads for bio-separation purposes. Biotechnol. Prog. 2008, 24, 409–416. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Peng, J.; Han, J.; Zhang, G.; Huang, Y.; Duan, M.; Liu, D.; Xiong, Y.; Xia, S.; Lai, W. A novel method based on fluorescent magnetic nanobeads for rapid detection of Escherichia coli O157:H7. Food Chem. 2019, 276, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Hu, S.; Xiong, Y.; Wei, H.; Xu, H.; Duan, H.; Lai, W. Application and development of superparamagnetic nanoparticles in sample pretreatment and immunochromatographic assay. TrAC Trends Anal. Chem. 2019, 114, 151–170. [Google Scholar] [CrossRef]
- Seo, S.M.; Cho, I.H.; Jeon, J.W.; Cho, H.K.; Oh, E.G.; Yu, H.S.; Shin, S.B.; Lee, H.J.; Paek, S.H. An ELISA-on-a-chip biosensor system coupled with immunomagnetic separation for the detection of vibrio parahaemolyticus within a single working day. J. Food Prot. 2010, 73, 1466–1473. [Google Scholar] [CrossRef]
- Sturbaum, G.D.; Klonicki, P.T.; Marshall, M.M.; Jost, B.H.; Clay, B.L.; Sterling, C.R. Immunomagnetic separation (IMS)-fluorescent antibody detection and IMS-PCR detection of seeded Cryptosporidium parvum oocysts in natural waters and their limitations. Appl. Environ. Microb. 2002, 68, 2991–2996. [Google Scholar] [CrossRef] [Green Version]
- Reidt, U.; Geisberger, B.; Heller, C.; Friedberger, A. Automated immunomagnetic processing and separation of Legionella pneumophila with manual detection by sandwich ELISA and PCR amplification of the ompS gene. J. Lab. Autom. 2011, 16, 157–164. [Google Scholar] [CrossRef] [Green Version]
- Yang, W.; Gu, A.Z.; Zeng, S.Y.; Li, D.; He, M.; Shi, H.C. Development of a combined immunomagnetic separation and quantitative reverse transcription-PCR assay for sensitive detection of infectious rotavirus in water samples. J. Microbiol. Methods 2011, 84, 447–453. [Google Scholar] [CrossRef]
- Fu, Z.; Rogelj, S.; Kieft, T.L. Rapid detection of Escherichia coli O157:H7 by immunomagnetic separation and real-time PCR. Int. J. Food Microbiol. 2005, 99, 47–57. [Google Scholar] [CrossRef]
- He, J.; Huang, M.; Wang, D.; Zhang, Z.; Li, G. Magnetic separation techniques in sample preparation for biological analysis: A review. J. Pharm. Biomed. Anal. 2014, 101, 84–101. [Google Scholar] [CrossRef]
- Johnson, D.; Carter, M.D.; Crow, B.S.; Isenberg, S.L.; Graham, L.A.; Erol, H.A.; Watson, C.M.; Pantazides, B.G.; van der Schans, M.J.; Langenberg, J.P.; et al. Quantitation of ortho-cresyl phosphate adducts to butyrylcholinesterase in human serum by immunomagnetic-UHPLC-MS/MS. J. Mass Spectrom. 2015, 50, 683–692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, B.; Jin, X.; Wang, Q.; Li, Y.; Khan, W.Q. Synthesis of carbon coated iron nitride nanoparticles by using microwave plasma technique. Mater. Res. 2020, 7, 096103. [Google Scholar] [CrossRef]
- Ghosal, K.; Sarkar, K. Biomedical Applications of Graphene Nanomaterials and Beyond. ACS Biomater. Sci. Eng. 2018, 4, 2653–2703. [Google Scholar] [CrossRef] [PubMed]
- Meyer, J.C.; Geim, A.K.; Katsnelson, M.I.; Novoselov, K.S.; Booth, T.J.; Roth, S. The structure of suspended graphene sheets. Nature 2007, 446, 60–63. [Google Scholar] [CrossRef] [PubMed]
- Mu, E.F.; Gopinath, S.C.B. Feasibility of graphene in biomedical applications. Biomed. Pharmacother. 2017, 94, 354–361. [Google Scholar]
- Liu, K.; Zhang, J.J.; Cheng, F.F.; Zheng, T.T.; Wang, C.; Zhu, J.J. Green and facile synthesis of highly biocompatible graphene nanosheets and its application for cellular imaging and drug delivery. Mater. Chem. 2011, 21, 12034–12040. [Google Scholar] [CrossRef]
- Wang, J. Electrochemical glucose biosensors. Chem. Rev. 2008, 108, 814–825. [Google Scholar] [CrossRef]
- Sarkar, K.; Madras, G.; Chatterjee, K. Dendron conjugation to graphene oxide using click chemistry for efficient gene delivery. RSC Adv. 2015, 62, 50196–50211. [Google Scholar] [CrossRef] [Green Version]
