Optimization of Density Peak Clustering Algorithm Based on Improved Black Widow Algorithm
Abstract
:1. Introduction
- Intelligent Optimization with Sil Objective: By using an intelligent optimization algorithm with the Silhouette Coefficient as the objective, the BWDPC algorithm overcomes the problem of inaccurate density center selection in previous DPC algorithms, which could lead to chain errors in the clustering results.
- Improved Black Widow Algorithm: The traditional Black Widow Algorithm has been modified by incorporating search factors, making it more suitable for optimizing the DPC algorithm. Multiple rounds of swarm intelligence search have been conducted to address issues such as the algorithm’s limited search paths and slow convergence.
- Automatic Selection of “dc”: BWDPC requires only the initialization of “dc”, and then it automatically selects the appropriate “dc” value during the clustering process. This feature makes it well-suited for handling large-scale datasets.
2. Related Works
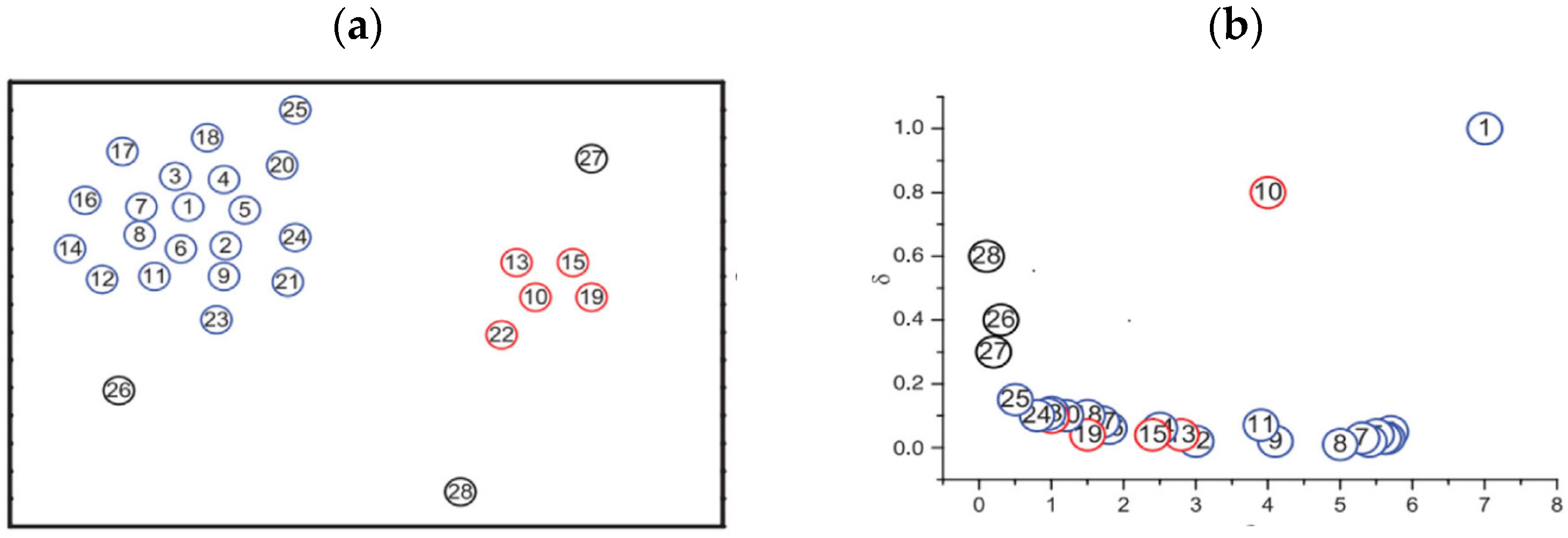
2.1. The DPC Algorithm
- Points around a clustering center have lower densities than the center itself.
- Clustering centers are farther away from other points with higher densities. The algorithm requires the input of a cutoff distance parameter dc. It then automatically selects clustering center points on the decision graph based on the given dc value. Afterward, using a one-step allocation strategy, it assigns the remaining points to the clusters represented by the identified centers to complete the clustering process.
2.1.1. The Relevant Parameters of the DPC Algorithm
- Calculate the local density () and relative distance (δ) of each sample point using Equations (1)–(5).
- Calculate the decision value for each sample point using Equation (6).
- Select the points with higher decision values as the cluster centers.
- Once the cluster centers are identified, allocate the remaining points in descending order of their local density. Each point is sequentially assigned to the cluster of the nearest preceding point in terms of relative distance.
