Peanut Allergy Diagnosis: Current Practices, Emerging Technologies, and Future Directions
Abstract
:1. Introduction
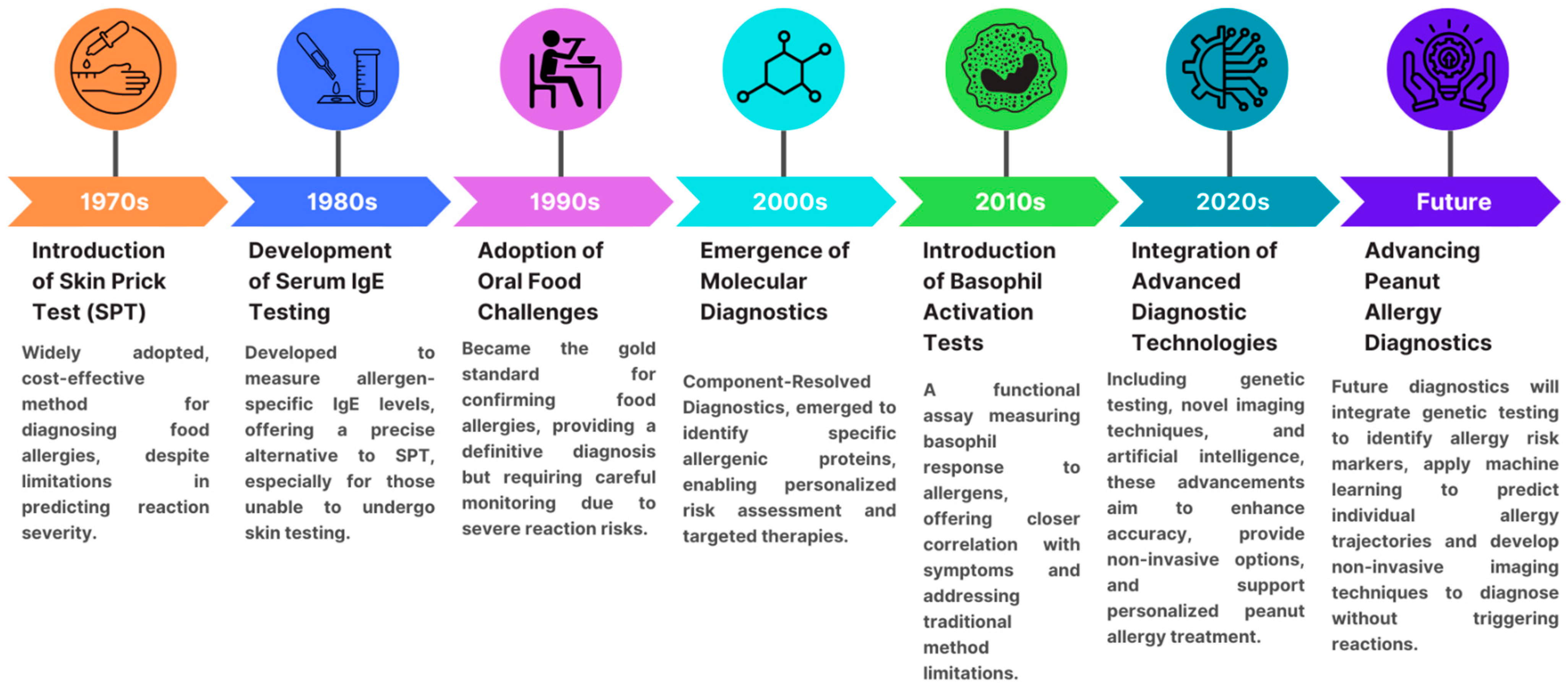
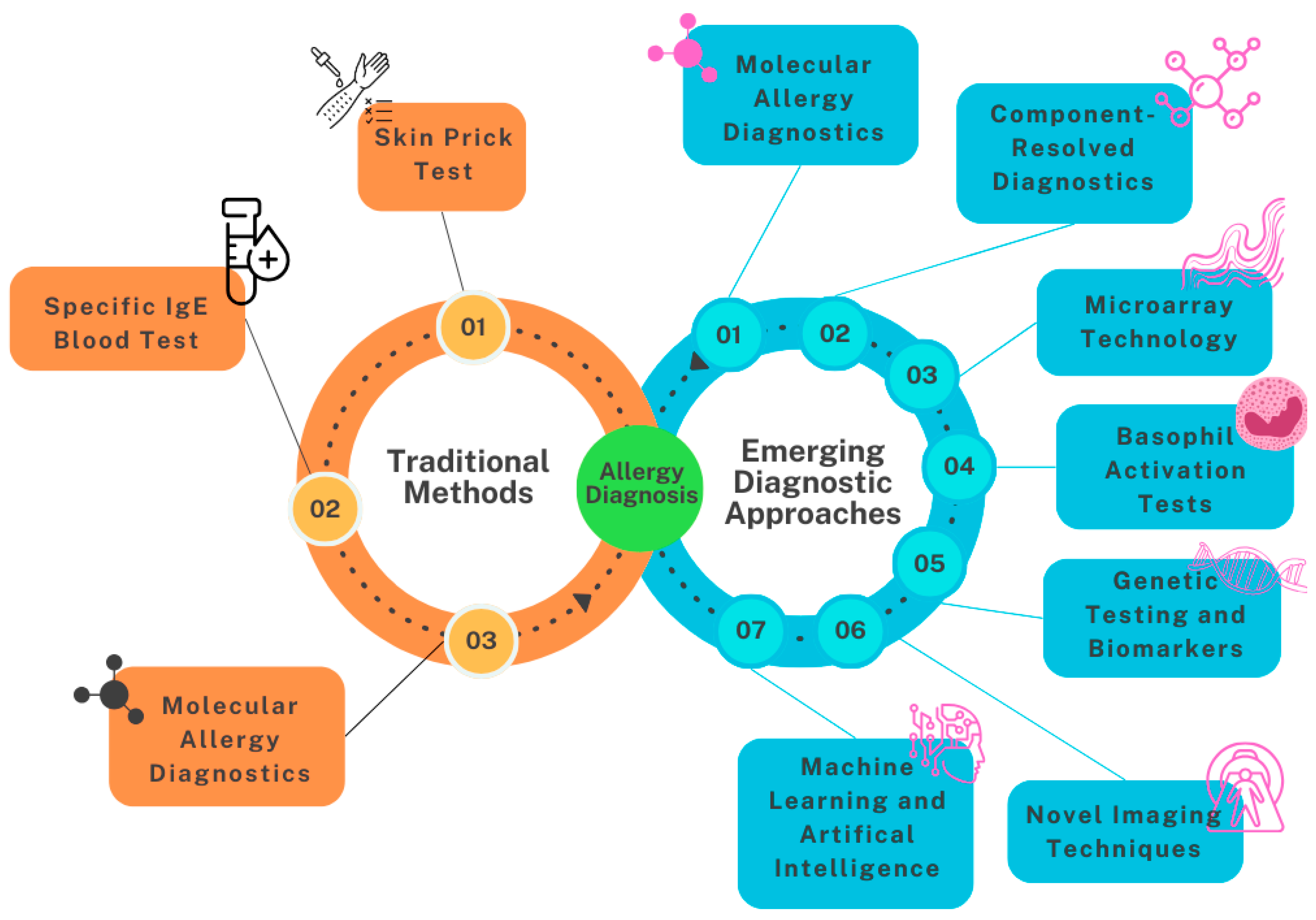
2. Diagnostic Methods for Peanut Allergy
3. Current Diagnostic Methods
3.1. Skin Prick Testing
3.2. Serum IgE Testing
3.3. Molecular Allergy Diagnosis
3.4. Oral Food Challenge
3.5. Bead-Based Epitope Assay
4. Emerging Diagnostic Approaches
4.1. Use of Microarrays for Allergen Components
4.2. Basophil Activation Test (BAT)
4.3. Mast Cell Activation Tests (MAT)
4.4. Genetic Testing
4.5. Novel Imaging Techniques
4.6. Machine Learning and AI in Allergy Diagnostics
4.7. Challenges and Limitations in Emerging Diagnostic Approaches
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Warren, C.; Lei, D.; Sicherer, S.; Schleimer, R.; Gupta, R. Prevalence and characteristics of peanut allergy in US adults. J. Allergy Clin. Immunol. 2021, 147, 2263–2270.e5. [Google Scholar] [CrossRef]
- Nowak-Wegrzyn, A.; Hass, S.L.; Donelson, S.M.; Robison, D.; Cameron, A.; Etschmaier, M.; Duhig, A.; McCann, W.A. The Peanut Allergy Burden Study: Impact on the quality of life of patients and caregivers. World Allergy Organ. J. 2021, 14, 100512. [Google Scholar] [CrossRef]
- Gupta, R.S.; Warren, C.M.; Smith, B.M.; Jiang, J.; Blumenstock, J.A.; Davis, M.M.; Schleimer, R.P.; Nadeau, K.C. Prevalence and Severity of Food Allergies Among US Adults. JAMA Netw. Open 2019, 2, e185630. [Google Scholar] [CrossRef]
- Midun, E.; Radulovic, S.; Brough, H.; Caubet, J.-C. Recent advances in the management of nut allergy. World Allergy Organ. J. 2021, 14, 100491. [Google Scholar] [CrossRef]
- Khan, M.; Banerjee, S.; Muskawad, S.; Maity, R.; Chowdhury, S.R.; Ejaz, R.; Kuuzie, E.; Satnarine, T. The Impact of Artificial Intelligence on Allergy Diagnosis and Treatment. Curr. Allergy Asthma Rep. 2024, 24, 361–372. [Google Scholar] [CrossRef]
- Eckman, J.; Saini, S.S.; Hamilton, R.G. Diagnostic evaluation of food-related allergic diseases. Allergy, Asthma Clin. Immunol. 2009, 5, 2. [Google Scholar] [CrossRef]
- Sicherer, S.H.; Sampson, H.A. Food allergy. J. Allergy Clin. Immunol. 2010, 125 (Suppl. S2), S116–S125. [Google Scholar] [CrossRef]
- Anyane-Yeboa, A.; Wang, W.; Kavitt, R.T. The Role of Allergy Testing in Eosinophilic Esophagitis. Gastroenterol. Hepatol. 2018, 14, 463–469. [Google Scholar]
- Sporik, R.; Hill, D.J.; Hosking, C.S. Specificity of allergen skin testing in predicting positive open food challenges to milk, egg and peanut in children. Clin. Exp. Allergy 2000, 30, 1541–1546. [Google Scholar] [CrossRef]
- Foong, R.-X.; Dantzer, J.A.; Wood, R.A.; Santos, A.F. Improving Diagnostic Accuracy in Food Allergy. J. Allergy Clin. Immunol. Pract. 2021, 9, 71–80. [Google Scholar] [CrossRef]
- Hill, D.J.; Heine, R.G.; Hosking, C.S. The diagnostic value of skin prick testing in children with food allergy. Pediatr. Allergy Immunol. 2004, 15, 435–441. [Google Scholar] [CrossRef]
