Skeletal Muscle Segmentation at the Level of the Third Lumbar Vertebra (L3) in Low-Dose Computed Tomography: A Lightweight Algorithm
Abstract
1. Introduction
- (1)
- A lightweight image-processing algorithm is proposed for the automated segmentation of skeletal muscles at the L3 level in low-dose CT images.
- (2)
- The proposed algorithm is developed using a small, unlabeled dataset and can be efficiently run on a laptop without a graphic processing unit (GPU) device.
- (3)
- The proposed algorithm is validated on both plain (i.e., non-contrast) and contrast-enhanced L3 CT images.
- (4)
- The results indicate that the segmentation accuracy of the proposed algorithm is comparable to that of AutoMATICA, and close to the reference determined with the inter-observer variation.
2. Materials and Methods
2.1. Patients
2.2. Image Acquisition
2.3. Data Partitioning
2.4. Gold Standard
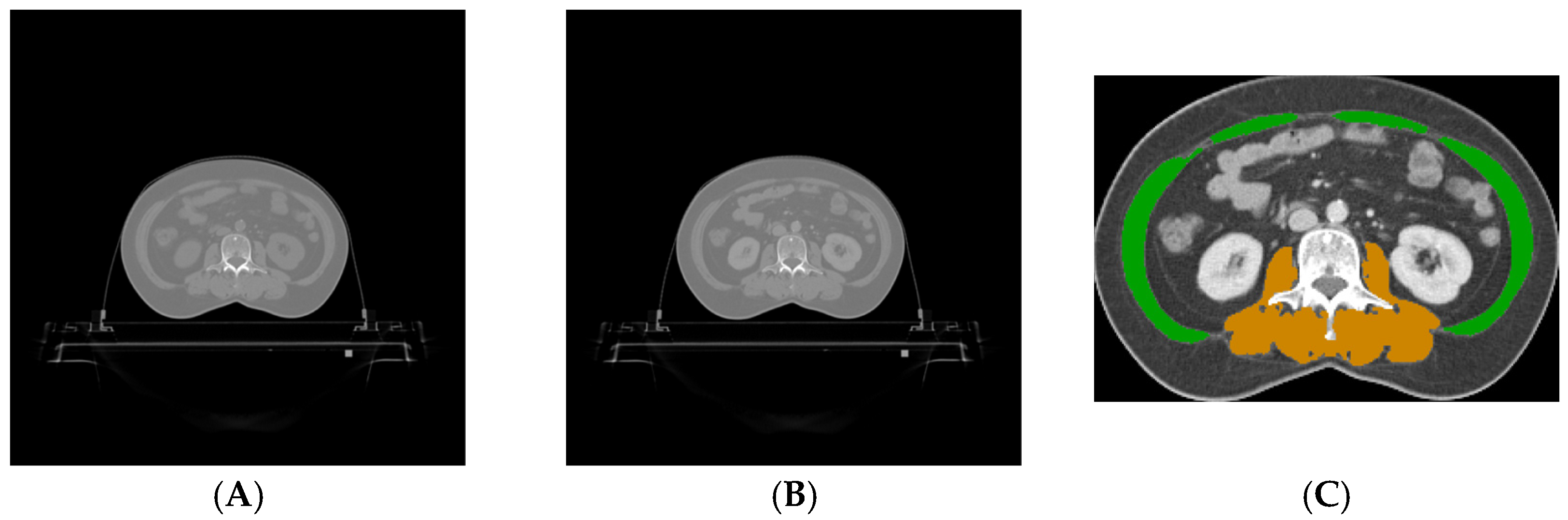
2.5. Skeletal Muscle Segmentation
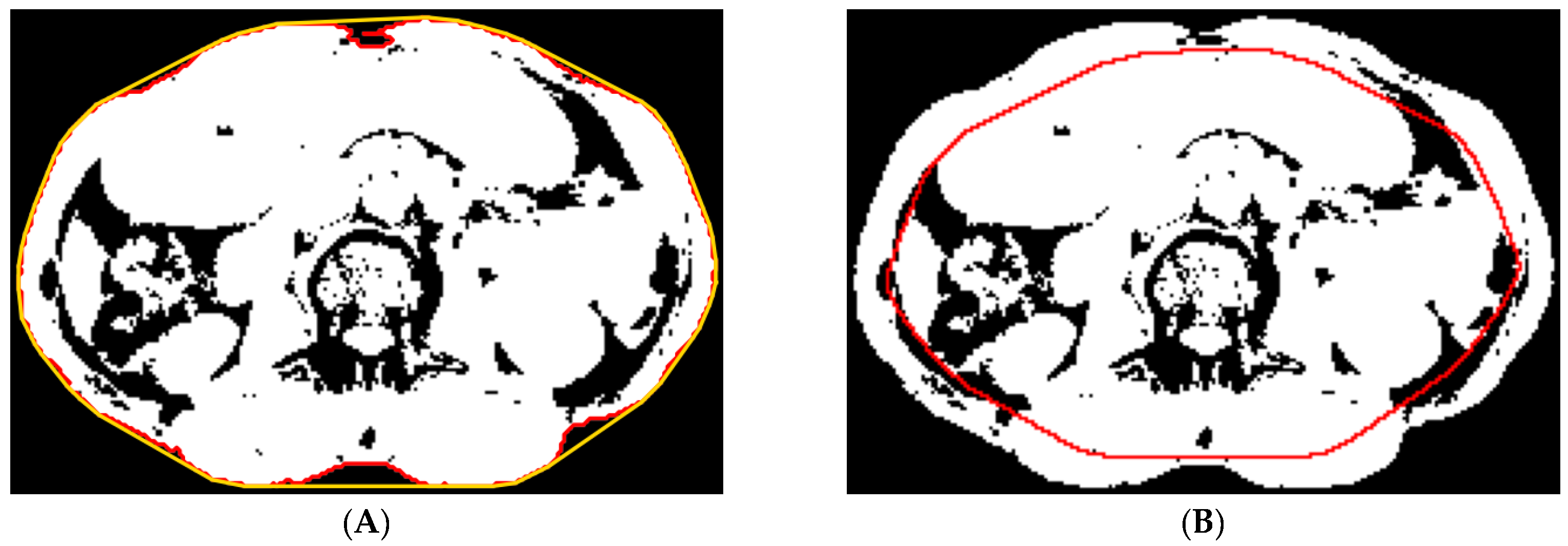
2.5.1. Preprocessing
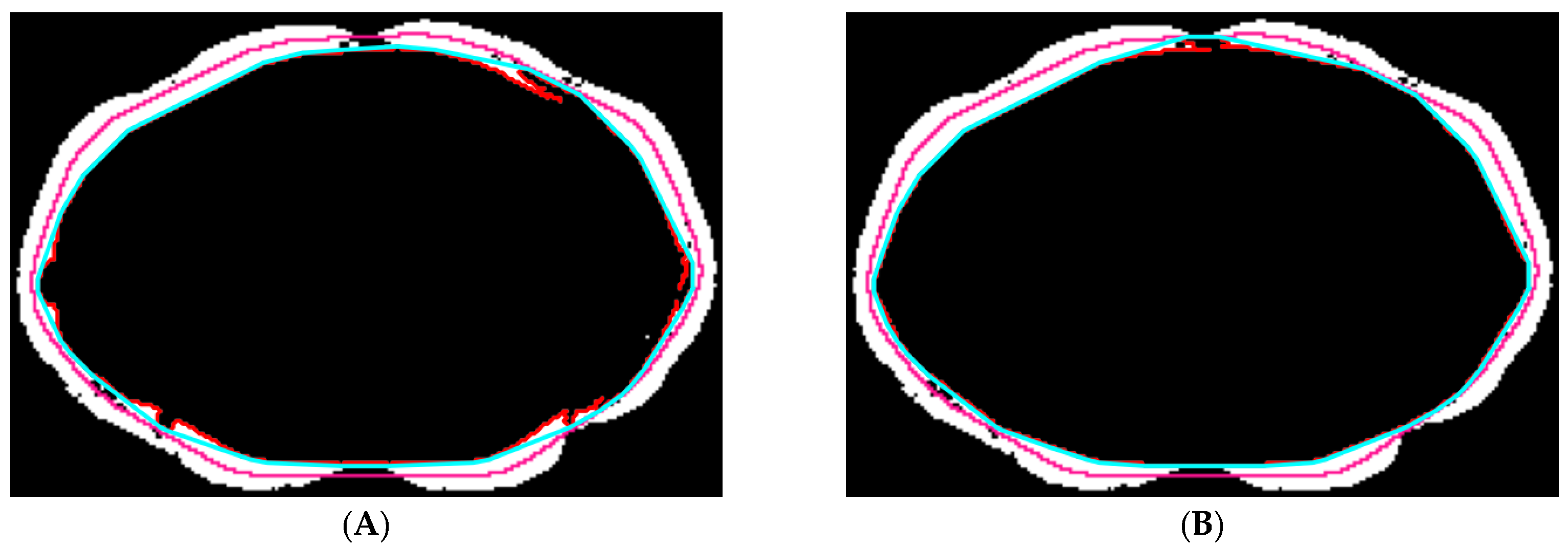
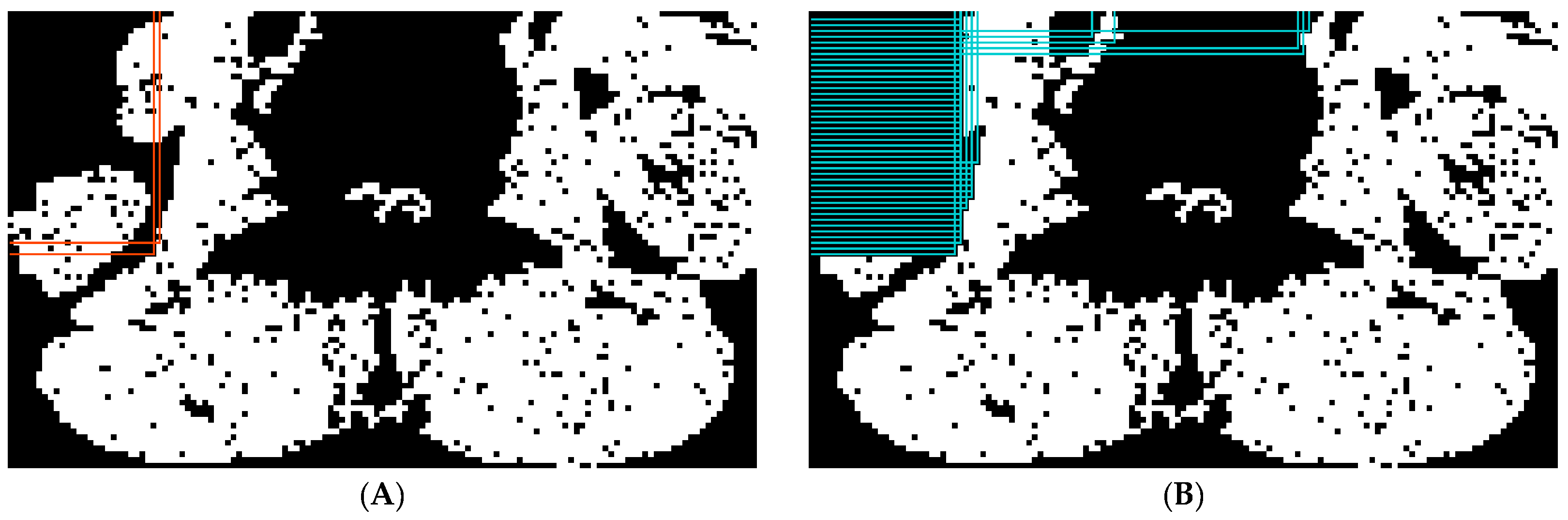
2.5.2. Abdominal Muscle Segmentation
2.5.3. Paraspinal Muscle Segmentation
2.6. Comparison Study
2.7. Performance Evaluation
2.8. Statistical Analysis
3. Results
3.1. Segmentation Accuracy Comparison with AutoMATICA
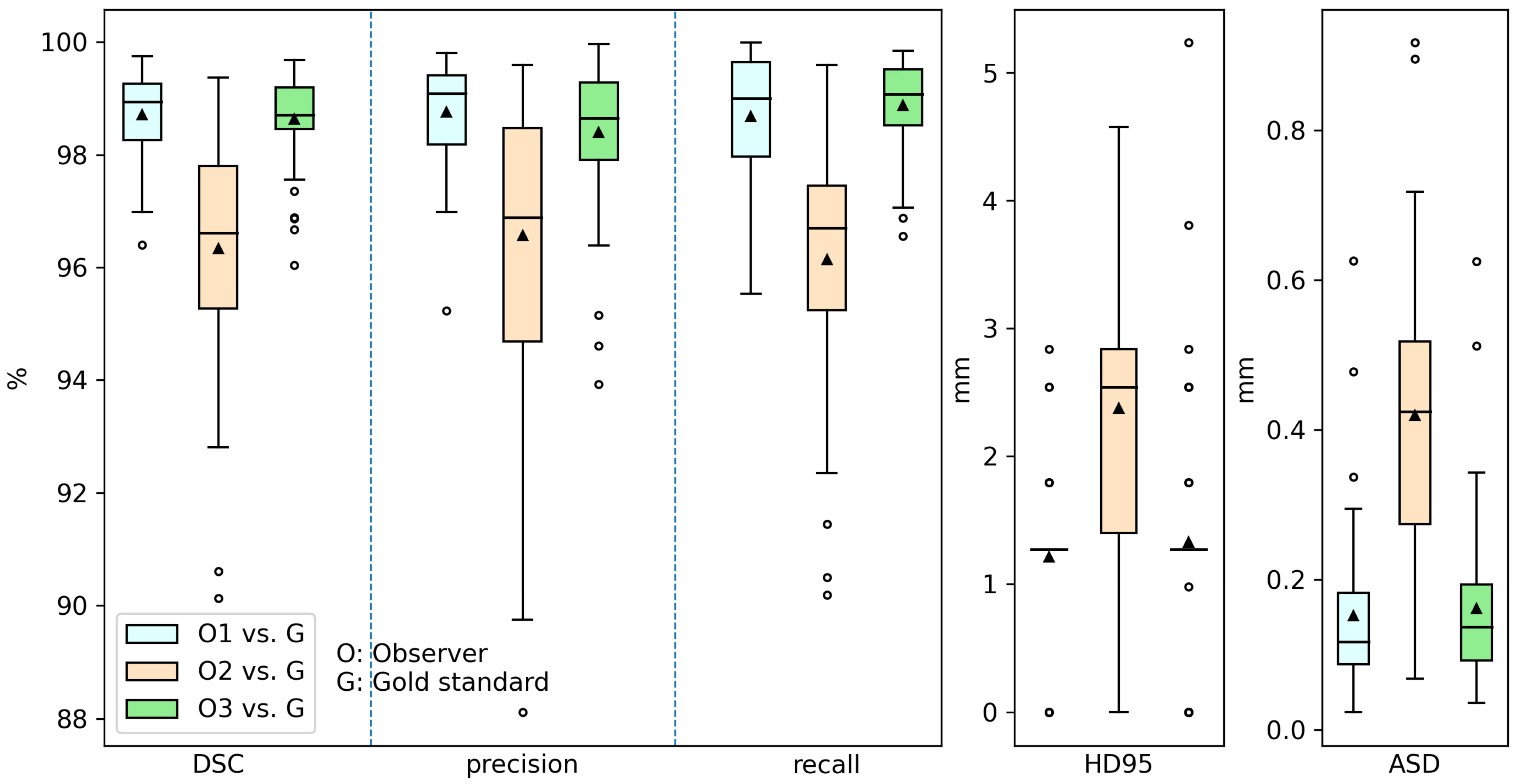
3.2. Inter-Observer Variation
3.3. Time Cost
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Unsal, D.; Mentes, B.; Akmansu, M.; Uner, A.; Oguz, M.; Pak, Y. Evaluation of nutritional status in cancer patients receiving radiotherapy: A prospective study. Am. J. Clin. Oncol. 2006, 29, 183–188. [Google Scholar] [CrossRef] [PubMed]
- McMillan, D.C. Systemic inflammation, nutritional status and survival in patients with cancer. Curr. Opin. Clin. Nutr. Metab. Care 2009, 12, 223–226. [Google Scholar] [CrossRef] [PubMed]
- Portal, D.; Hofstetter, L.; Eshed, I.; Dan-Lantsman, C.; Sella, T.; Urban, D.; Onn, A.; Bar, J.; Segal, G. L3 skeletal muscle index (L3SMI) is a surrogate marker of sarcopenia and frailty in non-small cell lung cancer patients. Cancer Manag. Res. 2019, 11, 2579. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Xie, H.; Gong, Y.; Kuang, J.; Yan, L.; Ruan, G.; Gao, F.; Gan, J. The value of L3 skeletal muscle index in evaluating preoperative nutritional risk and long-term prognosis in colorectal cancer patients. Sci. Rep. 2020, 10, 8153. [Google Scholar] [CrossRef]
