Abstract
Computed tomography (CT) is used in a wide range of medical imaging diagnoses. However, the reconstruction of CT images from raw projection data is inherently complex and is subject to artifacts and noise, which compromises image quality and accuracy. In order to address these challenges, deep learning developments have the potential to improve the reconstruction of computed tomography images. In this regard, our research aim is to determine the techniques that are used for 3D deep learning in CT reconstruction and to identify the training and validation datasets that are accessible. This research was performed on five databases. After a careful assessment of each record based on the objective and scope of the study, we selected 60 research articles for this review. This systematic literature review revealed that convolutional neural networks (CNNs), 3D convolutional neural networks (3D CNNs), and deep learning reconstruction (DLR) were the most suitable deep learning algorithms for CT reconstruction. Additionally, two major datasets appropriate for training and developing deep learning systems were identified: 2016 NIH-AAPM-Mayo and MSCT. These datasets are important resources for the creation and assessment of CT reconstruction models. According to the results, 3D deep learning may increase the effectiveness of CT image reconstruction, boost image quality, and lower radiation exposure. By using these deep learning approaches, CT image reconstruction may be made more precise and effective, improving patient outcomes, diagnostic accuracy, and healthcare system productivity.
1. Introduction
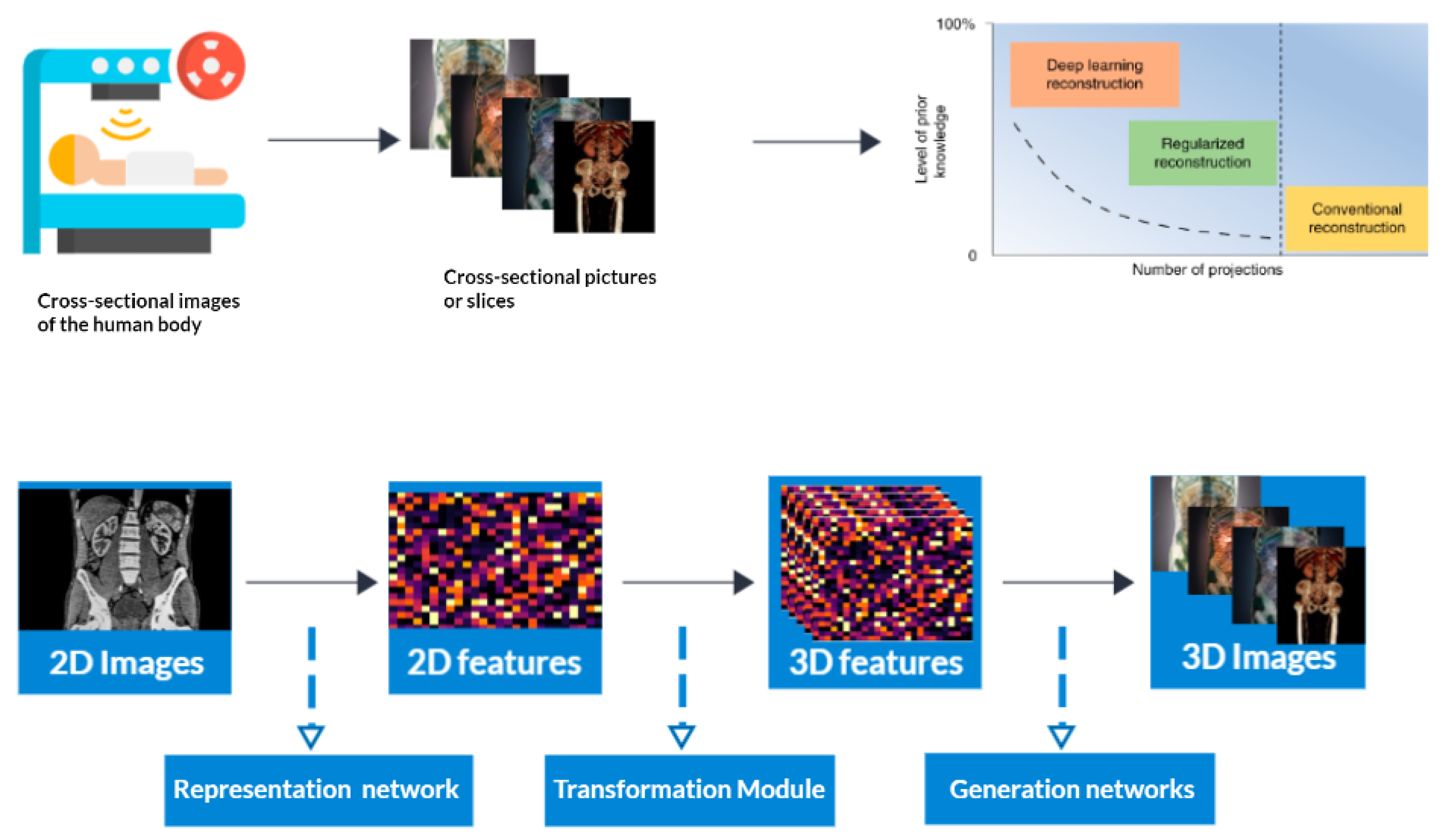
Computed tomography (CT) is a medical imaging method that offers cross-sectional images of the body of a person, which allows for the visualization of organs and tissues aiding in the diagnosis and management of a wide range of medical conditions, including cancer [1,2], heart disease [3], and neurological disorders [4]. CT refers to a computerized X-ray imaging procedure. In this method, a patient is swiftly rotated around a narrow beam, creating signals processed by the machine’s computer to produce cross-sectional pictures or slices. Once the scanner has gathered a number of these slices, which are known as tomography pictures, they are stacked together to create three-dimensional representations of the patient [5,6,7,8,9]. These three-dimensional representations are