- Toews, A.D.; Ray, R.B.; Goines, N.D.; Bouldin, T.W. Increased synthesis of membrane macromolecules is an early response of retinal neurons to trimethyltin intoxication. Brain Res. 1986, 398, 298–304. [Google Scholar] [CrossRef]
- Roberts, G.; Lack, G. The Avon Longitudinal Study of Pa Diagnosing peanut allergy with skin prick and specific IgE testing. J. Allergy Clin. Immunol. 2005, 115, 1291–1296. [Google Scholar] [CrossRef]
- Tourlas, K.; Burman, D. Allergy Testing. Prim. Care 2016, 43, 363–374. [Google Scholar] [CrossRef]
- Werther, R.L.; Choo, S.; Lee, K.J.; Poole, D.; Allen, K.J.; Tang, M.L. Variability in Skin Prick Test Results Performed by Multiple Operators Depends on the Device Used. World Allergy Organ. J. 2012, 5, 200–204. [Google Scholar] [CrossRef]
- Kraft, M.T.; Coletta, C.; Mikhail, I.; Prince, B.T.; Scherzer, R.; Eisner, M.; Mustillo, P. The sensitivity of food allergy skin testing soon after food-induced anaphylaxis. J. Allergy Clin. Immunol. Pract. 2024, 12, 1384–1386.e2. [Google Scholar] [CrossRef]
- Alnæs, M. Anaphylaxis following prick-by-prick testing with peanut. Clin. Case Rep. 2020, 8, 2366–2368. [Google Scholar] [CrossRef]
- Kansen, H.M.; van Erp, F.C.; Meijer, Y.; Gorissen, D.M.; Stadermann, M.; van Velzen, M.F.; Keusters, W.R.; Frederix, G.W.; Knulst, A.C.; van der, C.K.; et al. Diagnostic accuracy of Ara h 2 for detecting peanut allergy in children. Clin. Exp. Allergy 2021, 51, 1069–1079. [Google Scholar] [CrossRef]
- Hemmings, O.; Du Toit, G.; Radulovic, S.; Lack, G.; Santos, A.F. Ara h 2 is the dominant peanut allergen despite similarities with Ara h 6. J. Allergy Clin. Immunol. 2020, 146, 621–630.e5. [Google Scholar] [CrossRef]
- Ji, C.; Huang, Y.; Yeung, L.H.; Hemmings, O.; Jama, Z.; Kwok, M.; Lack, G.; Santos, A.F. Ara h 2-Specific IgE Presence Rather Than Its Function Is the Best Predictor of Mast Cell Activation in Children. J. Allergy Clin. Immunol. Pract. 2023, 11, 1154–1161.e3. [Google Scholar] [CrossRef]
- Wood, R.A.; Segall, N.; Ahlstedt, S.; Williams, P.B. Accuracy of IgE antibody laboratory results. Ann. Allergy Asthma Immunol. 2007, 99, 34–41. [Google Scholar] [CrossRef]
- Anvari, S.; Miller, J.; Yeh, C.-Y.; Davis, C.M. IgE-Mediated Food Allergy. Clin. Rev. Allergy Immunol. 2019, 57, 244–260. [Google Scholar] [CrossRef]
- Klemans, R.J.; Otte, D.; Knol, M.; Knol, E.F.; Meijer, Y.; Gmelig-Meyling, F.H.; Bruijnzeel-Koomen, C.A.; Knulst, A.C.; Pasmans, S.G. The diagnostic value of specific IgE to Ara h 2 to predict peanut allergy in children is comparable to a validated and updated diagnostic prediction model. J. Allergy Clin. Immunol. 2013, 131, 157–163. [Google Scholar] [CrossRef]
- LaHood, N.A.; Patil, S.U. Food Allergy Testing. Clin. Lab. Med. 2019, 39, 625–642. [Google Scholar] [CrossRef]
- Kulis, M.; Wright, B.L.; Jones, S.M.; Burks, A.W. Diagnosis, management, and investigational therapies for food allergies. Gastroenterology 2015, 148, 1132–1142. [Google Scholar] [CrossRef]
- Sampson, H.A. Update on food allergy. J. Allergy Clin. Immunol. 2004, 113, 805–819, quiz 820. [Google Scholar] [CrossRef]