- Derstine, B.A.; Holcombe, S.A.; Ross, B.E.; Wang, N.C.; Su, G.L.; Wang, S.C. Optimal body size adjustment of L3 CT skeletal muscle area for sarcopenia assessment. Sci. Rep. 2021, 11, 279. [Google Scholar] [CrossRef]
- Go, S.I.; Park, M.J.; Park, S.; Kang, M.H.; Kim, H.G.; Kang, J.H.; Kim, J.H.; Lee, G.W. Cachexia index as a potential biomarker for cancer cachexia and a prognostic indicator in diffuse large B-cell lymphoma. J. Cachexia Sarcopenia Muscle 2021, 12, 2211–2219. [Google Scholar] [CrossRef]
- Fang, Z.; Du, F.; Shang, L.; Liu, J.; Ren, F.; Liu, Y.; Wu, H.; Liu, Y.; Li, P.; Li, L. CT assessment of preoperative nutritional status in gastric cancer: Severe low skeletal muscle mass and obesity-related low skeletal muscle mass are unfavorable factors of postoperative complications. Expert Rev. Gastroenterol. Hepatol. 2021, 15, 317–324. [Google Scholar] [CrossRef]
- Berkelhammer, C.H.; Leiter, L.A.; Jeejeebhoy, K.N.; Detsky, A.S.; Oreopoulos, D.G.; Uldall, P.R.; Baker, J.P. Skeletal muscle function in chronic renal failure: An index of nutritional status. Am. J. Clin. Nutr. 1985, 42, 845–854. [Google Scholar] [CrossRef]
- Mourtzakis, M.; Prado, C.M.M.; Lieffers, J.R.; Reiman, T.; McCargar, L.J.; Baracos, V.E. A practical and precise approach to quantification of body composition in cancer patients using computed tomography images acquired during routine care. Appl. Physiol. Nutr. Metab. 2008, 33, 997–1006. [Google Scholar] [CrossRef]
- Di Sebastiano, K.M.; Mourtzakis, M. A critical evaluation of body composition modalities used to assess adipose and skeletal muscle tissue in cancer. Appl. Physiol. Nutr. Metab. 2012, 37, 811–821. [Google Scholar] [CrossRef]
- Aredes, M.A.; da Camara, A.O.; de Paula, N.S.; Fraga, K.Y.D.; do Carmo, M.d.G.T.; Chaves, G.V. Efficacy of ω-3 supplementation on nutritional status, skeletal muscle, and chemoradiotherapy toxicity in cervical cancer patients: A randomized, triple-blind, clinical trial conducted in a middle-income country. Nutrition 2019, 67, 110528. [Google Scholar] [CrossRef] [PubMed]