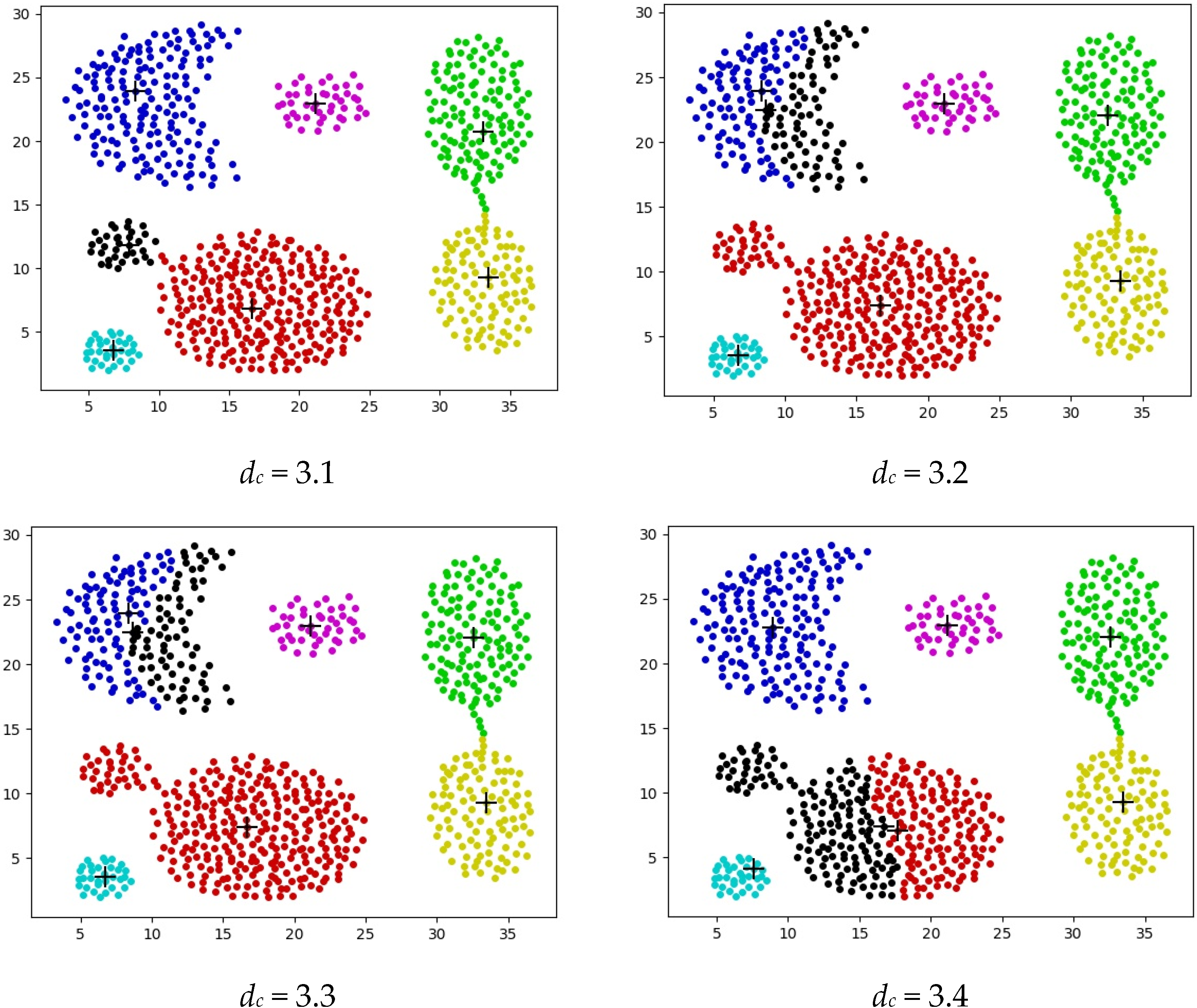
2.1.2. The Limitations of DPC
2.2. BWOA Algorithm
2.2.1. Spider Movement
2.2.2. Pheromone
2.3. Abbreviations and Their Descriptions
- (1)
- Silhouette Coefficient (Sil):
- (2)
- Fowlkes–Mallows Index (FMI):
- (3)
- The Adjusted Rand Index (RI):
- (4)
- Adjusted Mutual Information (AMI):
3. The Clustering of BWDPC
3.1. The Shortcomings of the BWOA
3.2. The RBWOA Model
3.3. Regarding the Pseudocode for BWDPC
| Algorithm 1: RBWOA algorithm |
8. fit = Sil // Calculate the fitness value “fit” using Sil 9. pheromone = (fitmax − fit) / (fitmax − fitmin) // Calculate the fitness value “fit” using Sil is less than or equal to 0, it indicates the current population Si has been searched, and iterate to the next population Si + 1. 14. u = 15. |
3.4. Algorithm Flow Steps
| Algorithm 2: BWDPC algorithm |
| Input: Experimental Dataset X = {x1, x2, …, xn} Output: Clustering Results C = {c1, c2, …, cm}, m Is the Number of Data Cluster Results 1. Set the population size S and the maximum number of iterations T for the BWDPC algorithm 2. Data preprocessing: Calculate the distance matrix for all data points and determine the range of dc values 3. Enter S into BWDPC and set the output x of BWDPC to dc 4. Substitute dc into equations 4 and 5 to calculate the local density i and for all points 5. Formula (6) is employed to calculate γi, and the initial m points with the highest γi values are automatically chosen as the cluster centers 6. Introduce the evaluation metric Sil as the objective function for BWDPC and record the dc value d* corresponding to the maximum Sil 7. Check if the termination condition is met. If t > T, end the iteration and proceed to step 8. If not, go back to step 3 for further optimization 9. Output the optimal dc value and obtain the final clustering results to complete the clustering process |