created by applying an algorithm to the raw data, resulting in image slices that are then reconstructed into a 3D volume [10,11]. This process is called CT reconstruction (Figure 1). Figure 1 is attributed to the work by Liyue Shen in his paper titled ‘Patient-specific reconstruction of volumetric computed tomography images from a single projection view via deep learning’ [12].
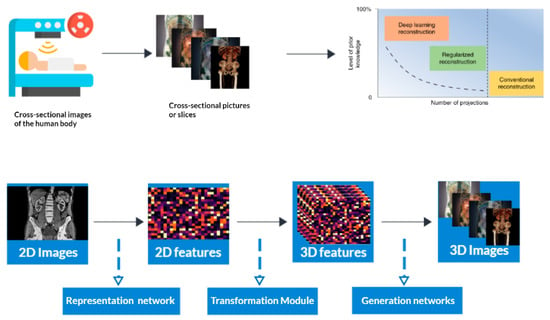
Figure 1.
3D Reconstruction and enhanced visualization: CT scan data transformed into a detailed 3D image using deep learning techniques.
Computed tomography (CT) initially relied on traditional computationally intensive analytical techniques used in image reconstruction, such as filtered back-projection (FBP). However, these methods often had limitations, including image noise and other artifacts that compromised the quality and efficiency of the reconstructed images. To address these challenges deep learning emerged as a powerful tool in CT image reconstruction. By leveraging deep neural networks, deep learning algorithms have significantly improved the quality and efficiency of CT image reconstruction.
In addition to 2D image reconstruction, deep learning has also been extended to 3D image reconstruction in CT. By incorporating volumetric information, deep learning algorithms can generate more precise and detailed 3D images, further enhancing the diagnostic capabilities of CT scans [13,14,15].
What exactly is 3D deep learning? It is a sort of machine learning that examines and interprets 3D data using AI neural networks. It requires preparing neural networks to discover intricate correlations and characteristics in 3D datasets. We know that machine learning algorithms [16,17] typically operate on 2D data, but deep learning allows for the analysis of 3D data more intuitively and efficiently. 3D deep learning algorithms [18,19,20,21,22,23] can extract features such as shapes, textures, and volumes from 3D data and can be used for a wide range of applications, such as medical imaging, robotics, and virtual reality. Even though computed tomography reconstruction is a well-established technique for generating high-quality 3D images of the body, there are still several gaps in knowledge and research that require filling. Past research studies have explored aspects of CT reconstruction, but the emergence of 3D deep learning algorithms is a novel approach to enhancing image quality and efficiency.