- Kwong, K.Y.; Lu, Y.Z. Cost of Serum Versus Skin Allergy Testing Among Medicare Fee-for-Service Beneficiaries in the United States. J. Health Econ. Outcomes Res. 2023, 10, 14–21. [Google Scholar] [CrossRef]
- Fuhrmann, V.; Huang, H.-J.; Akarsu, A.; Shilovskiy, I.; Elisyutina, O.; Khaitov, M.; van Hage, M.; Linhart, B.; Focke-Tejkl, M.; Valenta, R.; et al. From Allergen Molecules to Molecular Immunotherapy of Nut Allergy: A Hard Nut to Crack. Front. Immunol. 2021, 12, 742732. [Google Scholar] [CrossRef]
- Matricardi, P.M.; Kleine-Tebbe, J.; Hoffmann, H.J.; Valenta, R.; Hilger, C.; Hofmaier, S.; Aalberse, R.C.; Agache, I.; Asero, R.; Ballmer-Weber, B.; et al. EAACI Molecular Allergology User’s Guide. Pediatr. Allergy Immunol. 2016, 27 (Suppl. S23), 1–250. [Google Scholar] [CrossRef]
- Chinthrajah, R.S.; Hernandez, J.D.; Boyd, S.D.; Galli, S.J.; Nadeau, K.C. Molecular and cellular mechanisms of food allergy and food tolerance. J. Allergy Clin. Immunol. 2016, 137, 984–997. [Google Scholar] [CrossRef]
- Połomska, J.; Dydak, P.; Sozańska, B.; Sikorska-Szaflik, H. Peanut Allergy and Component-Resolved Diagnostics Possibilities—What Are the Benefits? Nutrients 2023, 15, 5132. [Google Scholar] [CrossRef]
- Valcour, A.; Jones, J.E.; Lidholm, J.; Borres, M.P.; Hamilton, R.G. Sensitization profiles to peanut allergens across the United States. Ann. Allergy Asthma Immunol. 2017, 119, 262–266.e1. [Google Scholar] [CrossRef]
- Vereda, A.; van Hage, M.; Ahlstedt, S.; Ibañez, M.D.; Cuesta-Herranz, J.; van Odijk, J.; Wickman, M.; Sampson, H.A. Peanut allergy: Clinical and immunologic differences among patients from 3 different geographic regions. J. Allergy Clin. Immunol. 2010, 127, 603–607. [Google Scholar] [CrossRef]
- van Erp, F.C.; Klemans, R.J.B.; Meijer, Y.; van der Ent, C.K.; Knulst, A.C. Using Component-Resolved Diagnostics in the Management of Peanut-Allergic Patients. Curr. Treat. Options Allergy 2016, 3, 169–180. [Google Scholar] [CrossRef]
- Sampson, H.A.; van Wijk, R.G.; Bindslev-Jensen, C.; Sicherer, S.; Teuber, S.S.; Burks, A.W.; Dubois, A.E.J.; Beyer, K.; Eigenmann, P.A.; Spergel, J.M.; et al. Standardizing double-blind, placebo-controlled oral food challenges: American Academy of Allergy, Asthma & Immunology–European Academy of Allergy and Clinical Immunology PRACTALL consensus report. J. Allergy Clin. Immunol. 2012, 130, 1260–1274. [Google Scholar] [CrossRef]
- Perry, T.T.; Matsui, E.C.; Conover-Walker, M.K.; Wood, R.A. Risk of oral food challenges. J. Allergy Clin. Immunol. 2004, 114, 1164–1168. [Google Scholar] [CrossRef]
- Upton, J.; Alvaro, M.; Nadeau, K. A perspective on the pediatric death from oral food challenge reported from the Allergy Vigilance Network. Allergy 2019, 74, 1035–1036. [Google Scholar] [CrossRef]
- Bird, J.A.; Leonard, S.; Groetch, M.; Assa’Ad, A.; Cianferoni, A.; Clark, A.; Crain, M.; Fausnight, T.; Fleischer, D.; Green, T.; et al. Conducting an Oral Food Challenge: An Update to the 2009 Adverse Reactions to Foods Committee Work Group Report. J. Allergy Clin. Immunol. Pract. 2020, 8, 75–90.e17. [Google Scholar] [CrossRef]