- Bamba, S.; Inatomi, O.; Takahashi, K.; Morita, Y.; Imai, T.; Ohno, M.; Kurihara, M.; Takebayashi, K.; Kojima, M.; Iida, H.; et al. Assessment of Body Composition From CT Images at the Level of the Third Lumbar Vertebra in Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2021, 27, 1435–1442. [Google Scholar] [CrossRef] [PubMed]
- Weston, A.D.; Korfiatis, P.; Kline, T.L.; Philbrick, K.A.; Kostandy, P.; Sakinis, T.; Sugimoto, M.; Takahashi, N.; Erickson, B.J. Automated abdominal segmentation of CT scans for body composition analysis using deep learning. Radiology 2019, 290, 669–679. [Google Scholar] [CrossRef]
- Paris, M.T.; Tandon, P.; Heyland, D.K.; Furberg, H.; Premji, T.; Low, G.; Mourtzakis, M. Automated body composition analysis of clinically acquired computed tomography scans using neural networks. Clin. Nutr. 2020, 39, 3049–3055. [Google Scholar] [CrossRef] [PubMed]
- Burns, J.E.; Yao, J.; Chalhoub, D.; Chen, J.J.; Summers, R.M. A Machine Learning Algorithm to Estimate Sarcopenia on Abdominal CT. Acad. Radiol. 2020, 27, 311–320. [Google Scholar] [CrossRef]
- Blanc-Durand, P.; Schiratti, J.-B.; Schutte, K.; Jehanno, P.; Herent, P.; Pigneur, F.; Lucidarme, O.; Benaceur, Y.; Sadate, A.; Luciani, A. Abdominal musculature segmentation and surface prediction from CT using deep learning for sarcopenia assessment. Diagn. Interv. Imaging 2020, 101, 789–794. [Google Scholar] [CrossRef]
- Castiglione, J.; Somasundaram, E.; Gilligan, L.A.; Trout, A.T.; Brady, S. Automated Segmentation of Abdominal Skeletal Muscle on Pediatric CT Scans Using Deep Learning. Radiol. Artif. Intell. 2021, 3, e200130. [Google Scholar] [CrossRef]
- Teplyakova Anastasia, R.; Shershnev Roman, V.; Starkov Sergey, O.; Agababian Tatev, A.; Kukarskaya Valeria, A. Segmentation of muscle tissue in computed tomography images at the level of the L3 vertebra. J. Sci. Tech. Inf. Technol. Mech. Opt. 2024, 153, 124. [Google Scholar] [CrossRef]
- Delrieu, L.; Blanc, D.; Bouhamama, A.; Reyal, F.; Pilleul, F.; Racine, V.; Hamy, A.S.; Crochet, H.; Marchal, T.; Heudel, P.E. Automatic deep learning method for third lumbar selection and body composition evaluation on CT scans of cancer patients. Front. Nucl. Med. 2024, 3, 1292676. [Google Scholar] [CrossRef]
- Hsu, T.-M.H.; Schawkat, K.; Berkowitz, S.J.; Wei, J.L.; Makoyeva, A.; Legare, K.; DeCicco, C.; Paez, S.N.; Wu, J.S.; Szolovits, P. Artificial intelligence to assess body composition on routine abdominal CT scans and predict mortality in pancreatic cancer—A recipe for your local application. Eur. J. Radiol. 2021, 142, 109834. [Google Scholar] [CrossRef]