This planned literature review aims to show the problems and opportunities in CT reconstruction that arise when 3D deep learning is used in CT reconstruction, especially when CT images are rebuilt from raw projection data into 3D data. This paper also addresses the requirement of collecting and integrating existing research, which helped the authors highlight the potential applications, challenges, and advancements of 3D deep learning in CT reconstruction. This study involved a review of the most advanced methods for 3D deep learning for CT reconstruction, its effectiveness, and its efficiency in producing high-quality 3D images. The review follows the guidelines of PRISMA [24,25,26,27] (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) and also incorporates the Kitchenhand and Charters [28,29] methodology, which is specifically adapted to investigate 3D deep learning in CT reconstruction.
This SLR seeks to add to the existing literature by giving a clear picture of how 3D deep learning techniques can be used to improve and make CT image reconstruction work better. These discoveries have enhanced medical imaging research and the use of 3D deep learning algorithms for CT reconstruction. Exploring these insights may lead to improved image quality, reduced radiation exposure, and enhanced diagnostic accuracy by reconstructing CT images more precisely and effectively. This organized literature review looks into the most up-to-date and effective training and validation datasets and methods for 3D deep learning in computed tomography reconstruction. It intends to highlight the challenges faced in 3D deep learning for CT image reconstruction and to identify potential applications, improvements, and limitations. The use of 3D deep learning methods improves the quality and efficiency of CT image reconstruction by utilizing deep neural networks, resulting in exact 3D representations. By offering vital insights into their efficacy, 3D deep learning algorithms help to improve diagnostic accuracy, advance medical imaging research, and improve patient care.
2. Methods
2.1. Research Objective
The objective of this work is to examine the use of 3D deep learning in computed tomography reconstruction through an extensive survey of the literature. A systematic literature review was utilized as the research technique in this study, which is a structured and thorough method of discovering, evaluating, and analyzing published information to look into certain research concerns. We follow the well-organized recommendations provided by PRISMA [25] in this study. We also incorporate the Kitchenham and Charters [28] methodology, as well as an extension for 3D deep learning in computed tomography reconstruction investigations. In this systematic literature review, our aim is to address the following questions:
- RQ1
- What are the current state-of-the-art methods in 3D deep learning in computed tomography reconstruction?
- RQ2
- What datasets are available for training and validating 3D deep learning in computed tomography reconstruction?
This review of the literature is meant to find the best 3D deep learning methods for reconstructing computed tomography images and to find datasets that can be used to train and test these models. The purpose of the first study question is to examine the most cutting-edge techniques for 3D deep learning in CT reconstruction. The creation of deep learning algorithms is a revolutionary strategy for improving picture efficiency and quality, even though CT reconstruction is a well-established method for creating high-quality 3D images of the human body. Both scholars and practitioners may obtain insight into the most recent possibilities and breakthroughs in applications in the area by knowing the most cutting-edge approaches and techniques in 3D deep learning for CT reconstruction. Our objective was to compile current and pertinent knowledge on techniques and datasets related to 3D deep learning in computed tomography reconstruction using these eligibility criteria.
2.2. Data Sources and Searches
2.2.1. Search String
We created a search method to find all published materials that were related to 3D deep learning in computed tomography reconstruction. By defining the population, intervention, and results in the first place, we were able to identify the keywords. As previously stated, we addressed the following study questions: “What are the state-of-the-art methods in 3D deep learning?” and “What datasets are available for 3D deep learning in computed tomography reconstruction?” Next, we determined other ways to spell the primary concepts as well as synonyms (keywords), such as “3D deep learning” and “computed tomography” for “3D reconstruction”. We checked the keywords in pertinent publications and combined the search phrases using Boolean operators like AND, OR, and NOT. (“3D deep learning” OR “deep learning”) AND (“computed tomography” OR “CT” OR “tomography reconstruction” OR “reconstruction”) AND (“3D reconstruction” OR “image reconstruction”) were the keywords we used to conduct our search as outlined in Table 1.