- Turner, P.J.; Jerschow, E.; Umasunthar, T.; Lin, R.; Campbell, D.E.; Boyle, R.J. Fatal Anaphylaxis: Mortality Rate and Risk Factors. J. Allergy Clin. Immunol. Pract. 2017, 5, 1169–1178. [Google Scholar] [CrossRef]
- Suárez-Fariñas, M.; Suprun, M.; Kearney, P.; Getts, R.; Grishina, G.; Hayward, C.; Luta, D.; Porter, A.; Witmer, M.; du Toit, G.; et al. Accurate and reproducible diagnosis of peanut allergy using epitope mapping. Allergy 2021, 76, 3789–3797. [Google Scholar] [CrossRef]
- Huang, H.-J.; Campana, R.; Akinfenwa, O.; Curin, M.; Sarzsinszky, E.; Karsonova, A.; Riabova, K.; Karaulov, A.; Niespodziana, K.; Elisyutina, O.; et al. Microarray-Based Allergy Diagnosis: Quo Vadis? Front. Immunol. 2020, 11, 594978. [Google Scholar] [CrossRef]
- Jakob, T.; Forstenlechner, P.; Matricardi, P.; Kleine-Tebbe, J. Molecular allergy diagnostics using multiplex assays: Methodological and practical considerations for use in research and clinical routine. Allergo J. Int. 2015, 24, 320–332. [Google Scholar] [CrossRef]
- Hamilton, R.G. Microarray Technology Applied to Human Allergic Disease. BioTech 2017, 6, 3. [Google Scholar] [CrossRef]
- Borges, J.-P.; Gironde, C.; Trouilh, L.; Berteloite, B.; Apoil, A.P.; Rouge, P.; Barre, A.; Trevisiol, E. Allergenarrays: Low-cost food allergy diagnosis using allergen extracts-based microarrays. Clin. Transl. Allergy 2014, 4, P37. [Google Scholar] [CrossRef]
- Hemmings, O.; Kwok, M.; McKendry, R.; Santos, A.F. Basophil Activation Test: Old and New Applications in Allergy. Curr. Allergy Asthma Rep. 2018, 18, 77. [Google Scholar] [CrossRef]
- Jaumdally, H.; Kwok, M.; Jama, Z.; Hesse-Lamptey, R.; McKendry, R.; Galvez, O.; Daniel, Y.; Santos, A.F. Basophil activation test has high reproducibility and is feasible in the clinical setting. Pediatr. Allergy Immunol. 2022, 33, e13870. [Google Scholar] [CrossRef]
- Mari, A.; Iacovacci, P.; Afferni, C.; Barletta, B.; Tinghino, R.; Di Felice, G.; Pini, C. Specific IgE to cross-reactive carbohydrate determinants strongly affect the in vitro diagnosis of allergic diseases. J. Allergy Clin. Immunol. 1999, 103, 1005–1011. [Google Scholar] [CrossRef]
- Glaumann, S.; Nopp, A.; Johansson, S.G.O.; Rudengren, M.; Borres, M.P.; Nilsson, C. Basophil allergen threshold sensitivity, CD-sens, IgE-sensitization and DBPCFC in peanut-sensitized children. Allergy 2012, 67, 242–247. [Google Scholar] [CrossRef]
- Duan, L.; Celik, A.; Hoang, J.A.; Schmidthaler, K.; So, D.; Yin, X.; Ditlof, C.M.; Ponce, M.; Upton, J.E.; Lee, J.; et al. Basophil activation test shows high accuracy in the diagnosis of peanut and tree nut allergy: The Markers of Nut Allergy Study. Allergy 2021, 76, 1800–1812. [Google Scholar] [CrossRef]
- Chirumbolo, S. Immunotherapy in allergy and cellular tests. Hum. Vaccines Immunother. 2014, 10, 1595–1610. [Google Scholar] [CrossRef]
- Hoffmann, H.J.; Santos, A.F.; Mayorga, C.; Nopp, A.; Eberlein, B.; Ferrer, M.; Rouzaire, P.; Ebo, D.G.; Sabato, V.; Sanz, M.L.; et al. The clinical utility of basophil activation testing in diagnosis and monitoring of allergic disease. Allergy 2015, 70, 1393–1405. [Google Scholar] [CrossRef]