- Nowak, S.; Theis, M.; Wichtmann, B.D.; Faron, A.; Froelich, M.F.; Tollens, F.; Geißler, H.L.; Block, W.; Luetkens, J.A.; Attenberger, U.I.; et al. End-to-end automated body composition analyses with integrated quality control for opportunistic assessment of sarcopenia in CT. Eur. Radiol. 2022, 32, 3142–3151. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhou, J.; Chen, S.; Liu, L. Muscle segmentation of L3 slice in abdomen CT images based on fully convolutional networks. In Proceedings of the 2019 Ninth International Conference on Image Processing Theory, Tools and Applications (IPTA), Istanbul, Turkey, 6–9 November 2019; IEEE: Piscataway, NJ, USA, 2019. [Google Scholar]
- Lee, H.; Troschel, F.M.; Tajmir, S.; Fuchs, G.; Mario, J.; Fintelmann, F.J.; Do, S. Pixel-Level Deep Segmentation: Artificial Intelligence Quantifies Muscle on Computed Tomography for Body Morphometric Analysis. J. Digit. Imaging 2017, 30, 487–498. [Google Scholar] [CrossRef] [PubMed]
- Park, H.J.; Shin, Y.; Park, J.; Kim, H.; Lee, I.S.; Seo, D.-W.; Huh, J.; Lee, T.Y.; Park, T.; Lee, J.; et al. Development and Validation of a Deep Learning System for Segmentation of Abdominal Muscle and Fat on Computed Tomography. Korean J. Radiol. 2020, 21, 88. [Google Scholar] [CrossRef]
- Ha, J.; Park, T.; Kim, H.-K.; Shin, Y.; Ko, Y.; Kim, D.W.; Sung, Y.S.; Lee, J.; Ham, S.J.; Khang, S.; et al. Development of a fully automatic deep learning system for L3 selection and body composition assessment on computed tomography. Sci. Rep. 2021, 11, 21656. [Google Scholar] [CrossRef] [PubMed]
- Dabiri, S.; Popuri, K.; Cespedes Feliciano, E.M.; Caan, B.J.; Baracos, V.E.; Beg, M.F. Muscle segmentation in axial computed tomography (CT) images at the lumbar (L3) and thoracic (T4) levels for body composition analysis. Comput. Med. Imaging Graph. 2019, 75, 47–55. [Google Scholar] [CrossRef]
- Dabiri, S.; Popuri, K.; Ma, C.; Chow, V.; Feliciano, E.M.C.; Caan, B.J.; Baracos, V.E.; Beg, M.F. Deep learning method for localization and segmentation of abdominal CT. Comput. Med. Imaging Graph. 2020, 85, 101776. [Google Scholar] [CrossRef]
