Safety Assessment of Recycled Plastics from Post-Consumer Waste with a Combination of a Miniaturized Ames Test and Chromatographic Analysis
Abstract
:1. Introduction
2. Results
2.1. Results from the Miniaturized Ames Test Analysis
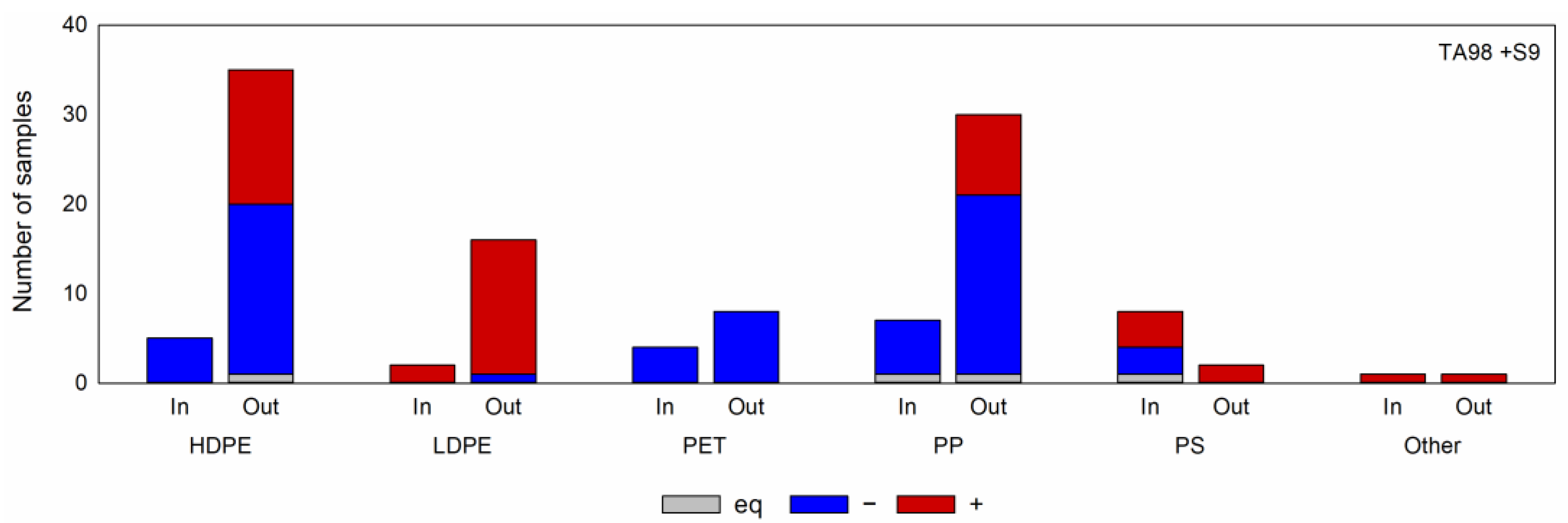
2.1.1. Overview of the Screening Results
2.1.2. Comparison of Different Polymer Types
2.1.3. Comparison of Different Processing Stages
2.1.4. Comparison of Different Polymer Batches
2.2. Correlation of Miniaturized Ames Test Results with Analytical Data
3. Discussion
3.1. Interpretation of the Miniaturized Ames Test Results
3.2. Identification of the Sources for DNA-Reactive, Mutagenic Activity
3.3. Potential Impact on Plastic Recycling for Food Contact
3.4. Outlook and Future Perspective
4. Materials and Methods
4.1. Sample Material
4.2. Evaluation of DNA-Reactive, Mutagenic Effects with the Miniaturized Ames Test
4.2.1. Sample Extraction and Pre-Concentration
4.2.2. The Miniaturized Ames Test
4.3. Correlation Analysis of Miniaturized Ames Test and Chromatographic Analysis Data
4.3.1. Chromatographic Analysis
4.3.2. Principal Component Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Franz, R.; Welle, F. Recycling of Post-Consumer Packaging Materials into New Food Packaging Applications—Critical Review of the European Approach and Future Perspectives. Sustainability 2022, 14, 824. [Google Scholar] [CrossRef]
- Kato, L.S.; Conte-Junior, C.A. Safety of Plastic Food Packaging: The Challenges about Non-Intentionally Added Substances (NIAS) Discovery, Identification and Risk Assessment. Polymers 2021, 13, 2077. [Google Scholar] [CrossRef] [PubMed]
- Horodytska, O.; Cabanes, A.; Fullana, A. Non-intentionally added substances (NIAS) in recycled plastics. Chemosphere 2020, 251, 126373. [Google Scholar] [CrossRef] [PubMed]
- More, S.J.; Bampidis, V.; Benford, D.; Bragard, C.; Halldorsson, T.I.; Hernández-Jerez, A.F.; Hougaard Bennekou, S.; Koutsoumanis, K.P.; Machera, K.; Naegeli, H.; et al. Guidance on the use of the Threshold of Toxicological Concern approach in food safety assessment. EFS2 2019, 17, e05708. [Google Scholar] [CrossRef]
- Kroes, R.; Renwick, A.G.; Cheeseman, M.; Kleiner, J.; Mangelsdorf, I.; Piersma, A.; Schilter, B.; Schlatter, J.; van Schothorst, F.; Vos, J.G.; et al. Structure-based thresholds of toxicological concern (TTC): Guidance for application to substances present at low levels in the diet. Food Chem. Toxicol. 2004, 42, 65–83. [Google Scholar] [CrossRef] [PubMed]
- EFSA Panel on Food Contact Materials, Enzymes, Flavourings and Processing Aids (CEF). Scientific Opinion on the criteria to be used for safety evaluation of a mechanical recycling process to produce recycled PET intended to be used for manufacture of materials and articles in contact with food. EFS2 2011, 9, e2184. [Google Scholar] [CrossRef]