3.5. Algorithm Time Complexity
4. Experimental Results and Analysis
4.1. Experimental Dataset and Experimental Environment
4.2. Experiments on Synthetic Datasets
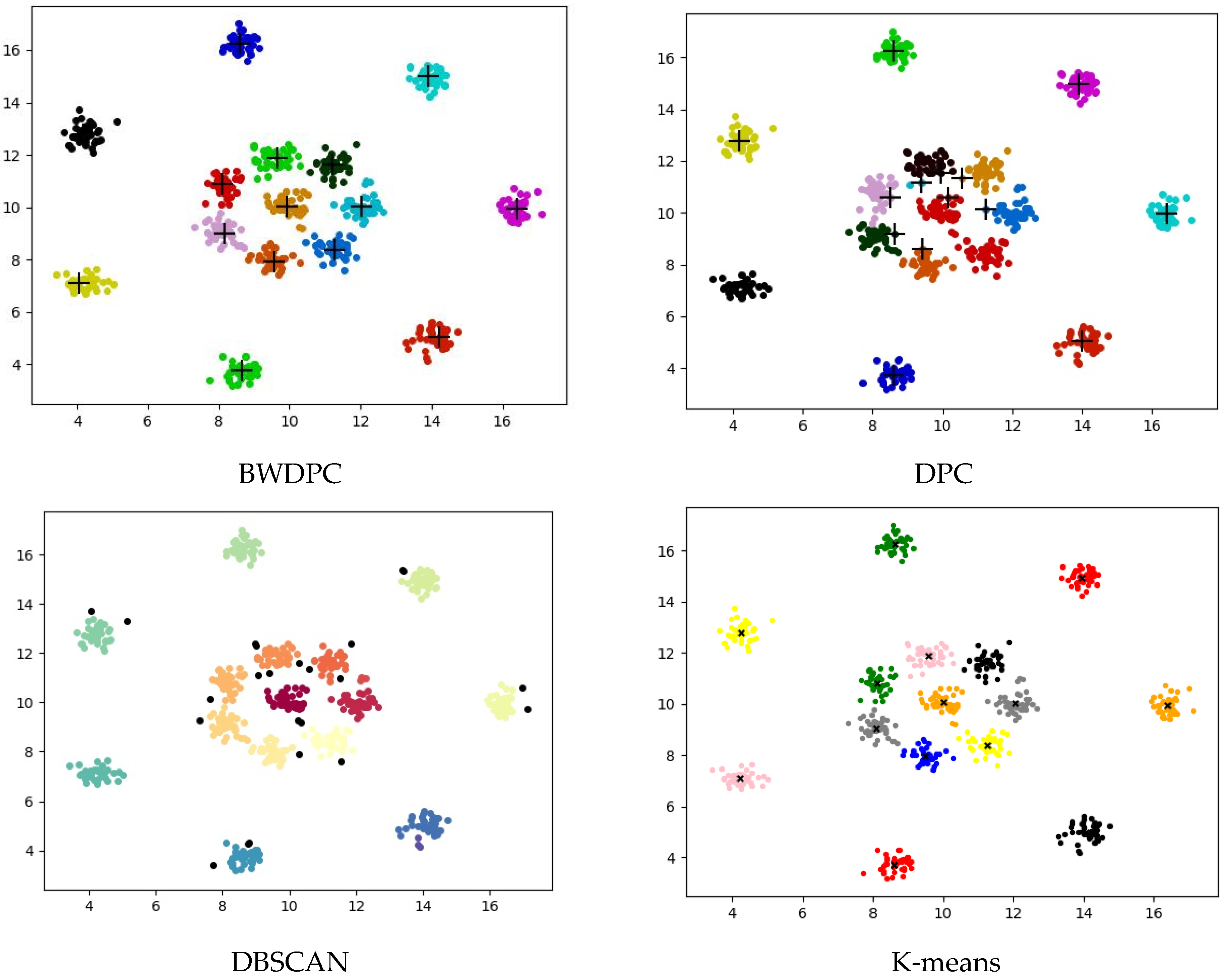
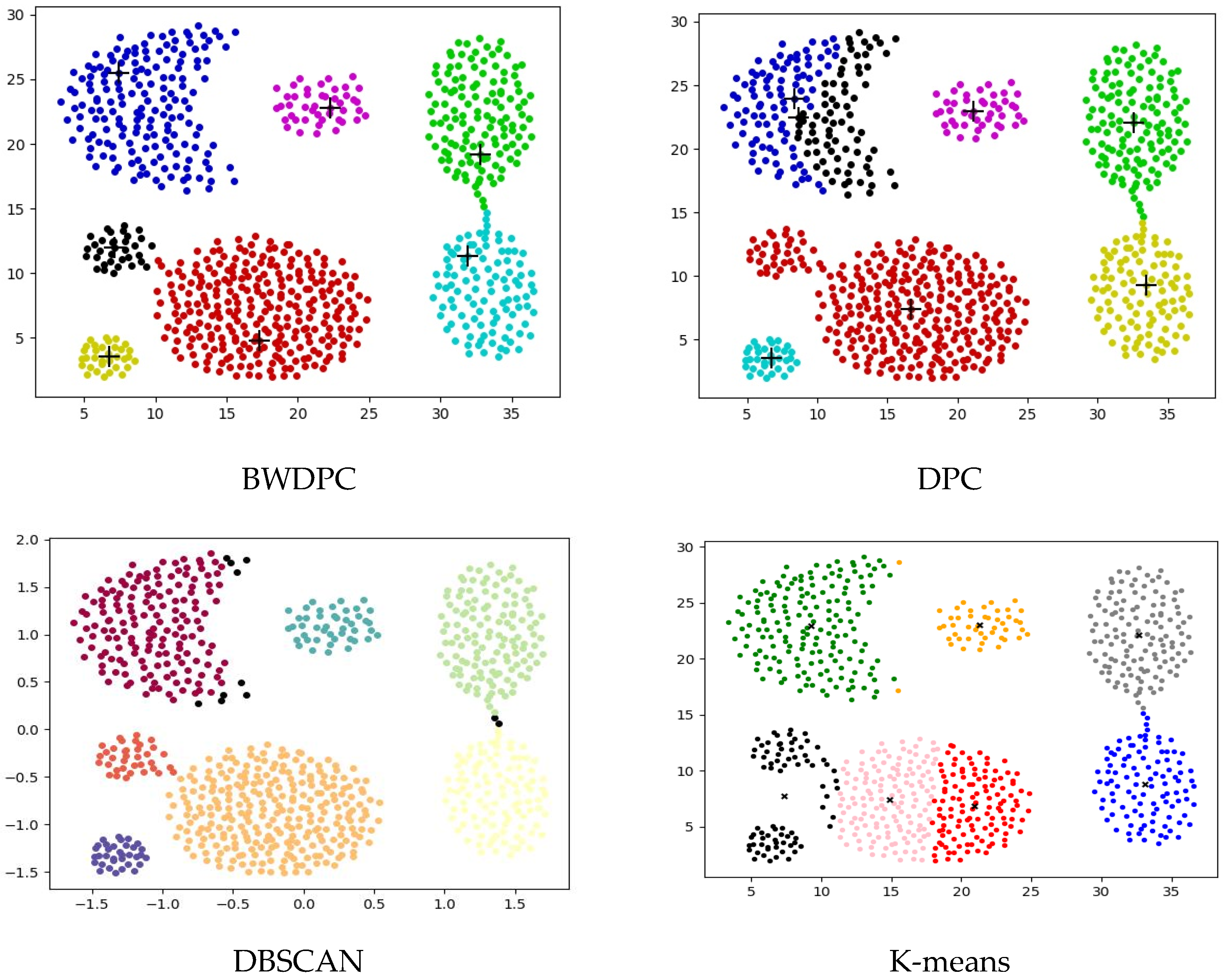
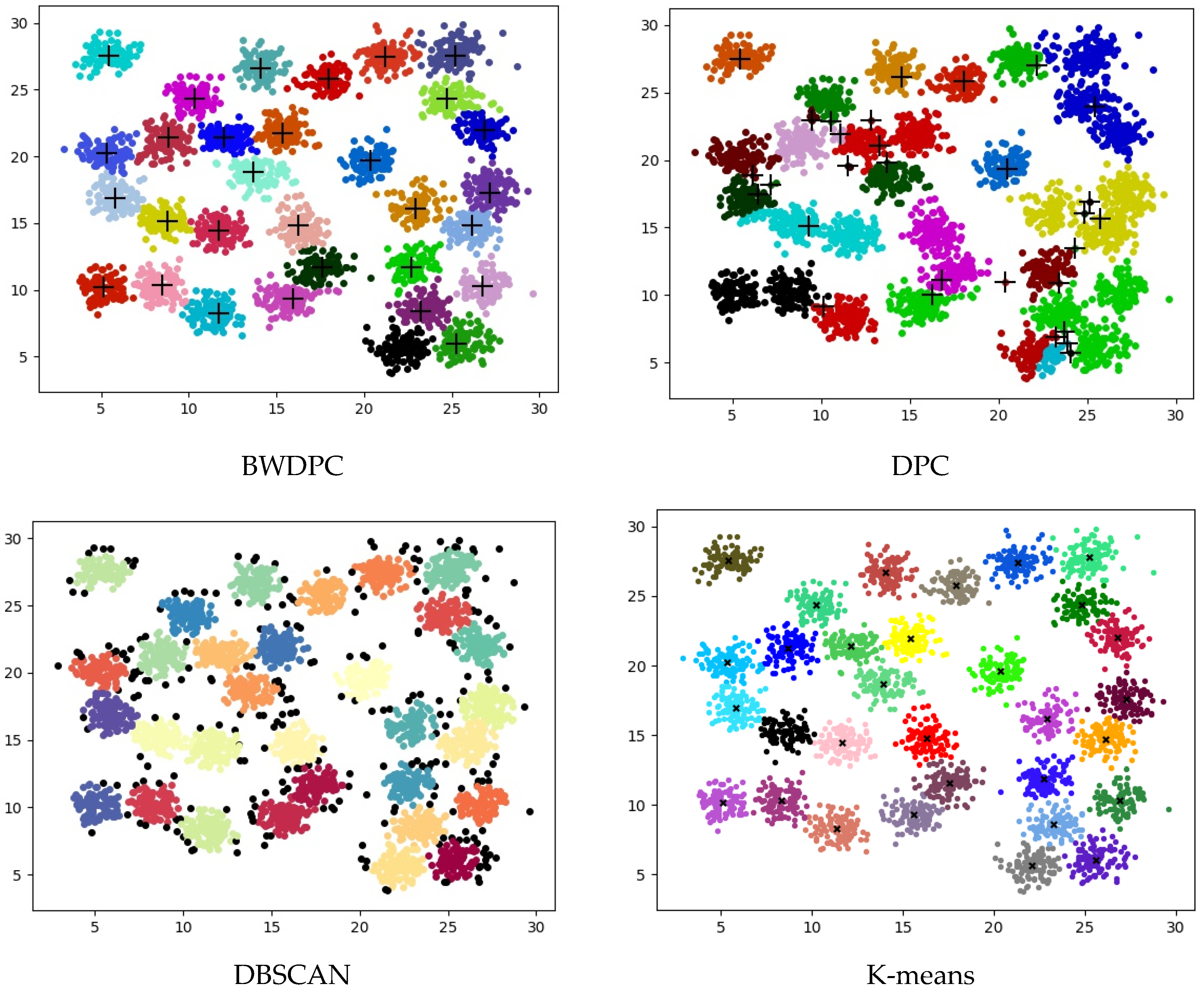
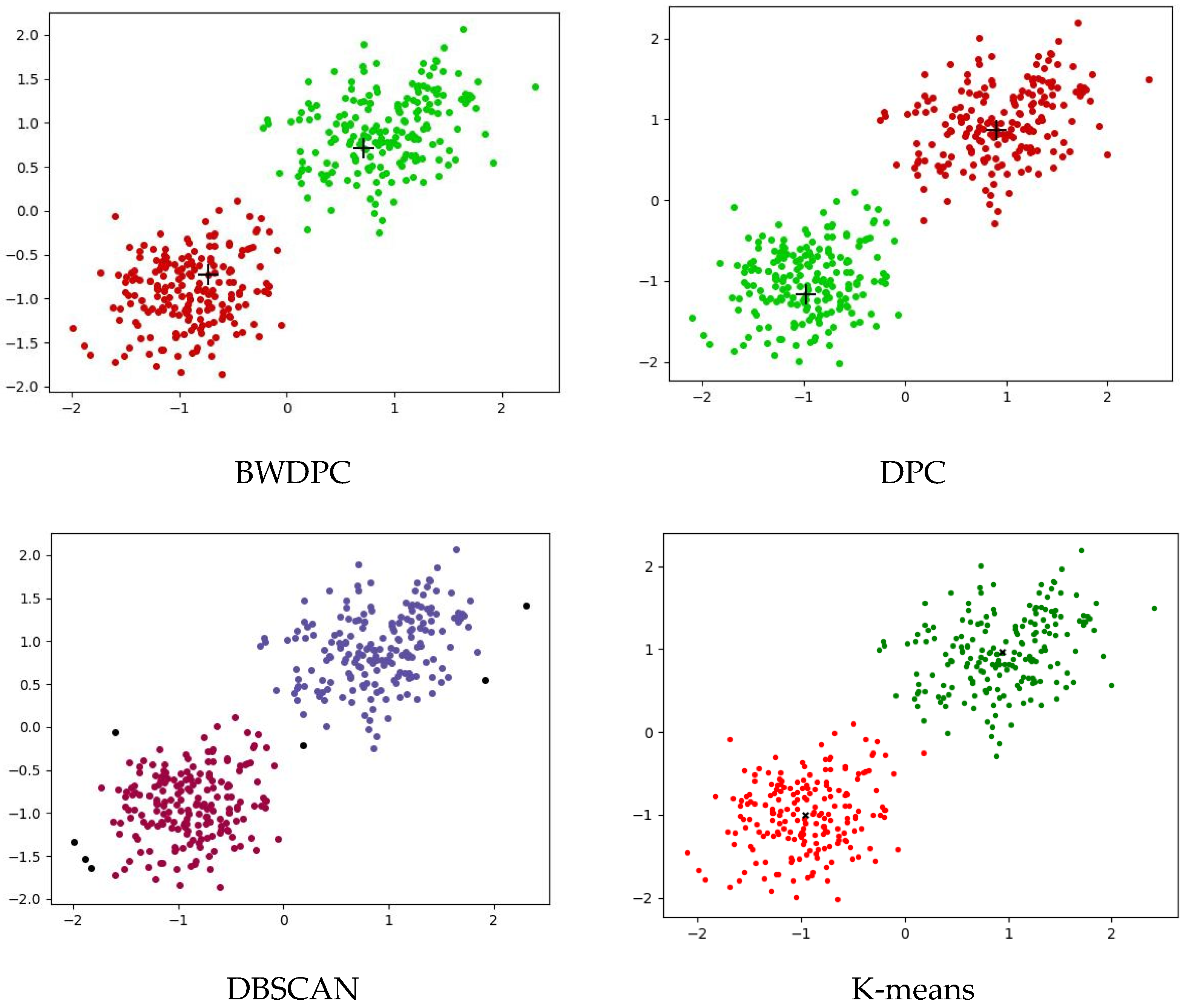
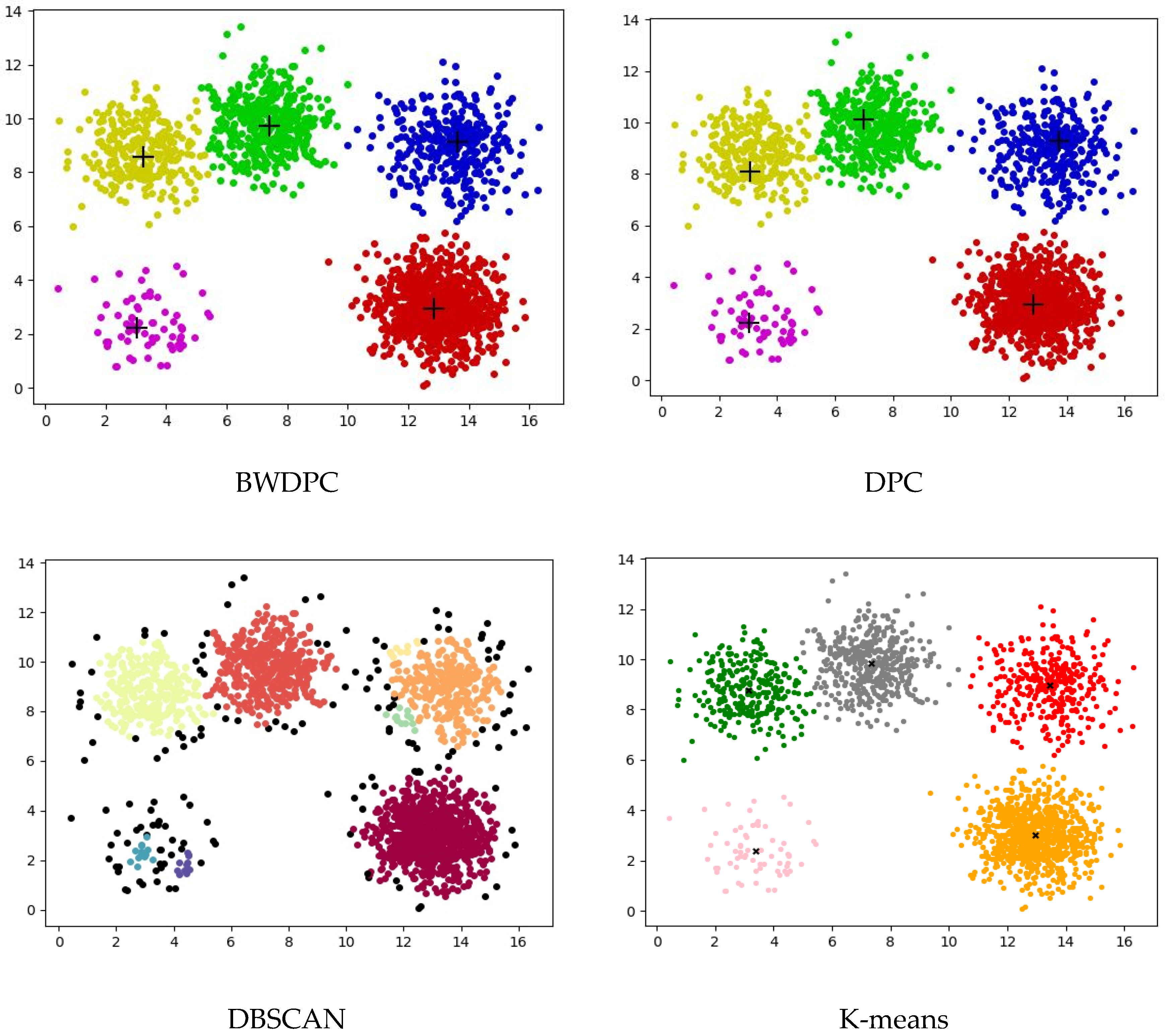
4.3. Experiments on Real-World Datasets
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Ding, S.F.; Du, W.; Xu, X.; Shi, T.; Wang, Y.; Li, C. An improved density peaks clustering algorithm based on natural neighbor with a merging strategy. Inf. Sci. 2023, 624, 252–276. [Google Scholar] [CrossRef]
- Guan, J.; Li, S.; He, X.; Chen, J. Clustering by fast detection of main density peaks within a peak digraph. Inf. Sci. 2023, 628, 504–521. [Google Scholar] [CrossRef]
- Shi, T.H.; Ding, S.; Xu, X.; Ding, L. A community detection algorithm based on Quasi-Laplacian centrality peaks clustering. Appl. Intell. 2021, 51, 1–16. [Google Scholar] [CrossRef]
- Gao, M.; Shi, G.-Y. Ship-handling behavior pattern recognition using AIS sub-trajectory clustering analysis based on the T-SNE and spectral clustering algorithms. Ocean Eng. 2020, 205, 106919. [Google Scholar] [CrossRef]
- Yan, X.Q.; Ye, Y.; Qiu, X.; Yu, H. Synergetic information bottleneck for joint multi-view and ensemble clustering. Inf. Fusion 2020, 56, 15–27. [Google Scholar] [CrossRef]