- Zhang, G.; Yang, Y.; Xu, S.; Nan, Y.; Lv, C.; Wei, L.; Qian, T.; Han, J.; Xie, G. Autonomous localization and segmentation for body composition quantization on abdominal CT. Biomed. Signal Process. Control 2022, 71, 103172. [Google Scholar] [CrossRef]
- Zhang, L.; Li, J.; Yan, J.; Zhang, L.; Gong, L.-b. A Deep Learning Body Compositions Assessment Application with L3 CT Images: Multiple Validations. Res. Sq. 2024. [Google Scholar] [CrossRef]
- Kamiya, N.; Zhou, X.; Chen, H.; Muramatsu, C.; Hara, T.; Yokoyama, R.; Kanematsu, M.; Hoshi, H.; Fujita, H. Automated segmentation of recuts abdominis muscle using shape model in X-ray CT images. In Proceedings of the 2011 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Boston, MA, USA, 30 August–3 September 2011; IEEE: Piscataway, NJ, USA, 2011. [Google Scholar]
- Kamiya, N.; Zhou, X.; Chen, H.; Muramatsu, C.; Hara, T.; Yokoyama, R.; Kanematsu, M.; Hoshi, H.; Fujita, H. Automated segmentation of psoas major muscle in X-ray CT images by use of a shape model: Preliminary study. Radiol. Phys. Technol. 2012, 5, 5–14. [Google Scholar] [CrossRef]
- Chung, H.; Cobzas, D.; Birdsell, L.; Lieffers, J.; Baracos, V. Automated segmentation of muscle and adipose tissue on CT images for human body composition analysis. In Proceedings of the Medical Imaging 2009: Visualization, Image-Guided Procedures, and Modeling, Lake Buena Vista, FL, USA, 7–12 February 2009; SPIE: Edinburgh, UK, 2009. [Google Scholar]
- Popuri, K.; Cobzas, D.; Esfandiari, N.; Baracos, V.; Jägersand, M. Body composition assessment in axial CT images using FEM-based automatic segmentation of skeletal muscle. IEEE Trans. Med. Imaging 2015, 35, 512–520. [Google Scholar] [CrossRef]
- Meesters, S.; Yokota, F.; Okada, T.; Takaya, M.; Tomiyama, N.; Yao, J.; Liguraru, M.; Summers, R.M.; Sato, Y. Multi atlas-based muscle segmentation in abdominal CT images with varying field of view. In Proceedings of the International Forum on Medical Imaging in Asia (IFMIA), Daejon, Republic of Korea, 16–17 November 2012. [Google Scholar]
- Polan, D.F.; Brady, S.L.; Kaufman, R.A. Tissue segmentation of Computed Tomography images using a Random Forest algorithm: A feasibility study. Phys. Med. Biol. 2016, 61, 6553–6569. [Google Scholar] [CrossRef] [PubMed]