- Franz, R.; Welle, F. Contamination Levels in Recollected PET Bottles from Non-Food Applications and their Impact on the Safety of Recycled PET for Food Contact. Molecules 2020, 25, 4998. [Google Scholar] [CrossRef]
- EFSA Panel on Food Contact Materials, Enzymes, Flavourings and Processing Aids (CEF). Scientific Opinion on the safety assessment of the processes ‘Biffa Polymers’ and ‘CLRrHDPE’ used to recycle high-density polyethylene bottles for use as food contact material. EFS2 2015, 13, e4016. [Google Scholar] [CrossRef]
- O’ Brien, S.; Briard, M.; Pelsy, F. A European Strategy for Plastics in the Circular Economy: Local and Regional Dimension; Milieu Ltd.: Bruxelles, Belgium, 2018. [Google Scholar] [CrossRef]
- Proposal for a Regulation of the European Parliament and of the Council on Packaging and Packaging Waste, Amending Regulation (EU) 2019/1020 and Directive (EU) 2019/904, and Repealing Directive 94/62/EC. 2022. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX%3A52022PC0677 (accessed on 30 March 2023).
- Commission Regulation (EU) 2022/1616 of 15 September 2022 on Recycled Plastic Materials and Articles Intended to Come into Contact with Foods, and Repealing Regulation (EC) No 282/2008. 2022. Available online: https://eur-lex.europa.eu/eli/reg/2022/1616/oj (accessed on 30 March 2023).
- Commission Regulation (EU) No 10/2011 of 14 January 2011 on Plastic Materials and Articles Intended to Come into Contact with Food. 2011. Available online: https://eur-lex.europa.eu/legal-content/EN/ALL/?uri=celex%3A32011R0010 (accessed on 30 March 2023).
- Koster, S.; Rennen, M.; Leeman, W.; Houben, G.; Muilwijk, B.; van Acker, F.; Krul, L. A novel safety assessment strategy for non-intentionally added substances (NIAS) in carton food contact materials. Food Addit. Contam. Part A 2014, 31, 422–443. [Google Scholar] [CrossRef] [PubMed]
- Rennen, M.A.J.; Koster, S.; Krul, C.A.M.; Houben, G.F. Application of the threshold of toxicological concern (TTC) concept to the safety assessment of chemically complex food matrices. Food Chem. Toxicol. 2011, 49, 933–940. [Google Scholar] [CrossRef]
- Schilter, B.; Burnett, K.; Eskes, C.; Geurts, L.; Jacquet, M.; Kirchnawy, C.; Oldring, P.; Pieper, G.; Pinter, E.; Tacker, M.; et al. Value and limitation of in vitro bioassays to support the application of the threshold of toxicological concern to prioritise unidentified chemicals in food contact materials. Food Addit. Contam. Part A 2019, 36, 1903–1936. [Google Scholar] [CrossRef] [PubMed]
- Adahchour, M.; Vreuls, J.J.; van Hattum, A.G.M. Concentration Techniques for Genotoxicity Testing of Effluents. (IVM Report; No. E-01/08). Institute for Environmental Studies. 2001. Available online: https://research.vu.nl/ws/portalfiles/portal/1784423/e01-08.gentox.pdf (accessed on 17 October 2023).