Table 1.
Database search strings.
2.2.2. Resources to Be Searched
With our current keyword list, we worked to compile all the literature that is relevant to the 3D deep learning research issues for CT reconstruction. Our selection process involved picking five databases to ensure a comprehensive search: Elsevier, Springer, MDPI, IEEE Xplore, and the Nature Publishing Group. These databases were picked because they are known for having the most representative sources for research in the fields of medicine and artificial intelligence. They are also often used in other systematic literature reviews because they have large collections of literature that are relevant to the research questions, such as journals, articles, conferences, and books. By choosing these sources, we sought to compile as many articles as we could to guarantee a thorough evaluation of the literature.
2.2.3. Overview of the Search Process
We performed four basic stages to compile all the pertinent research on 3D deep learning for computed tomography reconstruction. First, we gathered primary research from the online libraries listed in the section utilizing the search phrases there. Search results were produced by the digital libraries. There were 774 publications found that were published between 2013 and 2023. In the first instance, we included every article that was cited in the original papers we picked. The second step was to remove papers that were unnecessary based on exclusion criteria. We picked the articles that satisfied the inclusion criteria from those that did not. In the second instance, we included every paper that cited the initial papers we chose. As a result of the fourth exclusion/inclusion criterion, a final selection either included or excluded the candidate. The title was the first thing we considered. The paper was skipped if it was clear that it was off-limits. As an alternative, we checked the abstract, introduction, and conclusion of each article that was thought to be possibly helpful to see if it was relevant to our study. Third, we conducted a snowballing search for any possibly misplaced papers. Given the set of sources that were eventually located using a search string and supplemented with those obtained by snowballing and manual search, for a set made up of 60 sources, 54 papers were determined to pass all phases as outlined in Table 2. After selecting the final articles for the systematic literature review, we utilized the last filtering phase, the quality assessment, to ensure that each publication provided the necessary information to answer our study questions.

Table 2.
Search results and information sources.
2.3. Eligibility Criteria
2.3.1. Inclusion Criteria
The reliability of 3D deep learning in CT reconstruction was considered in this review. It included both supervised and unsupervised techniques. We were looking for research that detailed 3D deep learning’s accuracy, precision, or effectiveness in computed tomography reconstruction. Additionally, we sought to include research that offered unique data, including modeling studies, observational studies, or clinical trials. We were also interested in papers that summarized the state-of-the-art methods in 3D deep learning in computed tomography reconstruction. We only took into account research that was printed in English-language journals to guarantee uniformity.
2.3.2. Exclusion Criteria
Studies that primarily examined conventional image-processing methods were not included in this review, however. Additionally, we did not include studies that used imaging techniques other than deep learning. Studies that had not undergone peer review or were not available in English were also excluded from this review. Additionally, editorials, letters, or conference presentations were not taken into consideration for inclusion.
2.4. Quality of Evidence
In a systematic evaluation of 3D deep learning in computed tomography reconstruction, the quality of evidence is defined as the robustness and reliability of the conclusions drawn from the reviewed research. Some of the factors used to assess the quality of the evidence include the study design, bias risk, consistency of results, accuracy of estimations, and relevance and applicability of the included studies to the research topic. We evaluated the quality of the 3D deep learning in computed tomography reconstructions using inquiries:
- Is the concept of 3D deep learning clearly defined?
- Is the method for 3D deep learning clearly defined?
- Are the state-of-the-art metrics explicitly reported?
When discussing the strength and reliability of the results drawn from the reviewed research on 3D deep learning in computed tomography reconstruction, the term “quality of evidence” is used. It reflects how confident we are in the veracity and applicability of the research to various contexts. Several factors are considered when judging the quality of the evidence. These include the research design, any possible biases, how consistent the results are, how accurate the estimates are, and how relevant and useful the studies included are.