- Morris, K.; McNicholas, P.D. Clustering, classification, discriminant analysis, and dimension reduction via generalized hyperbolic mixtures. Comput. Stat. Data Anal. 2016, 97, 133–150. [Google Scholar] [CrossRef]
- Lv, Z.; Di, L.; Chen, C.; Zhang, B.; Li, N. A Fast Density Peak Clustering Method for Power Data Security Detection Based on Local Outlier Factors. Processes 2023, 11, 2036. [Google Scholar] [CrossRef]
- Guha, S.; Rastogi, R.; Shim, K. CURE: An efficient clustering algorithm for large databases. ACM Sigmod Rec. 1998, 27, 73–84. [Google Scholar] [CrossRef]
- Zhang, T.; Ramakrishnan, R.; Livny, M. BIRCH: An efficient data clustering method for very large databases. ACM Sigmod Rec. 1996, 25, 103–114. [Google Scholar] [CrossRef]
- MacQueen, J. Some Methods for Classification and Analysis of Multivariate Observations. In Proceedings of the Fifth Berkeley Symposium on Mathematical Statistics and Probability; University of California Press: Los Angeles, CA, USA, 1967; Volume 1, pp. 281–297. [Google Scholar]
- Park, H.S.; Jun, C.-H. A simple and fast algorithm for K-medoids clustering. Expert Syst. Appl. 2009, 36, 3336–3341. [Google Scholar] [CrossRef]
- Dempster, A.P.; Lanird, N.M.; Rubin, D.B. Maximum likelihood from incomplete data via the EM algorithm. J. R. Stat. Soc. Ser. B Methodol. 1977, 39, 1–22. [Google Scholar] [CrossRef]
- Ester, M.; Kriegel, H.-P.; Sander, J.; Xu, X. A density-based algorithm for discovering clusters in large spatial databases with noise. Kdd 1996, 96, 226–231. [Google Scholar]
- Ankerst, M.; Breunig, M.M.; Kriegel, H.-P.; Sander, J. OPTICS: Ordering points to identify the clustering structure. ACM Sigmod 1999, 28, 49–60. [Google Scholar] [CrossRef]