- Nerín, C.; Bourdoux, S.; Faust, B.; Gude, T.; Lesueur, C.; Simat, T.; Stoermer, A.; van Hoek, E.; Oldring, P. Guidance in selecting analytical techniques for identification and quantification of non-intentionally added substances (NIAS) in food contact materials (FCMS). Food Addit. Contam. Part A 2022, 39, 620–643. [Google Scholar] [CrossRef] [PubMed]
- Severin, I.; Souton, E.; Dahbi, L.; Chagnon, M.C. Use of bioassays to assess hazard of food contact material extracts: State of the art. Food Chem. Toxicol. 2017, 105, 429–447. [Google Scholar] [CrossRef]
- Pinter, E.; Welle, F.; Mayrhofer, E.; Pechhacker, A.; Motloch, L.; Lahme, V.; Grant, A.; Tacker, M. Circularity Study on PET Bottle-To-Bottle Recycling. Sustainability 2021, 13, 7370. [Google Scholar] [CrossRef]
- Koster, S.; Bani-Estivals, M.H.; Bonuomo, M.; Bradley, E.; Chagnon, M.C.; Garcia, M.L.; Godts, F.; Gude, T.; Helling, R.; Paseiro-Losada, P.; et al. Guidance on Best Practices on the Risk Assessment of Non Intentionally Added Substances (NIAS) in Food Contact Materials and Articles; ILSI Europe: Brussels, Belgium, 2015; ISBN 9789078637424. [Google Scholar]
- ICH Guideline M7(R1) on Assessment and Control of DNA Reactive (Mutagenic) Impurities in Pharmaceuticals to Limit Potential Carcinogenic Risk. 2019. Available online: https://www.ema.europa.eu/en/documents/scientific-guideline/ich-guideline-m7r1-assessment-control-dna-reactive-mutagenic-impurities-pharmaceuticals-limit_en.pdf (accessed on 30 March 2023).
- Rainer, B.; Pinter, E.; Czerny, T.; Riegel, E.; Kirchnawy, C.; Marin-Kuan, M.; Schilter, B.; Tacker, M. Suitability of the Ames test to characterise genotoxicity of food contact material migrates. Food Addit. Contam. Part A 2018, 35, 2230–2243. [Google Scholar] [CrossRef] [PubMed]
- Pinter, E.; Rainer, B.; Czerny, T.; Riegel, E.; Schilter, B.; Marin-Kuan, M.; Tacker, M. Evaluation of the Suitability of Mammalian In Vitro Assays to Assess the Genotoxic Potential of Food Contact Materials. Foods 2020, 9, 237. [Google Scholar] [CrossRef]
- Flückiger-Isler, S.; Kamber, M. Direct comparison of the Ames microplate format (MPF) test in liquid medium with the standard Ames pre-incubation assay on agar plates by use of equivocal to weakly positive test compounds. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2012, 747, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Rainer, B.; Pinter, E.; Prielinger, L.; Coppola, C.; Marin-Kuan, M.; Schilter, B.; Apprich, S.; Tacker, M. Direct Comparison of the Lowest Effect Concentrations of Mutagenic Reference Substances in Two Ames Test Formats. Toxics 2021, 9, 152. [Google Scholar] [CrossRef]
- OECD. Test No. 471: Bacterial Reverse Mutation Test, OECD Guidelines for the Testing of Chemicals, Section 4; OECD Publishing: Paris, France, 2020; ISBN 9789264071247. [Google Scholar] [CrossRef]
- Rainer, B.; Mayrhofer, E.; Redl, M.; Dolak, I.; Mislivececk, D.; Czerny, T.; Kirchnawy, C.; Marin-Kuan, M.; Schilter, B.; Tacker, M. Mutagenicity assessment of food contact material migrates with the Ames MPF assay. Food Addit. Contam. Part A 2019, 36, 1419–1432. [Google Scholar] [CrossRef] [PubMed]
- Williams, R.V.; DeMarini, D.M.; Stankowski, L.F.; Escobar, P.A.; Zeiger, E.; Howe, J.; Elespuru, R.; Cross, K.P. Are all bacterial strains required by OECD mutagenicity test guideline TG471 needed? Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2019, 848, 503081. [Google Scholar] [CrossRef]
- Rung, C.; Welle, F.; Gruner, A.; Springer, A.; Steinmetz, Z.; Munoz, K. Identification and Evaluation of (Non-)Intentionally Added Substances in Post-Consumer Recyclates and Their Toxicological Classification. Recycling 2023, 8, 24. [Google Scholar] [CrossRef]
- Hoffmann, G.R.; Fuchs, R.P. Mechanisms of frameshift mutations: Insight from aromatic amines. Chem. Res. Toxicol. 1997, 10, 347–359. [Google Scholar] [CrossRef]
- Pinter, E.; Friedl, C.; Irnesberger, A.; Czerny, T.; Piwonka, T.; Peñarroya, A.; Tacker, M.; Riegel, E. HepGentox: A novel promising HepG2 reportergene-assay for the detection of genotoxic substances in complex mixtures. PeerJ 2021, 9, e11883. [Google Scholar] [CrossRef] [PubMed]
- Oda, Y. Development and progress for three decades in umu test systems. Genes Environ. 2016, 38, 24. [Google Scholar] [CrossRef] [PubMed]
- Hochegger, A.; Wagenhofer, R.; Savić, S.; Mayrhofer, E.; Washüttl, M.; Leitner, E. Combination of Multidimensional Instrumental Analysis and the Ames Test for the Toxicological Evaluation of Mineral Oil Aromatic Hydrocarbons. J. Agric. Food Chem. 2022, 70, 16401–16409. [Google Scholar] [CrossRef] [PubMed]
- Hamel, A.; Roy, M.; Proudlock, R. The Bacterial Reverse Mutation Test. Genet. Toxicol. Test. 2016, 79–138. [Google Scholar] [CrossRef]
- Xenometrix. Ames MPFTM Penta 2. Microplate Format Mutagenicity Assay. S.typhimuriumTA98, TA100, TA1535, TA1537 and E.coli WP2 uvrA[pKM101]. Instructions for Use. 2.0. 2018. Available online: https://www.xenometrix.ch/shop/mediafiles/Xeno%20Dateien/Short%20Protocol/Ames/Ames%20MPF%20Short%20Protocol_Penta2.pdf (accessed on 10 July 2023).