- Santos, A.F.; Du Toit, G.; Douiri, A.; Radulovic, S.; Stephens, A.; Turcanu, V.; Lack, G. Distinct parameters of the basophil activation test reflect the severity and threshold of allergic reactions to peanut. J. Allergy Clin. Immunol. 2015, 135, 179–186. [Google Scholar] [CrossRef]
- Noriega, D.B.; Teodorowicz, M.; Savelkoul, H.; Ruinemans-Koerts, J. The Basophil Activation Test for Clinical Management of Food Allergies: Recent Advances and Future Directions. J. Asthma Allergy 2021, 14, 1335–1348. [Google Scholar] [CrossRef]
- Piletta-Zanin, A.; Ricci, C.; Santos, A.F.; Eigenmann, P.A. BAT and MAT for diagnosis of peanut allergy: A systematic review and meta-analysis. Pediatr. Allergy Immunol. 2024, 35, e14140. [Google Scholar] [CrossRef]
- Santos, A.F.; Couto-Francisco, N.; Bécares, N.; Kwok, M.; Bahnson, H.T.; Lack, G. A novel human mast cell activation test for peanut allergy. J. Allergy Clin. Immunol. 2018, 142, 689–691.e9. [Google Scholar] [CrossRef]
- Bachmeier-Zbären, N.; Celik, A.; van Brummelen, R.; Roos, N.; Steinmann, M.; Hoang, J.A.; Yin, X.; Ditlof, C.M.; Duan, L.; Upton, J.E.M.; et al. Clinical utility analysis of the Hoxb8 mast cell activation test for the diagnosis of peanut allergy. Allergy 2024, 80, 215–226. [Google Scholar] [CrossRef]
- Brown, S.J.; Asai, Y.; Cordell, H.J.; Campbell, L.E.; Zhao, Y.; Liao, H.; Northstone, K.; Henderson, J.; Alizadehfar, R.; Ben-Shoshan, M.; et al. Loss-of-function variants in the filaggrin gene are a significant risk factor for peanut allergy. J. Allergy Clin. Immunol. 2011, 127, 661–667. [Google Scholar] [CrossRef]
- Donovan, G.R.; Manolios, N.; Weiner, J.M.; Grennan, D.; Huang, Q.; Dunckley, H.; Baldo, B.A. A family study of allergy: Segregation with HLA but not with T-cell receptor genes. J. Allergy Clin. Immunol. 1996, 97, 712–713. [Google Scholar] [CrossRef]
- Kostara, M.; Chondrou, V.; Sgourou, A.; Douros, K.; Tsabouri, S. HLA Polymorphisms and Food Allergy Predisposition. J. Pediatr. Genet. 2020, 9, 77–86. [Google Scholar] [CrossRef]
- Rodríguez, E.; Baurecht, H.; Herberich, E.; Wagenpfeil, S.; Brown, S.J.; Cordell, H.J.; Irvine, A.D.; Weidinger, S. Meta-analysis of filaggrin polymorphisms in eczema and asthma: Robust risk factors in atopic disease. J. Allergy Clin. Immunol. 2009, 123, 1361–1370.e7. [Google Scholar] [CrossRef]
- Clark, A.; Mangat, J.; King, Y.; Islam, S.; Anagnostou, K.; Foley, L.; Deighton, J.; Ewan, P. Thermographic imaging during nasal peanut challenge may be useful in the diagnosis of peanut allergy. Allergy 2012, 67, 574–576. [Google Scholar] [CrossRef]
- Joshi, A.A.; Peczuh, M.W.; Kumar, C.V.; Rusling, J.F. Ultrasensitive carbohydrate-peptide SPR imaging microarray for diagnosing IgE mediated peanut allergy. Analyst 2014, 139, 5728–5733. [Google Scholar] [CrossRef]
- Rath, T.; Dieterich, W.; Kätscher-Murad, C.; Neurath, M.F.; Zopf, Y. Cross-sectional imaging of intestinal barrier dysfunction by confocal laser endomicroscopy can identify patients with food allergy in vivo with high sensitivity. Sci. Rep. 2021, 11, 12777. [Google Scholar] [CrossRef]