For this, we had to address the above specific questions, such as the clarity of the definitions for 3D deep learning and computed tomography reconstruction, the explicit description of the method for 3D deep learning, and the reporting of cutting-edge techniques and metrics. The quality of the evidence for 3D deep learning in computed tomography reconstructions was assessed in this evaluation. The research utilized data from 18 papers on CT imaging. Additionally, we included one paper each from the HRCT and MRI modalities, along with two papers from the X-rays modality, which allowed us to explore the effectiveness of 3D deep learning techniques in medical imaging by analyzing various imaging modalities.
2.5. Data Extraction
The information required to address the research questions was eventually retrieved from the chosen papers. The type of 3D deep learning technique utilized for computed tomography reconstruction, the unique application domain, the dataset details, and the performance measures were all taken into account when creating the data extraction form. A further field was included to allow for the reporting of any study limitations. We started by extracting the appropriate features and evaluating the research’s shortcomings. The constraints of the investigations could be accurately and thoroughly evaluated using this cooperative method. Additionally, adding a section for restrictions made it easier to spot possible research gaps and topics for additional study. Overall, this part emphasizes the authors’ meticulous and stringent data extraction methodology, which is crucial for guaranteeing the validity and trustworthiness of a systematic literature review [30].
3. Background
Computed tomography (CT) is a vital medical imaging technology that revolutionizes healthcare by providing high-resolution images of internal body structures, making it an essential tool in fields like radiology, oncology, and surgery. CT imaging uses X-ray technology to scan a patient. During the CT imaging process, the patient is positioned on a motorized examination table that passes through a CT scanner. The scanner emits narrow X-ray beams, which are measured by detectors on the opposite side of the patient. The data collected are X-ray projections or profiles. CT reconstruction involves various techniques and methods to generate cross-sectional images from X-ray projection data, including the following:
3.1. Tomography Reconstruction
To understand 3D deep learning for computed tomography reconstruction, it is essential to understand the basic tomography reconstruction principles. Tomography is a medical imaging technique that captures cross-sectional images of the human body using X-rays or other imaging modalities. The reconstruction process transforms this data into detailed, two-dimensional (2D) or three-dimensional (3D) images representing the object’s internal structures. For turning raw projection data into useful images, it is important to use traditional tomography reconstruction methods, such as filtered back projection (FBP) and iterative reconstruction (IR) algorithms. FBP filters and back-projects data, but has limitations in sparse or irregularly sampled scenarios. Iterative reconstruction methods, on the other hand, involve an iterative optimization process to refine the image, offering advantages in handling noisy data and irregular sampling but often requiring increased computational demands. Artifacts, noise, and the requirement for a substantial amount of data can compromise the accuracy of traditional tomography reconstruction methods, leading to a reduction in image quality. This effect is particularly pronounced when employing low-dose CT or sparse-view CT, as discussed in [31].
3.2. Filtered Back Projection (FBP)
Filtered back projection (FBP) [32,33] plays a pivotal role in CT image reconstruction, and has revolutionized the field of medical imaging. CT imaging aims to create precise and informative images reflecting internal anatomical structures and pathological conditions. X-ray projection data is collected and then put through mathematical operations such as filtering and back projection to create cross-sectional images in two dimensions. These images are crucial for clinical interpretation, allowing physicians to visualize and analyze anatomical structures, detect abnormalities, and guide medical interventions. FBP’s [34] computational efficiency and straightforward mathematical foundation make it ideal for real-time diagnostic applications. Despite its historical importance, FBP has limitations, especially in addressing complex data corrections like scatter and beam-hardening artifacts. These limitations have spurred ongoing research and innovation in CT imaging, leading to the development of advanced reconstruction methods like iterative algorithms and deep-learning-based techniques. Filtered back projection (FBP) assumes consistent X-ray attenuation within the scanned object, which may not be consistent in some cases. It may not fully utilize raw data information, leading to potential image artifacts, and is less suitable for complex data corrections.