- European Chemicals Agency (ECHA). Registration Dossier for 3,3’,3’’,5,5’,5’’-hexa-tert-butyl-α,α’,α’’-(mesitylene-2,4,6-triyl)tri-p-cresol, CAS Number: 1709-70-2. Available online: https://echa.europa.eu/cs/registration-dossier/-/registered-dossier/14678/7/7/1 (accessed on 10 July 2023).
- Johnson, W.; Bergfeld, W.F.; Belsito, D.V.; Hill, R.A.; Klaassen, C.D.; Liebler, D.C.; Marks, J.G.; Shank, R.C.; Slaga, T.J.; Snyder, P.W.; et al. Safety Assessment of Pentaerythrityl Tetra-Di -t- Butyl Hydroxyhydrocinnamate as Used in Cosmetics. Int. J. Toxicol. 2018, 37, 80S–89S. [Google Scholar] [CrossRef]
- European Chemicals Agency (ECHA). Registration Dossier for Pentaerythritol tetrakis(3-(3,5-di-tert-butyl-4-hydroxyphenyl)propionate), CAS Number: 6683-19-8. Available online: https://echa.europa.eu/cs/registration-dossier/-/registered-dossier/15308/7/7/2 (accessed on 10 July 2023).
- European Chemicals Agency (ECHA). Registration dossier for Tris(2,4-ditert-butylphenyl) phosphite, CAS Number: 31570-04-4. Available online: https://echa.europa.eu/cs/registration-dossier/-/registered-dossier/15253/7/7/2 (accessed on 10 July 2023).
- Sigma Aldrich: Data Sheet for Oleamide. Available online: https://www.sigmaaldrich.com/DE/en/product/sigma/o2136 (accessed on 3 January 2022).
- Elsner, P.; Eyerer, P.; Hirth, T. Die Kunststoffe und ihre Eigenschaften; 6. neu bearb. und erw. Auflage, S. 157; Springer: Berlin/Heidelberg, Germany; New York, NY, USA, 2005; ISBN 3-540-21410-0. [Google Scholar]
- Politano, V.T.; McGinty, D.; Lewis, E.M.; Hoberman, A.M.; Diener, R.M.; Api, A.M. Evaluation of the developmental toxicity of 4-tert-butylcyclohexyl acetate in Sprague-Dawley rats. Int. J. Toxicol. 2012, 31, 477–482. [Google Scholar] [CrossRef]
- Bhatia, S.P.; Jones, L.; Letizia, C.S.; Api, A.M. Fragrance material review on 4-tert-butylcyclohexyl acetate. Food Chem. Toxicol. 2008, 46 (Suppl. S12), S36–S41. [Google Scholar] [CrossRef]
- Sigma Aldrich: Data Sheet for 4-tert-Butylcyclohexyl Acetate. Available online: https://www.sigmaaldrich.com/DE/en/sds/ALDRICH/W507318 (accessed on 3 April 2023).