- Peppers, B.P.; Jhaveri, D.; Van Heeckeren, R.; Fletcher, D.; Sutton, M.; Hostoffer, R.W.; Bonfield, T. Stratification of peanut allergic murine model into anaphylaxis severity risk groups using thermography. J. Immunol. Methods 2018, 459, 29–34. [Google Scholar] [CrossRef]
- Manzano-Szalai, K.; Pali-Schöll, I.; Krishnamurthy, D.; Stremnitzer, C.; Flaschberger, I.; Jensen-Jarolim, E. Anaphylaxis Imaging: Non-Invasive Measurement of Surface Body Temperature and Physical Activity in Small Animals. PLoS ONE 2016, 11, e0150819. [Google Scholar] [CrossRef]
- van Breugel, M.; Fehrmann, R.S.N.; Bügel, M.; Rezwan, F.I.; Holloway, J.W.; Nawijn, M.C.; Fontanella, S.; Custovic, A.; Koppelman, G.H. Current state and prospects of artificial intelligence in allergy. Allergy 2023, 78, 2623–2643. [Google Scholar] [CrossRef]
- Ferrante, G.; Licari, A.; Fasola, S.; Marseglia, G.L.; La Grutta, S. Artificial intelligence in the diagnosis of pediatric allergic diseases. Pediatr. Allergy Immunol. 2021, 32, 405–413. [Google Scholar] [CrossRef]
- Alag, A. Machine learning approach yields epigenetic biomarkers of food allergy: A novel 13-gene signature to diagnose clinical reactivity. PLoS ONE 2019, 14, e0218253. [Google Scholar] [CrossRef]
- Bahri, R.; Custovic, A.; Korosec, P.; Tsoumani, M.; Barron, M.; Wu, J.; Sayers, R.; Weimann, A.; Ruiz-Garcia, M.; Patel, N.; et al. Mast cell activation test in the diagnosis of allergic disease and anaphylaxis. J. Allergy Clin. Immunol. 2018, 142, 485–496.e16. [Google Scholar] [CrossRef]
- Chong, K.W.; Ruiz-Garcia, M.; Patel, N.; Boyle, R.J.; Turner, P.J. Reaction phenotypes in IgE-mediated food allergy and anaphylaxis. Ann. Allergy Asthma Immunol. 2020, 124, 473–478. [Google Scholar] [CrossRef]
- MacMath, D.; Chen, M.; Khoury, P. Artificial Intelligence: Exploring the Future of Innovation in Allergy Immunology. Curr. Allergy Asthma Rep. 2023, 23, 351–362. [Google Scholar] [CrossRef]
- Metwally, A.A.; Yu, P.S.; Reiman, D.; Dai, Y.; Finn, P.W.; Perkins, D.L. Utilizing longitudinal microbiome taxonomic profiles to predict food allergy via Long Short-Term Memory networks. PLoS Comput. Biol. 2019, 15, e1006693. [Google Scholar] [CrossRef]
- Suprun, M.; Sicherer, S.H.; Wood, R.A.; Jones, S.M.; Leung, D.Y.; Henning, A.K.; Dawson, P.; Burks, A.W.; Lindblad, R.; Getts, R.; et al. Early epitope-specific IgE antibodies are predictive of childhood peanut allergy. J. Allergy Clin. Immunol. 2020, 146, 1080–1088. [Google Scholar] [CrossRef]
- Tang, S.K.; Castaño, N.; Nadeau, K.C.; Galli, S.J. Can artificial intelligence (AI) replace oral food challenge? J. Allergy Clin. Immunol. 2024, 153, 666–668. [Google Scholar] [CrossRef]
| Diagnostic Method | Description | Advantages | Limitations | PPV/Sensitivity/Specificity | Duration | Expense | Sample Requirement |
|---|---|---|---|---|---|---|---|
| Skin Prick Test | A small puncture on the skin introduces allergens to observe for a reaction. | Quick, cost-effective, safe, and easy to perform. | Requires avoidance of certain medications, can produce false positives. | Sensitivity >90%; PPV ~95% (≥8 mm) | Immediate | + | No |