- Ding, S.F.; Li, C.; Xu, X.; Ding, L.; Zhang, J.; Guo, L.L.; Shi, T. A sampling-based density peaks clustering algorithm for large-scale data. Pattern Recognit. 2023, 136, 109238. [Google Scholar] [CrossRef]
- Quyang, T.; Witold, P.; Nick, J.; Pizzi, N.J. Rule-based modeling with DBSCAN-based information granules. IEEE Trans. Cybern. 2019, 51, 3653–3663. [Google Scholar]
- Smiti, A.; Elouedi, Z. Dbscan-gm: An improved clustering method based on gaussian means and dbscan techniques. In Proceedings of the 2012 IEEE 16th International Conference on Intelligent Engineering Systems (INES), Lisbon, Portugal, 13–15 June 2012; pp. 573–578. [Google Scholar]
- Tran, T.N.; Drab, K.; Daszykowski, M. Revised DBSCAN algorithm to cluster data with dense adjacent clusters. Chemom. Intell. Lab. Syst. 2013, 120, 92–96. [Google Scholar] [CrossRef]
- Rodriguez, A.; Alessandro, L. Clustering by fast search and find of density peaks. Science 2014, 344, 1492–1496. [Google Scholar] [CrossRef]
- Pizzagalli, D.U.; Gonzalez, S.F.; Krause, R. A trainable clustering algorithm based on shortest paths from density peaks. Sci. Adv. 2019, 5, eaax3770. [Google Scholar] [CrossRef]
- Guan, J.Y.; Li, S.; He, X.; Chen, J. Peak-graph-based fast density peak clustering for image segmentation. IEEE Signal Process. Lett. 2021, 28, 897–901. [Google Scholar] [CrossRef]
- Chen, H.; Zhou, Y.; Mei, K.; Wang, N.; Tang, M.; Cai, G. An Improved Density Peak Clustering Algorithm Based on Chebyshev Inequality and Differential Privacy. Appl. Sci. 2023, 13, 8674. [Google Scholar] [CrossRef]
- Wu, Z.; Tingting, S.; Yanbing, Z. Quantum Density Peak Clustering Algorithm. Entropy 2022, 24, 237. [Google Scholar] [CrossRef] [PubMed]
- Jiang, R.; Jianhuab, J. Density Peaks Clustering Algorithm Based on CDbw and ABC Optimization. J. Jilin Univ. Sci. Ed. 2018, 56, 1469–1475. [Google Scholar]