- Anlauf, M.; Paffrath, D.; Schwabe, U. Arzneiverordnungs-Report 2003: Aktuelle Daten, Kosten, Trends und Kommentare; [et autres], Cop; Springer: Berlin, Germany, 2004; ISBN 3-540-40188-1. [Google Scholar]
- Burdock, G.A.; Fenaroli, G. Flavor Ingredients, 5th ed.; CRC Press: Boca Raton, FL, USA; London, UK, 2004; ISBN 0-8493-3034-3. [Google Scholar]
- Hunnius, C.; Wachter, H.; Burger, A. Hunnius Pharmazeutisches Wörterbuch; 8. Aufl.; Walter de Gruyter: Berlin, Germany, 1998; ISBN 3-11-015792-6. [Google Scholar]
- Lutz, W.K.; Vamvakas, S.; Kopp-Schneider, A.; Schlatter, J.; Stopper, H. Deviation from additivity in mixture toxicity: Relevance of nonlinear dose-response relationships and cell line differences in genotoxicity assays with combinations of chemical mutagens and gamma-radiation. Environ. Health Perspect. 2002, 110 (Suppl. S6), 915–918. [Google Scholar] [CrossRef]
- Bernauer, U.; Bodin, L.; Chaudhry, Q.; Coenraads, P.J.; Dusinska, M.; Ezendam, J.; von Goetz, N. The SCCS Notes of Guidance for the testing of cosmetic ingredients and their safety evaluation, 11th revision, 30–31 March 2021, SCCS/1628/21. Regul. Toxicol. Pharmacol. 2021, 127, 105052. [Google Scholar] [CrossRef] [PubMed]
- Welle, F. Recycling of Post-Consumer Polystyrene Packaging Waste into New Food Packaging Applications—Part 2: Co-Extruded Functional Barriers. Recycling 2023, 8, 39. [Google Scholar] [CrossRef]
- Mortelmans, K.; Zeiger, E. The Ames Salmonella/microsome mutagenicity assay. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2000, 455, 29–60. [Google Scholar] [CrossRef]
- Kirkland, D.; Zeiger, E.; Madia, F.; Gooderham, N.; Kasper, P.; Lynch, A.; Morita, T.; Ouedraogo, G.; Parra Morte, J.M.; Pfuhler, S.; et al. Can in vitro mammalian cell genotoxicity test results be used to complement positive results in the Ames test and help predict carcinogenic or in vivo genotoxic activity? I. Reports of individual databases presented at an EURL ECVAM Workshop. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2014, 775–776, 55–68. [Google Scholar] [CrossRef]
- Forman, D. Ames, the Ames test, and the causes of cancer. BMJ 1991, 303, 428–429. [Google Scholar] [CrossRef]
- Ames, B.N.; Gold, L.S. Chemical carcinogenesis: Too many rodent carcinogens. Proc. Natl. Acad. Sci. USA 1990, 87, 7772–7776. [Google Scholar] [CrossRef]
- Ames, B.N.; Gold, L.S.; Willett, W.C. The causes and prevention of cancer. Proc. Natl. Acad. Sci. USA 1995, 92, 5258–5265. [Google Scholar] [CrossRef] [PubMed]
- Regulation (EC) No 648/2004 of the European Parliament and of the Council of 31 March 2004 on Detergents. 2004. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:32004R0648&from=DE (accessed on 19 April 2023).
- Regulation (EC) No 1272/2008 of the European Parliament and of the Council of 16 December 2008 on Classification, Labelling and Packaging of Substances and Mixtures, Amending and Repealing Directives 67/548/EEC and 1999/45/EC, and Amending Regulation (EC) No 1907/2006. 2008. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:02008R1272-20101201&from=EN (accessed on 19 April 2023).
- Regulation (EC) No 1223/2009 of the European Parliament and of the Council of 30 November 2009 on Cosmetic Products. 2009. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:32009R1223&from=DE (accessed on 19 April 2023).
- Goldman, R.; Shields, P.G. Food mutagens. J. Nutr. 2003, 133 (Suppl. S3), 965S–973S. [Google Scholar] [CrossRef] [PubMed]
- Levy, S.B. Tanning preparations. Dermatol. Clin. 2000, 18, 591–596. [Google Scholar] [CrossRef] [PubMed]
- The European Printing Ink Association: Printing Inks and Plastic Recycling—Q & A. Available online: https://www.eupia.org/wp-content/uploads/2022/09/Printing_inks_and_Plastic_Recycling_-_Q___A.pdf (accessed on 10 July 2023).
- The European Printing Ink Association: Inks and Coatings for High Temperature Applications. Available online: https://www.eupia.org/wp-content/uploads/2022/09/2016-02-24_EuPIA_Info_Note_Inks_and_Coatings_for_High_Temperature_Applications.pdf (accessed on 10 July 2023).