3.3. Iterative Reconstruction (IR)
Iterative reconstruction (IR) [35,36] techniques represent a revolutionary approach to reconstructing CT images, utilizing computational algorithms and iterative processes to improve image quality and reduce artifacts. IR is a paradigm shift in CT image reconstruction, focusing on a one-pass process rather than a one-pass reconstruction process. It employs an iterative approach, repeatedly refining the image based on a mathematical model that simulates the acquisition process. This process gradually converges towards a more accurate representation of the patient’s anatomy, reducing artifacts and improving image quality. IR [37] is particularly useful in scenarios with reduced radiation dose, limited projections, and prevalent noise or artifacts. It can produce high-quality images even with lower X-ray doses, mitigating health risks associated with ionizing radiation. IR’s advantages and limitations are discussed, along with its potential impact on clinical practice and its role in enhancing CT imaging. Recent advancements in computational techniques and hardware have rendered IR more accessible and effective for healthcare professionals, fundamentally reshaping the landscape of medical imaging and diagnosis.
3.4. Deep Learning Iterative Reconstruction (DLIR)
Deep learning iterative reconstruction (DLIR) is a revolutionary approach in medical imaging that combines deep learning with iterative reconstruction methods to produce high-quality, informative images with reduced radiation exposure. DLIR is a transformative approach that leverages deep neural networks to learn complex patterns and features from data, integrating them into the iterative reconstruction process. During each iteration, the deep learning model refines the reconstructed image, reducing artifacts and noise, and enhancing image quality. This iterative refinement process gradually converges towards a more precise representation of the patient’s anatomy, even in cases with limited data or low-dose scans. DLIR offers several advantages, including the potential to significantly reduce radiation exposure to patients without compromising image quality, making it particularly well-suited for pediatric imaging. It also excels in scenarios with challenging data, such as metal artifacts or limited projections, where traditional reconstruction methods may fall short. DLIR’s fundamental principles and mechanics will be explored, along with its advantages and limitations, its potential impact on clinical practice and healthcare, and its ongoing evolution. How DLIR is becoming increasingly accessible and efficient due to advancements in computational technology, reshaping the landscape of medical imaging and diagnosis, will also be examined.
3.5. Deep Learning Reconstruction (DLR)
Deep learning reconstruction (DLR) [38] is a revolutionary approach in medical imaging that improves the quality and speed of image reconstruction. It uses deep neural networks, a subset of artificial intelligence, to enhance the reconstruction of medical images, such as those obtained from CT scans or MRIs. Traditional methods, like filtered back projection (FBP) and iterative reconstruction (IR), have limitations, especially when dealing with noisy data or rapid reconstruction. DLR introduces a transformative approach by integrating deep neural networks into the image reconstruction process, which learns complex patterns and features from the acquired data. DLR can adapt and optimize the reconstruction process based on the specific data it processes, enhancing image quality by reducing artifacts, noise, and imperfections. This is particularly useful in real-time or near-real-time image-generation scenarios, such as interventional radiology or emergency medical situations. DLR’s [39] fundamental principles and mechanics are explored, along with its potential impact on clinical practice and healthcare. Advancements in computational technology have made DLR more accessible and efficient, ultimately reshaping the landscape of medical imaging and diagnosis.