- Charrière, K.; Boulouard, Q.; Artemova, S.; Vilotitch, A.; Ferretti, G.R.; Bosson, J.-L.; Moreau-Gaudry, A.; Giai, J.; Fontaine, E.; Bétry, C. A comparative study of two automated solutions for cross-sectional skeletal muscle measurement from abdominal computed tomography images. Med. Phys. 2023, 50, 4973–4980. [Google Scholar] [CrossRef] [PubMed]
- Rai, H.M.; Yoo, J.; Moqurrab, S.A.; Dashkevych, S. Advancements in traditional machine learning techniques for detection and diagnosis of fatal cancer types: Comprehensive review of biomedical imaging datasets. Measurement 2023, 225, 114059. [Google Scholar] [CrossRef]
- Bian, J.; Siewerdsen, J.H.; Han, X.; Sidky, E.Y.; Prince, J.L.; Pelizzari, C.A.; Pan, X. Evaluation of sparse-view reconstruction from flat-panel-detector cone-beam CT. Phys. Med. Biol. 2010, 55, 6575–6599. [Google Scholar] [CrossRef]
- Chen, H.; Zhang, Y.; Zhang, W.; Liao, P.; Li, K.; Zhou, J.; Wang, G. Low-dose CT via convolutional neural network. Biomed. Opt. Express 2017, 8, 679–694. [Google Scholar] [CrossRef]
- Chen, H.; Zhang, Y.; Kalra, M.K.; Lin, F.; Chen, Y.; Liao, P.; Zhou, J.; Wang, G. Low-dose CT with a residual encoder-decoder convolutional neural network. IEEE Trans. Med. Imaging 2017, 36, 2524–2535. [Google Scholar] [CrossRef]
- Sagara, Y.; Hara, A.K.; Pavlicek, W.; Silva, A.C.; Paden, R.G.; Wu, Q. Abdominal CT: Comparison of low-dose CT with adaptive statistical iterative reconstruction and routine-dose CT with filtered back projection in 53 patients. Am. J. Roentgenol. 2010, 195, 713–719. [Google Scholar] [CrossRef]
- Cao, L.; Liu, X.; Li, J.; Qu, T.; Chen, L.; Cheng, Y.; Hu, J.; Sun, J.; Guo, J. A study of using a deep learning image reconstruction to improve the image quality of extremely low-dose contrast-enhanced abdominal CT for patients with hepatic lesions. Br. J. Radiol. 2021, 94, 20201086. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.; Liu, J.; Lv, T.; Zhao, Q.; Zhang, Y.; Quan, G.; Feng, J.; Chen, Y.; Luo, L. Hybrid-Domain Neural Network Processing for Sparse-View CT Reconstruction. IEEE Trans. Radiat. Plasma Med. Sci. 2021, 5, 88–98. [Google Scholar] [CrossRef]
- Takahashi, E.A.; Takahashi, N.; Reisenauer, C.J.; Moynagh, M.R.; Misra, S. Body composition changes after left gastric artery embolization in overweight and obese individuals. Abdom. Radiol. 2019, 44, 2627–2631. [Google Scholar] [CrossRef]