| Serum IgE Testing | Measures allergen-specific IgE levels in the blood. | Can be performed even if on antihistamines, more precise. | May produce false positives, longer wait time for results. | Sensitivity ~95%; PPV ~95% (≥14 kU/L) | Days | ++ | Yes, serum required |
| Oral Food Challenge | Patient consumes suspected allergen under medical supervision. | Gold standard for diagnosis, directly tests clinical reactivity. | Time-consuming, high risk of severe reactions, stressful for patients. | Sensitivity/Specificity ~100% | 2–4 h | ++++ | No |
| Bead-Based Epitope Assay | Detects IgE binding to specific peanut epitopes using microbeads. | High sensitivity and specificity, less invasive than OFC. | Requires advanced lab facilities, higher cost than traditional methods. | Sensitivity 92%, Specificity 94%; PPV 91% | Hours | ++++ | Yes, serum or plasma sample required |
| Emerging Approach | Description | Advantages | Limitations | PPV/Sensitivity/Specificity | Duration | Expense | Sample Requirement |
|---|---|---|---|---|---|---|---|
| Molecular Allergy Diagnostics | Uses recombinant allergens to determine IgE sensitization at a molecular level. | Greater precision in identifying specific allergens, personalized treatment plans. | Higher cost, less widely available, requires specialized lab equipment. | PPV ~90%; Sensitivity ~93% | Hours | ++++ | Yes, serum required |
| Microarray Technology | Detects specific IgE antibodies to multiple allergens on a single chip. | High throughput, minimal serum required, detailed sensitization profile. | Higher costs compared to traditional methods, requires advanced technology. | Sensitivity ~90%; PPV ~85% | Hours | ++++ | Yes, serum required |
| Basophil Activation Test (BAT) | Measures basophil activation in response to allergens using flow cytometry. | Correlates closely with clinical symptoms, reduces false positives. | Requires specialized equipment and expertise, less widely available. | Sensitivity 92%, Specificity 94%; PPV 91% | 1–2 h | +++ | Yes, blood sample needed |
| Mast Cell Activation Test (MAT) | Measures mast cell activation after sensitization with patient sera. | High specificity (~98%), does not require fresh samples. | Underutilized, limited large-scale validation. | Sensitivity ~93%, Specificity ~96% | Hours | +++ | Yes, serum or plasma required |
| Genetic Testing | Identifies genetic predisposition to assess allergy severity. | Early identification of high-risk individuals, guides personalized care. | Still emerging, higher costs, not yet widely adopted in clinical practice. | Not widely standardized | Hours-Weeks | ++++ | Yes, genetic material needed |
| Novel Imaging Techniques | Non-invasive methods like thermographic imaging and SPRi to visualize allergic responses. | Safer, real-time insights, potential to replace riskier tests like OFC. | Still experimental, requires validation and standardization. | Sensitivity ~96%, Specificity ~90% | Minutes | ++++ | No |