- Golob, N.; Grahek, R.; Ross, M.; Roškar, R. Nitrocellulose blister material as a source of N-nitrosamine contamination of pharmaceutical drug products. Int. J. Pharm. 2022, 618, 121687. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.L.; Saleh, M.A. Thermal degradation of azobenzene dyes. Results Chem. 2020, 2, 100085. [Google Scholar] [CrossRef]
- Kuhnke, L.; Laak, A.T.; Göller, A.H. Mechanistic Reactivity Descriptors for the Prediction of Ames Mutagenicity of Primary Aromatic Amines. J. Chem. Inf. Model. 2019, 59, 668–672. [Google Scholar] [CrossRef]
- Yahagi, T.; Nagao, M.; Seino, Y.; Matsushima, T.; Sugimura, T. Mutagenicities of N-nitrosamines on Salmonella. Mutat. Res. Fundam. Mol. Mech. Mutagen. 1977, 48, 121–129. [Google Scholar] [CrossRef]
- Araki, A.; Muramatsu, M.; Matsushima, T. Comparison of mutagenicities of n-nitrosamines on Salmonella typhimurium TA100 and Escheria coli WP2 UVRA/PKM101 using rat and hamster liver S9. GANN Jpn. J. Cancer Res. 1984, 75, 8–16. [Google Scholar] [CrossRef]
- Bringezu, F.; Simon, S. Salmonella typhimurium TA100 and TA1535 and E. coli WP2 uvrA are highly sensitive to detect the mutagenicity of short Alkyl-N-Nitrosamines in the Bacterial Reverse Mutation Test. Toxicol. Rep. 2022, 9, 250–255. [Google Scholar] [CrossRef] [PubMed]
- Nielson, R.C. Extraction and Quantitation of Polyolefin Additives. J. Liq. Chromatogr. 1991, 14, 503–519. [Google Scholar] [CrossRef]
- Mayrhofer, E. Development of a Comprehensive Testing Strategy to Detect DNA-Reactive Effects of Food Contact Materials; Doctoral dissertation, Technische Universität Wien: Vienna, Austria, 2023. [Google Scholar] [CrossRef]
- ISO 11350:2012; Water Quality—Determination of the Genotoxicity of Water and Waste Water—Salmonella/Microsome Fluctuation Test (Ames Fluctuation Test). ISO: Geneva, Switzerland, 2012. Available online: https://www.iso.org/standard/50393.html (accessed on 30 March 2023).
- Shapiro, S.S.; Wilk, M.B. An Analysis of Variance Test for Normality (Complete Samples). Biometrika 1965, 52, 591. [Google Scholar] [CrossRef]
- Origin Pro: Origin(Pro), Version 2022b; OriginLab Corporation: Northampton, MA, USA, 2022.
- Fahrmeir, L.; Hamerle, A.; Tutz, G. Multivariate Statistische Verfahren; DE GRUYTER: Berlin, Germany, 1996; ISBN 978-3-11-013806-1. [Google Scholar]




| Code | Type | I/O | TA98 | TA100 | Code | Type | I/O | TA98 | TA100 | Code | Type | I/O | TA98 | TA100 | Code | Type | I/O | TA98 | TA100 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| −S9 | +S9 | −S9 | +S9 | −S9 | +S9 | −S9 | +S9 | −S9 | +S9 | −S9 | +S9 | −S9 | +S9 | −S9 | +S9 | ||||||||||||
| H1 | HDPE | I | nd | - | nd | nd | H11 | HDPE | O | -* | - | -* | - | H21 | HDPE | O | nd | + | nd | nd | H31 | HDPE | O | - | - | - | - |
| H2 | HDPE | I | nd | - | nd | nd | H12 | HDPE | O | -* | - | - | - | H22 | HDPE | O | nd | + | nd | nd | H32 | HDPE | O | - | - | - | - |
| H3 | HDPE | I | nd | - | nd | nd | H13 | HDPE | O | - | - | - | - | H23 | HDPE | O | -* | + | - | - | H33 | HDPE | O | - | - | - | - |
| H4 | HDPE | I | nd | - | nd | nd | H14 | HDPE | O | - | - | - | - | H24 | HDPE | O | nd | + | nd | nd | H34 | HDPE | O | nd | + | nd | nd |
| H5 | HDPE | I | - | - | - | - | H15 | HDPE | O | - | + | - | - | H25 | HDPE | O | nd | + | nd | nd | H35 | HDPE | O | - | - | - | - |
| H6 | HDPE | O | nd | - | nd | nd | H16 | HDPE | O | -* | + | - | - | H26 | HDPE | O | -* | + | - | - | H36 | HDPE | O | nd | eq | nd | nd |