- Pham, X.H.; Baek, A.; Kim, T.H.; Lee, S.H.; Rho, W.Y.; Chung, W.J.; Kim, D.E.; Jun, B.H. Graphene oxide conjugated magnetic beads for RNA extraction. Chem. Asian J. 2017, 12, 1883–1888. [Google Scholar] [CrossRef]
- Liang, T.; Li, J.; Liu, X.; Ma, Z.; Su, X.; Meng, X.; Zhanghuang, Z.; Wang, H.; Li, J.; Wang, Q.; et al. Preparation of CD3 Antibody-Conjugated, Graphene Oxide Coated Iron Nitride Magnetic Beads and Its Preliminary Application in T Cell Separation. Magnetochemistry 2021, 7, 58. [Google Scholar] [CrossRef]
- Bamrungsap, S.; Shukoor, M.I.; Chen, T.; Sefah, K.; Tan, W. Detection of lysozyme magnetic relaxation switches based on Aptamer-Functionalized superparamagnetic nanoparticles. Anal. Chem. 2011, 83, 7795–7799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, M.; Gupta, P.; Baronia, R.; Singh, P.; Karuppiah, S.; Shankar, R.; Dwivedi, P.D.; Singh, S.P. In vitro cytotoxicity of GO-DOx on FaDu squamous carcinoma cell lines. Int. J. Nanomed. 2018, 13, 107–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, R.; Guiney, L.M.; Chang, C.H.; Mansukhani, N.D.; Ji, Z.; Wang, X.; Liao, Y.P.; Jiang, W.; Sun, B.; Hersam, M.C.; et al. Surface Oxidation of Graphene Oxide Determines Membrane Damage, Lipid Peroxidation, and Cytotoxicity in Macrophages in a Pulmonary Toxicity Model. ACS Nano 2018, 12, 1390–1402. [Google Scholar] [CrossRef] [PubMed]
- Kudin, K.N.; Ozbas, B.; Schniepp, H.C.; Prud’Homme, R.K.; Aksay, I.A.; Car, R. Raman spectra of graphite oxide and functionalized graphene sheets. Nano Lett. 2008, 8, 36–41. [Google Scholar] [CrossRef]
- Nyakiti, L.O.; Myers-Ward, R.L.; Wheeler, V.D.; Imhoff, E.A.; Bezares, F.J.; Chun, H.; Caldwell, J.D.; Friedman, A.L.; Matis, B.R.; Baldwin, J.W.; et al. Bilayer graphene grown on 4H-SiC (0001) step-free mesas. Nano Lett. 2012, 12, 1749–1756. [Google Scholar] [CrossRef] [PubMed]
- Cançado, L.G.; Takai, K.; Enoki, T.; Endo, M.; Kim, Y.A.; Mizusaki, H.; Pimenta, M.A. General equation for the determination of the crystallite size La of nanographite by Raman spectroscopy. Appl. Phys. Lett. 2006, 88, 163106-1–163106-3. [Google Scholar] [CrossRef]
- Mohiuddin, T.M.G.; Lombardo, A.; Nair, R.R.; Bonetti, A.; Savini, G.; Jalil, R.; Ferrari, A.C. Uniaxial strain in graphene by Raman spectroscopy: G peak splitting, Grüneisen parameters, and sample orientation. Phys. Rev. B 2009, 79, 205433. [Google Scholar] [CrossRef]
- Ferrari, A.C.; Meyer, J.C.; Scardaci, V. Raman spectrum of graphene and graphene layers. Phys. Rev. Lett. 2006, 97, 187401. [Google Scholar] [CrossRef] [Green Version]
- Zunlon, K.; Joaquin, O.; Kun, Q. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature 2020, 588, 498–502. [Google Scholar]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Wang, M.; Jia, R.; Ma, Z.; Zhang, X.; Li, J.; Xiao, X.; Zhou, Y.; Wang, Q. Graphene-Coated Iron Nitride Streptavidin Magnetic Beads: Preparation and Application in SARS-CoV-2 Enrichment. Magnetochemistry 2022, 8, 41. https://doi.org/10.3390/magnetochemistry8040041
Li J, Wang M, Jia R, Ma Z, Zhang X, Li J, Xiao X, Zhou Y, Wang Q. Graphene-Coated Iron Nitride Streptavidin Magnetic Beads: Preparation and Application in SARS-CoV-2 Enrichment. Magnetochemistry. 2022; 8(4):41. https://doi.org/10.3390/magnetochemistry8040041
Chicago/Turabian StyleLi, Jianxing, Minglian Wang, Runqing Jia, Zhuang Ma, Xiaoxu Zhang, Jintao Li, Xiangqian Xiao, Yunzhi Zhou, and Qun Wang. 2022. "Graphene-Coated Iron Nitride Streptavidin Magnetic Beads: Preparation and Application in SARS-CoV-2 Enrichment" Magnetochemistry 8, no. 4: 41. https://doi.org/10.3390/magnetochemistry8040041
APA StyleLi, J., Wang, M., Jia, R., Ma, Z., Zhang, X., Li, J., Xiao, X., Zhou, Y., & Wang, Q. (2022). Graphene-Coated Iron Nitride Streptavidin Magnetic Beads: Preparation and Application in SARS-CoV-2 Enrichment. Magnetochemistry, 8(4), 41. https://doi.org/10.3390/magnetochemistry8040041