4. Results
4.1. Search and Study Selection
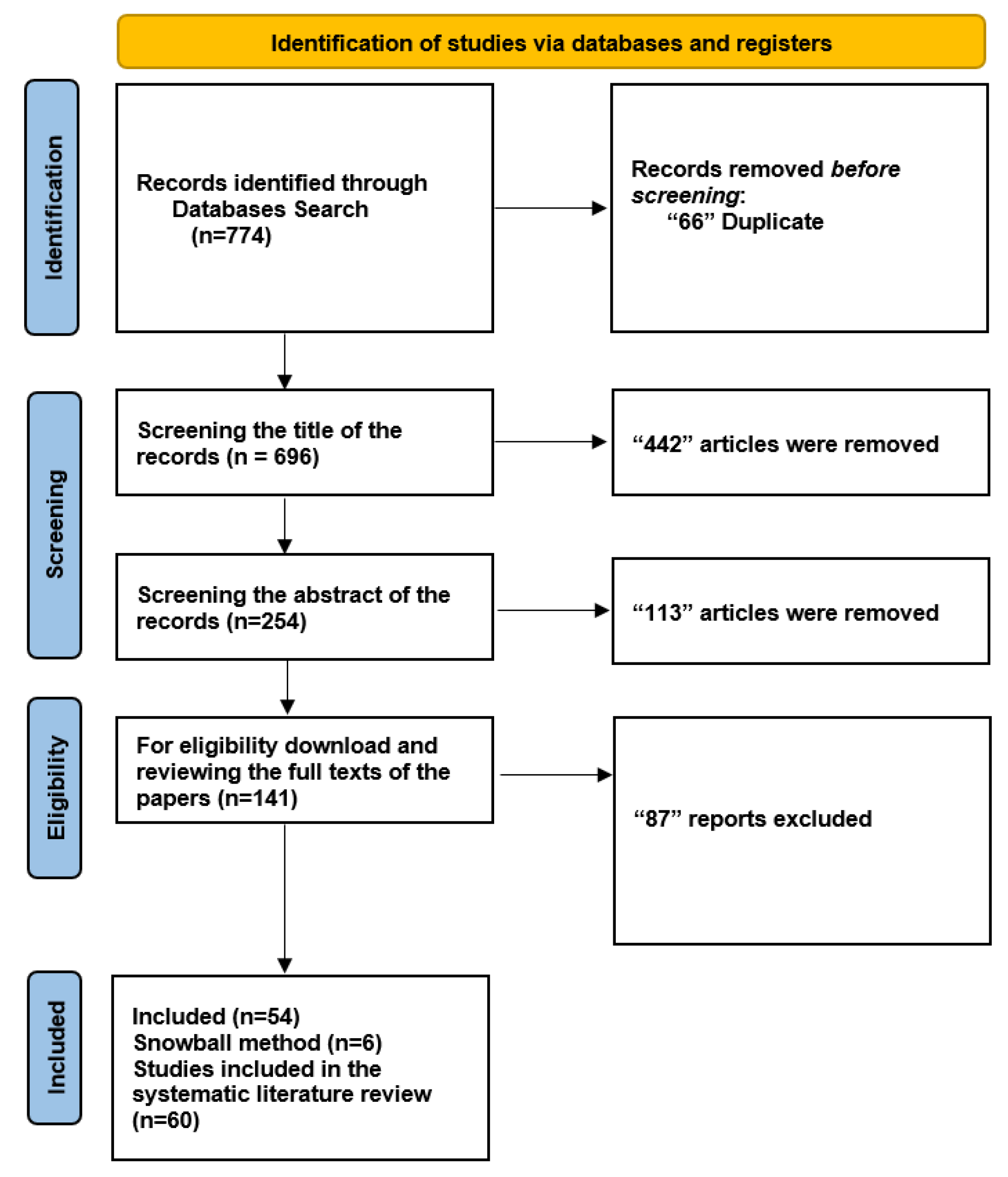
We initiated our search by exploring five databases to ensure a systematic literature review, resulting in the identification of 774 documents. A total of 66 duplicate entries were eliminated, leaving 696 unique records for further analysis. We carefully examined the titles of the 696 records during the screening phase and eliminated 442 items that did not fit our predetermined inclusion criteria. Subsequently, we thoroughly evaluated the abstracts of the remaining 254 records. Among these, 113 papers were disqualified for a variety of reasons, including not fitting the qualifying requirements or being deficient in pertinent data.
We collected and thoroughly read the complete texts of 141 articles to make sure they were eligible. Based on our predefined criteria, 87 reports were eliminated from this group. As a result, the 54 papers that met the inclusion requirements were incorporated into our systematic literature review, providing the information necessary for sensitivity and specificity assessments. We used a snowball strategy [40] in addition to our original search to find more connected publications. Utilizing this method, we identified an additional six publications, increasing the total number of included studies to 60. A thorough overview of the results is provided in Figure 2. The study also explores the relationship between CT image reconstruction and diagnostic accuracy, highlighting the impact of 3D deep learning techniques on image quality and diagnostic performance.