- Ackermans, L.L.G.C.; Volmer, L.; Wee, L.; Brecheisen, R.; Sánchez-González, P.; Seiffert, A.P.; Gómez, E.J.; Dekker, A.; Ten Bosch, J.A.; Olde Damink, S.M.W.; et al. Deep Learning Automated Segmentation for Muscle and Adipose Tissue from Abdominal Computed Tomography in Polytrauma Patients. Sensors 2021, 21, 2083. [Google Scholar] [CrossRef]
- Kalra, M.K.; Maher, M.M.; Sahani, D.V.; Blake, M.A.; Hahn, P.F.; Avinash, G.B.; Toth, T.L.; Halpern, E.; Saini, S. Low-dose CT of the abdomen: Evaluation of image improvement with use of noise reduction filters—Pilot study. Radiology 2003, 228, 251–256. [Google Scholar] [CrossRef]
- Hashimoto, F.; Kakimoto, A.; Ota, N.; Ito, S.; Nishizawa, S. Automated segmentation of 2D low-dose CT images of the psoas-major muscle using deep convolutional neural networks. Radiol. Phys. Technol. 2019, 12, 210–215. [Google Scholar] [CrossRef]
- Fisher, R.A.; Yates, F. Statistical Tables for Biological, Agricultural and Medical Research; Hafner Publishing Company: New York, NY, USA, 1953. [Google Scholar]
- Yushkevich, P.A.; Piven, J.; Hazlett, H.C.; Smith, R.G.; Ho, S.; Gee, J.C.; Gerig, G. User-guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. NeuroImage 2006, 31, 1116–1128. [Google Scholar] [CrossRef] [PubMed]
- Menze, B.H.; Jakab, A.; Bauer, S.; Kalpathy-Cramer, J.; Farahani, K.; Kirby, J.; Burren, Y.; Porz, N.; Slotboom, J.; Wiest, R.; et al. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans. Med. Imaging 2015, 34, 1993–2024. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Yue, H.; Du, Y.; Hou, S.; Du, W.; Peng, Y. Skeletal Muscle Segmentation at the Third Lumbar Vertebral Level in Radiotherapy CT Images. In Software Engineering, Artificial Intelligence, Networking and Parallel/Distributed Computing; Springer: Berlin/Heidelberg, Germany, 2022; pp. 77–88. [Google Scholar]
- Engelke, K.; Museyko, O.; Wang, L.; Laredo, J.-D. Quantitative analysis of skeletal muscle by computed tomography imaging—State of the art. J. Orthop. Transl. 2018, 15, 91–103. [Google Scholar] [CrossRef] [PubMed]
- Mitsiopoulos, N.; Baumgartner, R.N.; Heymsfield, S.B.; Lyons, W.; Gallagher, D.; Ross, R. Cadaver validation of skeletal muscle measurement by magnetic resonance imaging and computerized tomography. J. Appl. Physiol. 1998, 85, 115–122. [Google Scholar] [CrossRef]