- Li, F.; Yue, Q.; Pan, Z.; Sun, Y.; Yu, X. Dynamic particle swarm optimization algorithm based on automatic fast density peak clustering. J. Comput. Appl. 2023, 43, 154. [Google Scholar]
- Hayyolalam, V.; Kazem, A.A.P. Black widow optimization algorithm: A novel meta-heuristic approach for solving engineering optimization problems. Eng. Appl. Artif. Intell. 2020, 87, 103249. [Google Scholar] [CrossRef]
- Rousseeuw, P.J. Silhouettes: A graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math. 1987, 20, 53–65. [Google Scholar] [CrossRef]
- Fowlkes, E.B.; Mallows, C.L. A method for comparing two hierarchical clusterings. A Method Comp. Two Hierarchical Clust. 1983, 78, 553–569. [Google Scholar] [CrossRef]
- Vinh, N.X.; Epps, J.; Bailey, J. Information Theoretic Measures for Clusterings Comparison: Is A Correction for Chance Necessary? In Proceedings of the 26th Annual International Conference on Machine Learning, Montreal, QC, Canada, 14–18 June 2009; pp. 1073–1080. [Google Scholar]
- Han, J.; Moraga, C. The Influence of the Sigmoid Function Parameters on the Speed of Backpropagation Learning. In International Workshop on Artificial Neural Networks; Springer: Berlin/Heidelberg, Germany, 1995; pp. 195–201. [Google Scholar]
| Dataset | Instances | Attributes | Clusters |
|---|---|---|---|
| R15 | 600 | 2 | 15 |
| Aggregation | 788 | 2 | 7 |
| D31 | 3100 | 2 | 31 |
| Two_cluster | 400 | 2 | 2 |
| Five_cluster | 2000 | 2 | 5 |
| Flame | 240 | 2 | 2 |
| Iris | 150 | 4 | 3 |
| Wine | 178 | 13 | 3 |
| Sym | 350 | 2 | 3 |
| Waveform3 | 5000 | 21 | 3 |
| Segment | 2310 | 18 | 7 |
| Zoo | 266 | 2 | 3 |