| H7 | HDPE | O | - | - | - | - | H17 | HDPE | O | -* | - | - | - | H27 | HDPE | O | -* | + | - | - | H37 | HDPE | O | - | - | - | - |
| H8 | HDPE | O | - | - | - | - | H18 | HDPE | O | -* | - | - | - | H28 | HDPE | O | - | + | -* | - | H38 | HDPE | O | - | - | - | - |
| H9 | HDPE | O | - | - | - | - | H19 | HDPE | O | -* | + | -* | - | H29 | HDPE | O | -* | + | - | - | H39 | HDPE | O | - | - | - | - |
| H10 | HDPE | O | -* | - | - | - | H20 | HDPE | O | nd | + | nd | nd | H30 | HDPE | O | -* | - | - | - | H40 | HDPE | O | - | + | - | - |
| L1 | LDPE | I | nd | + | nd | nd | L6 | LDPE | O | eq | + | - | - | L11 | LDPE | O | nd | + | nd | nd | L16 | LDPE | O | - | + | - | - |
| L2 | LDPE | I | nd | + | nd | nd | L7 | LDPE | O | - | + | - | eq | L12 | LDPE | O | - | + | - | + | L17 | LDPE | O | + | + | - | - |
| L3 | LDPE | O | - | + | - | - | L8 | LDPE | O | - | + | - | + | L13 | LDPE | O | -* | + | - | + | L18 | LDPE | O | -* | - | - | - |
| L4 | LDPE | O | - | + | - | - | L9 | LDPE | O | - | + | + | + | L14 | LDPE | O | - | + | - | - | |||||||
| L5 | LDPE | O | - | + | - | - | L10 | LDPE | O | nd | + | nd | nd | L15 | LDPE | O | - | + | - | - | |||||||
| E1 | PET | I | nd | - | nd | nd | E4 | PET | I | -* | - | - | - | E7 | PET | O | - | - | - | - | E10 | PET | O | - | - | - | - |
| E2 | PET | I | nd | - | nd | nd | E5 | PET | O | - | - | - | - | E8 | PET | O | - | - | - | - | E11 | PET | O | -* | - | - | - |
| E3 | PET | I | - | - | - | - | E6 | PET | O | - | - | - | - | E9 | PET | O | - | - | - | - | E12 | PET | O | -* | - | - | - |
| P1 | PP | I | nd | - | nd | nd | P11 | PP | O | nd | + | nd | nd | P21 | PP | O | - | + | - | - | P31 | PP | O | + | + | - | - |
| P2 | PP | I | nd | - | nd | nd | P12 | PP | O | nd | - | nd | nd | P22 | PP | O | - | - | - | - | P32 | PP | O | + | + | - | - |
| P3 | PP | I | nd | - | nd | nd | P13 | PP | O | nd | - | nd | nd | P23 | PP | O | nd | - | nd | nd | P33 | PP | O | - | - | - | - |
| P4 | PP | I | - | - | - | - | P14 | PP | O | - | - | - | - | P24 | PP | O | - | - | - | - | P34 | PP | O | - | - | - | - |
| P5 | PP | I | - | - | - | - | P15 | PP | O | - | + | - | - | P25 | PP | O | - | - | - | - | P35 | PP | O | - | - | - | - |
| P6 | PP | I | - | - | - | - | P16 | PP | O | - | - | - | - | P26 | PP | O | - | - | - | - | P36 | PP | O | nd | + | nd | nd |
| P7 | PP | I | + | eq | - | - | P17 | PP | O | - | - | - | - | P27 | PP | O | - | - | - | - | P37 | PP | O | nd | - | nd | nd |
| P8 | PP | O | - | - | - | + | P18 | PP | O | - | eq | - | - | P28 | PP | O | nd | - | nd | nd | |||||||
| P9 | PP | O | - | - | - | - | P19 | PP | O | - | - | - | - | P29 | PP | O | - | - | - | - | |||||||
| P10 | PP | O | nd | + | nd | nd | P20 | PP | O | -* | + | - | - | P30 | PP | O | -* | + | -* | - | |||||||
| S1 | PS | I | nd | - | nd | nd | S5 | PS | I | - | eq | - | - | S9 | PS | O | - | + | - | - | O1 | PE | I | + | + | - | - |
| S2 | PS | I | - | + | - | - | S6 | PS | I | + | + | -* | -* | S10 | PS | O | - | + | - | - | O2 | PE/PP | O | - | + | - | eq |
| S3 | PS | I | + | + | - | - | S7 | PS | I | - | - | - | - | ||||||||||||||
| S4 | PS | I | - | + | - | - | S8 | PS | I | - | - | - | - | ||||||||||||||
| V | Substance (M in g/mol)–CAS | V | Substance (M in g/mol)–CAS |
|---|---|---|---|
| V1 | Irgafos 168 (646)–31570-04-4 | V22 | 2-Phenyl-1,2,3,4-tetrahydronaphthalene (208)–29422-13-7 |
| V2 | Oxidized Irgafos 168 (662)–95906-11-9 | V23 | N,N-dimethyltricyclo [5.3.1.04,9]undec-5-en-2-amine (191)–NA |