- Woolson, R.F. Wilcoxon signed-rank test. In Wiley Encyclopedia of Clinical Trials; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2007; pp. 1–3. [Google Scholar]





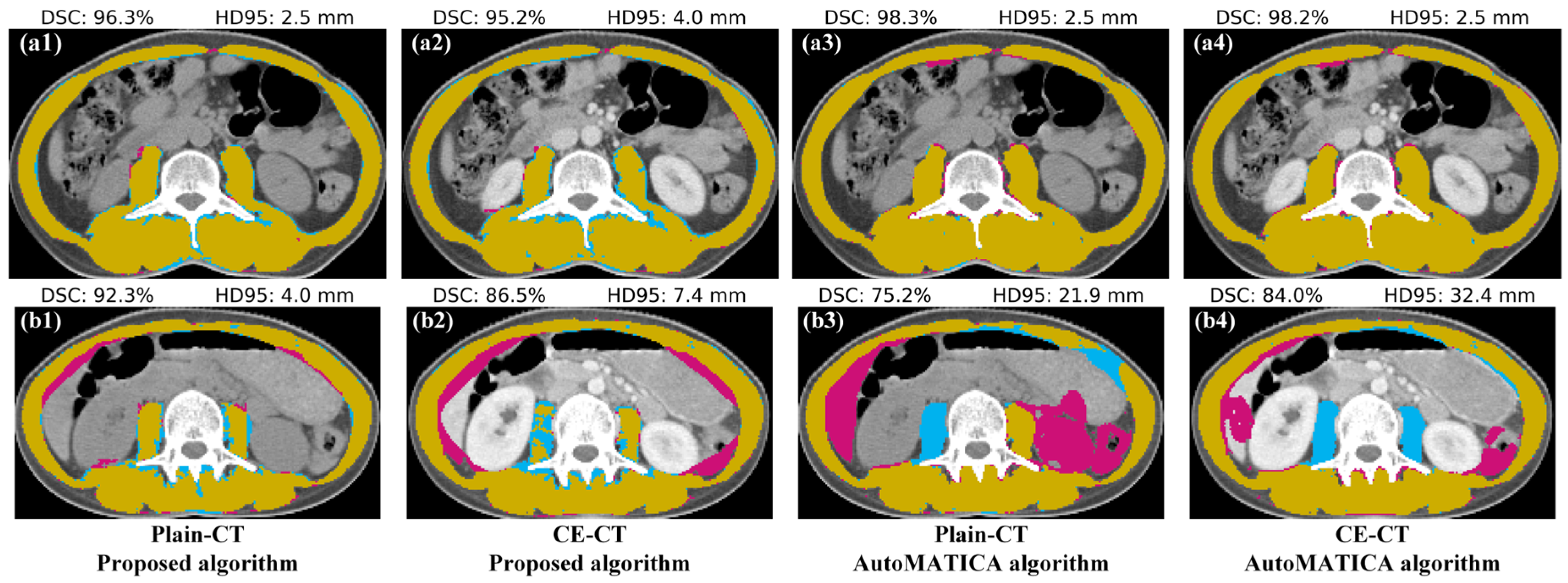
| Metrics † | Algorithm | Image Series | Summary | p-Value * | |
|---|---|---|---|---|---|
| Plain-CT | CE-CT | ||||
| DSC (%) | Proposed | 93.2 ± 1.6 | 93.2 ± 2.2 | 93.2 ± 1.9 | <0.01 |
| AutoMATICA | 94.0 ± 4.6 | 94.2 ± 3.4 | 94.1 ± 4.1 | ||
| precision (%) | Proposed | 97.0 ± 2.2 | 96.5 ± 3.5 | 96.7 ± 2.9 | <<0.01 |
| AutoMATICA | 92.5 ± 6.1 | 93.0 ± 4.8 | 92.7 ± 5.5 | ||
| recall (%) | Proposed | 89.7 ± 3.0 | 90.2 ± 2.9 | 90.0 ± 2.9 | <<0.01 |
| AutoMATICA | 95.7 ± 4.0 | 95.7 ± 4.0 | 95.7 ± 4.0 | ||
| HD95 (mm) | Proposed | 4.6 ± 1.1 | 4.9 ± 1.5 | 4.8 ± 1.3 | <0.01 |
| AutoMATICA | 6.9 ± 4.8 | 7.8 ± 6.5 | 7.4 ± 5.7 | ||
| ASD (mm) | Proposed | 0.8 ± 0.2 | 0.8 ± 0.3 | 0.8 ± 0.2 | >0.05 |
| AutoMATICA | 0.9 ± 0.6 | 0.9 ± 0.6 | 0.9 ± 0.6 | ||
| Items † | O1 | O2 | O3 | Proposed (CPU) * | AutoMATICA (CPU) * | AutoMATICA (GPU) * |
|---|---|---|---|---|---|---|
| Plain-CT | - | - | - | 289 ± 37 | 1416 ± 43 | 447 ± 39 |
| CE-CT | - | - | - | 316 ± 45 | 1681 ± 40 | 448 ± 42 |
| Summary | 334,444 | 212,222 | 455,556 | 303 ± 43 | 1548 ± 140 | 448 ± 40 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, X.; Du, Y.; Yue, H. Skeletal Muscle Segmentation at the Level of the Third Lumbar Vertebra (L3) in Low-Dose Computed Tomography: A Lightweight Algorithm. Tomography 2024, 10, 1513-1526. https://doi.org/10.3390/tomography10090111
Zhao X, Du Y, Yue H. Skeletal Muscle Segmentation at the Level of the Third Lumbar Vertebra (L3) in Low-Dose Computed Tomography: A Lightweight Algorithm. Tomography. 2024; 10(9):1513-1526. https://doi.org/10.3390/tomography10090111
Chicago/Turabian StyleZhao, Xuzhi, Yi Du, and Haizhen Yue. 2024. "Skeletal Muscle Segmentation at the Level of the Third Lumbar Vertebra (L3) in Low-Dose Computed Tomography: A Lightweight Algorithm" Tomography 10, no. 9: 1513-1526. https://doi.org/10.3390/tomography10090111
APA StyleZhao, X., Du, Y., & Yue, H. (2024). Skeletal Muscle Segmentation at the Level of the Third Lumbar Vertebra (L3) in Low-Dose Computed Tomography: A Lightweight Algorithm. Tomography, 10(9), 1513-1526. https://doi.org/10.3390/tomography10090111