| Dataset | BWDPC | DPC | DBSCAN | |
|---|---|---|---|---|
| dc | dc | eps | mpts | |
| R15 | 0.5533 | 2.0000 | 0.3200 | 3 |
| Aggregation | 3.1000 | 3.2000 | 1.7600 | 14 |
| D31 | 0.2121 | 2.4000 | 0.8000 | 24 |
| Two_cluster | 1.6802 | 1.6000 | 0.2500 | 2 |
| Five_cluster | 1.2126 | 1.3000 | 0.4200 | 7 |
| Flame | 0.3447 | 4.0000 | 1.4800 | 16 |
| Iris | 0.2932 | 3.0000 | 1.6300 | 2 |
| Wine | 32.3152 | 3.6000 | 4.3000 | 2 |
| Segment | 3.6890 | 1.6000 | 0.1000 | 2 |
| Waveform3 | 3.7420 | 0.1200 | 3.6000 | 2 |
| Zoo | 6.9286 | 1.6000 | 2.5000 | 6 |
| Sym | 0.0784 | 0.5000 | 0.1000 | 6 |
| Dataset | Metric | BWDPC | DPC | DBSCAN | K-Means |
|---|---|---|---|---|---|
| R15 | FMI | 0.9932 | 0.9220 | 0.9394 | 0.9932 |
| ARI | 0.9913 | 9.9144 | 0.9347 | 0.9927 | |
| AMI | 0.9967 | 0.9672 | 0.9450 | 0.9938 | |
| Aggregation | FMI | 0.9982 | 0.8796 | 0.9110 | 0.8159 |
| ARI | 0.9978 | 0.8454 | 0.8853 | 0.7624 | |
| AMI | 0.9956 | 0.9152 | 0.8865 | 0.8776 | |
| D31 | FMI | 0.9390 | 0.6040 | 0.7901 | 0.9538 |
| ARI | 0.9370 | 0.5369 | 0.7826 | 0.9522 | |
| AMI | 0.9558 | 0.8325 | 0.8882 | 0.9653 | |
| Two_cluster | FMI | 0.9950 | 0.9950 | 0.9651 | 0.9950 |
| ARI | 0.9900 | 0.9900 | 0.9315 | 0.9900 | |
| AMI | 0.9772 | 0.9772 | 0.8833 | 0.9772 | |
| Five_cluster | FMI | 0.9921 | 0.9316 | 0.9387 | 0.9940 |
| ARI | 0.9905 | 0.9033 | 0.9137 | 0.9915 | |
| AMI | 0.9754 | 0.8823 | 0.8480 | 0.9809 | |
| Flame | FMI | 0.7942 | 0.6002 | 0.7511 | 0.7363 |
| ARI | 0.5734 | −0.0302 | 0.5453 | 0.4534 | |
| AMI | 0.5672 | 0.1064 | 0.5985 | 0.3969 |
| Dataset | Metric | BWDPC | DPC | DBSCAN | K-Means |
|---|---|---|---|---|---|
| Iris | FMI | 0.8407 | 0.7567 | 0.7714 | 0.5835 |