| Machine Learning and AI | Analyzes large datasets to predict allergy risks and outcomes. | Enhances diagnostic accuracy, supports personalized treatment strategies. | Requires high-quality datasets, challenges with data privacy and reliability. | Dependent on algorithm validation | Variable | ++ | No |
| Diagnostic Method | Clinical Considerations | Potential Limitations |
|---|---|---|
| Skin Prick Test (SPT) | Should be avoided if the patient is on antihistamines, has severe eczema, or recent anaphylaxis. | False positives/negatives, affected by skin conditions, requires medication avoidance. |
| Serum IgE Testing | Useful when SPT is not feasible, interpretation requires clinical correlation. | False positives, long wait time for results, variable insurance coverage. |
| Oral Food Challenge (OFC) | Best conducted when other tests are inconclusive, requires careful monitoring. | High risk of severe reactions, not suitable for all patients, resource intensive. |
| Bead-Based Epitope Assay (BBEA) | May be used for detailed risk assessment, potentially replacing OFC for some patients. | Higher cost, less accessible, requires specialized lab settings. |
| Molecular Diagnostics | Ideal for personalized treatment plans, understanding specific allergenic components. | Costly, requires access to advanced diagnostic laboratories. |
| Basophil Activation Test (BAT) | Valuable for complex cases with ambiguous results, correlates with clinical symptoms. | Requires flow cytometry expertise, higher cost, less accessible. |
| Genetic Testing | Helps in early identification of at-risk individuals, supports personalized care. | Emerging technology, not yet widely available or adopted, higher cost. |
| Novel Imaging Techniques | Non-invasive and safer alternative to OFC, especially in high-risk patients. | Still in experimental stages, needs further validation and standardization. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Satnarine, T.; Makkoukdji, N.; Pundit, V.; Vignau, A.; Sharma, P.; Warren, D.; Kleiner, G.; Gans, M. Peanut Allergy Diagnosis: Current Practices, Emerging Technologies, and Future Directions. Allergies 2025, 5, 4. https://doi.org/10.3390/allergies5010004
Satnarine T, Makkoukdji N, Pundit V, Vignau A, Sharma P, Warren D, Kleiner G, Gans M. Peanut Allergy Diagnosis: Current Practices, Emerging Technologies, and Future Directions. Allergies. 2025; 5(1):4. https://doi.org/10.3390/allergies5010004
Chicago/Turabian StyleSatnarine, Travis, Nadia Makkoukdji, Valishti Pundit, Alexia Vignau, Pranav Sharma, Duenna Warren, Gary Kleiner, and Melissa Gans. 2025. "Peanut Allergy Diagnosis: Current Practices, Emerging Technologies, and Future Directions" Allergies 5, no. 1: 4. https://doi.org/10.3390/allergies5010004
APA StyleSatnarine, T., Makkoukdji, N., Pundit, V., Vignau, A., Sharma, P., Warren, D., Kleiner, G., & Gans, M. (2025). Peanut Allergy Diagnosis: Current Practices, Emerging Technologies, and Future Directions. Allergies, 5(1), 4. https://doi.org/10.3390/allergies5010004