| V3 | Irganox 245 (587)–36443-68-2 | V24 | 1,3,5-Triphenylcyclohexane (312)–28336-57-4 |
| V4 | Irganox 1010 (1178)–6683-19-8 | V25 | 2-Methylanthraquinone (222)–84-54-8 |
| V5 | Irganox 1076 (531)–2082-79-3 | V26 | (2E)-2-(3-Oxo-1-benzothiophen-2-ylidene)-1-benzothiophen-3-one (296)–522-75-8 |
| V6 | Irganox 1330 (775)–1709-70-2 | V27 | 4-Isopropyl-N-(2-(2-methyl-1 H-indol-3-yl)-ethyl)-benzenesulfoamide (356)–NA |
| V7 | Limonene (136)–138-86-3 | V28 | 2,2’-Thiobis(6-tert-butyl-p-cresol) (358)–90-66-4 |
| V8 | Acetaldehyde (44)–75-07-0 | V29 | Bumetrizole (316)–3896-11-5 |
| V9 | 1,3-Dioxolane (74)–646-06-0 | V30 | (Z)-Docos-13-enamide (337)–112-84-5 |
| V10 | Ethylene glycol (62)–107-21-1 | V31 | Octocrylene (361)–6197-30-4 |
| V11 | Oxidized Irgafos 168 (662)–95906-11-9 | V32 | 1-Phenyl-1,2,3,4-tetrahydronaphthalene (208)–3018-20-0 |
| V12 | Irgafos 168 (646)–31570-04-4 | V33 | Triphenyl phosphate (326)–115-86-6 |
| V13 | (1-Methyl-2,2-diphenylcyclopropyl)sulfanylbenzene (317)–56728-02-0 | V34 | Tributyl citrate (360)–77-94-1 |
| V14 | (2,3-Diphenylcyclopropyl)methyl phenyl sulfoxide (332)–131758-71-9 | V35 | Tinuvin 770 (480)–52829-07-9 |
| V15 | 1,3-Diphenylpropane (196)–1081-75-0 | V36 | 2-Butyl-5-hexyloctahydro-1H-indene (264)–55044-33-2 |
| V16 | (3-Cyclopent-2-en-1-yl-2-methyl-1-phenylprop-1-enyl)benzene (274)–NA | V37 | S-[(E)-1,3-Diphenylbut-2-enyl] N,N-dimethylcarbamothioate (311)–NA |
| V17 | Di-p-xylylene (208)–1633-22-3 | V38 | 4-tert-Butylcyclohexyl acetate (198)–10411-92-4 |
| V18 | 4-Cyano-1,2,3,4-tetrahydro-1-naphthaleneacetonitrile (210)–57964-40-6 | V39 | Oleamide (282)–301-02-0 |
| V19 | 4-(1-Cyanoethyl)-1,2,3,4-tetrahydronaphthalene-1-carbonitrile (210)–57964-39-3 | V40 | Docosanamide (340)–3061-75-4 |
| V20 | 3,6,13,16-Tetraoxatricyclo [16.2.2.2(8,11)]tetracosa-1(20),8,10,18,21,23-hexaene-2,7,12,17-tetrone (384)–24388-68-9 | V41 | 1-Iodo-2-methylundecane (296)–73105-67-6 |
| V21 | 1,2-Diphenylpropane (196)–1081-75-0 | V42 | Bornyl acetate (196)–76-49-3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mayrhofer, E.; Prielinger, L.; Sharp, V.; Rainer, B.; Kirchnawy, C.; Rung, C.; Gruner, A.; Juric, M.; Springer, A. Safety Assessment of Recycled Plastics from Post-Consumer Waste with a Combination of a Miniaturized Ames Test and Chromatographic Analysis. Recycling 2023, 8, 87. https://doi.org/10.3390/recycling8060087
Mayrhofer E, Prielinger L, Sharp V, Rainer B, Kirchnawy C, Rung C, Gruner A, Juric M, Springer A. Safety Assessment of Recycled Plastics from Post-Consumer Waste with a Combination of a Miniaturized Ames Test and Chromatographic Analysis. Recycling. 2023; 8(6):87. https://doi.org/10.3390/recycling8060087
Chicago/Turabian StyleMayrhofer, Elisa, Lukas Prielinger, Victor Sharp, Bernhard Rainer, Christian Kirchnawy, Christian Rung, Anita Gruner, Mladen Juric, and Arielle Springer. 2023. "Safety Assessment of Recycled Plastics from Post-Consumer Waste with a Combination of a Miniaturized Ames Test and Chromatographic Analysis" Recycling 8, no. 6: 87. https://doi.org/10.3390/recycling8060087
APA StyleMayrhofer, E., Prielinger, L., Sharp, V., Rainer, B., Kirchnawy, C., Rung, C., Gruner, A., Juric, M., & Springer, A. (2023). Safety Assessment of Recycled Plastics from Post-Consumer Waste with a Combination of a Miniaturized Ames Test and Chromatographic Analysis. Recycling, 8(6), 87. https://doi.org/10.3390/recycling8060087