| ARI | 0.7592 | 0.5609 | 0.5681 | 0.3711 | |
| AMI | 0.8032 | 0.7050 | 0.7316 | 0.4227 | |
| Wine | FMI | 0.5834 | 0.5674 | 0.5354 | 0.5039 |
| ARI | 0.3715 | 0.3016 | −0.0003 | 0.2536 | |
| AMI | 0.4131 | 0.4169 | 0.0502 | 0.3600 | |
| Segment | FMI | 0.4618 | 0.3808 | 0.3047 | 0.4370 |
| ARI | 0.3204 | 0.2013 | 0.0001 | 0.3133 | |
| AMI | 0.4927 | 0.3920 | 0.0780 | 0.4518 | |
| Waveform3 | FMI | 0.5338 | 0.5011 | 0.4152 | 0.5039 |
| ARI | 0.2817 | 0.1584 | 0.0029 | 0.2536 | |
| AMI | 0.3512 | 0.2467 | 0.0587 | 0.3620 | |
| Zoo | FMI | 0.8136 | 0.5076 | 0.4839 | 0.7741 |
| ARI | 0.7197 | 0.3796 | 0.0075 | 0.7087 | |
| AMI | 0.7598 | 0.5687 | 0.0079 | 0.7527 | |
| Sym | FMI | 0.8333 | 0.8239 | 0.4793 | 0.7369 |
| ARI | 0.7357 | 0.7178 | 0.2888 | 0.5335 | |
| AMI | 0.7727 | 0.7626 | 0.4719 | 0.5645 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, H.; Wu, H.; Wei, X.; Zhou, Y. Optimization of Density Peak Clustering Algorithm Based on Improved Black Widow Algorithm. Biomimetics 2024, 9, 3. https://doi.org/10.3390/biomimetics9010003
Huang H, Wu H, Wei X, Zhou Y. Optimization of Density Peak Clustering Algorithm Based on Improved Black Widow Algorithm. Biomimetics. 2024; 9(1):3. https://doi.org/10.3390/biomimetics9010003
Chicago/Turabian StyleHuang, Huajuan, Hao Wu, Xiuxi Wei, and Yongquan Zhou. 2024. "Optimization of Density Peak Clustering Algorithm Based on Improved Black Widow Algorithm" Biomimetics 9, no. 1: 3. https://doi.org/10.3390/biomimetics9010003
APA StyleHuang, H., Wu, H., Wei, X., & Zhou, Y. (2024). Optimization of Density Peak Clustering Algorithm Based on Improved Black Widow Algorithm. Biomimetics, 9(1), 3. https://doi.org/10.3390/biomimetics9010003