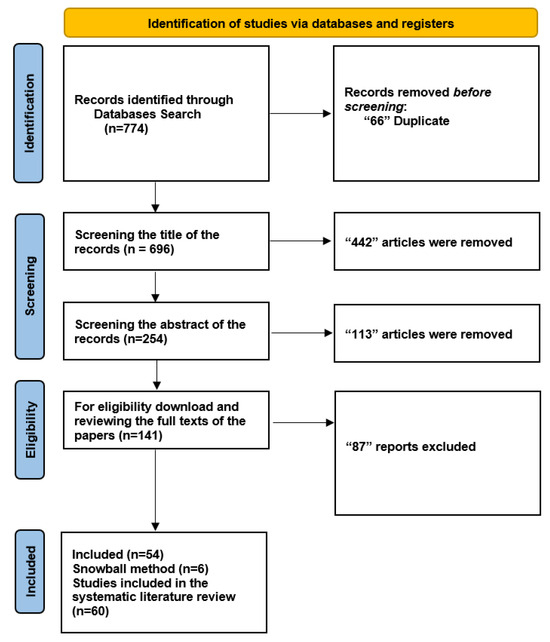
Figure 2.
Flowchart: The study involved a comprehensive search of over 60,000 abstracts and ultimately selected 10 studies for meta-analysis and 2 studies for qualitative synthesis, with a focus on the application of 3D deep learning in computed tomography reconstruction, and presents the findings in tables summarizing diagnostic accuracy metrics for imaging and several specialties.
4.2. RQ1 What Are the Current State-of-the-Art Methods in 3D Deep Learning in Computed Tomography Reconstruction?
Understanding the current state-of-the-art methodologies in 3D deep learning in computed tomography reconstruction is essential for researchers and practitioners alike. The primary aim of this systematic literature review is to provide a review of the current state-of-the-art methods in 3D deep learning in computed tomography reconstruction. The review was carried out by searching through relevant journals and choosing studies based on predefined inclusion criteria. This ensured that all of the most recent developments were covered.
The presented Table 3 provides an overview of 3D deep learning methods for computed tomography reconstruction, encompassing various methodologies and their outcomes in medical image analysis, serving as a valuable resource for researchers, clinicians, and anyone interested in the intersection of deep learning and medical imaging. It showcases the diverse range of state-of-the-art methods and their potential to impact various medical applications, while offering a global perspective on their development and adoption.

Table 3.
Overview of state-of-the-art 3D deep learning methods in computed tomography reconstruction.
Table 3 presents a comprehensive overview of recent advancements in medical imaging, particularly in the field of computed tomography (CT) reconstruction. Researchers worldwide have contributed to this field, showcasing diverse methodologies and their respective population performance metrics. Some notable studies include those by Setio, Li, Meng, Wang, Gruetzema, Gu, Yu, Ren, and Xuhua. These studies have achieved impressive accuracy rates, sensitivity, and specificity in CT detection, improved reconstruction metrics, and enhanced segmentation accuracy. In other research, CNN has been used in CT scans to identify pneumothorax with high sensitivity and specificity and to improve reconstruction metrics. Annarumma achieved 100% sensitivity and 96% specificity for lung nodule classification, while Lee H in the UK focused on CNN for radiograph triaging. The diversification extends beyond CT, with studies exploring DNN-MPRAGE in MRI and comparing DLIR-H in CT. These studies contribute to the evolving landscape of medical imaging, pushing the boundaries in accuracy, speed, and radiation reduction.
4.2.1. Low-Dose CT Reconstruction
The popularity of low-dose CT imaging has grown significantly owing to its effectiveness in reducing radiation exposure. Studies by Li, Meng [42] and Uthoff [49] illustrate the effectiveness of 3D CNNs in achieving high sensitivity and specificity, ensuring accurate diagnostic outcomes. These advancements not only improve patient safety but also demonstrate deep learning’s potential to maintain diagnostic accuracy even with reduced radiation doses.
4.2.2. Sparse-View CT Reconstruction
Researchers such as Park, Sungeun [55] and Higaki, Toru [38] have explored novel techniques, including deep learning algorithms and cycleGAN, to address challenges in sparse-view CT reconstruction. These methods effectively reconstruct images from limited data views, providing a potential solution to the problem.
4.2.3. Country-Based Analysis
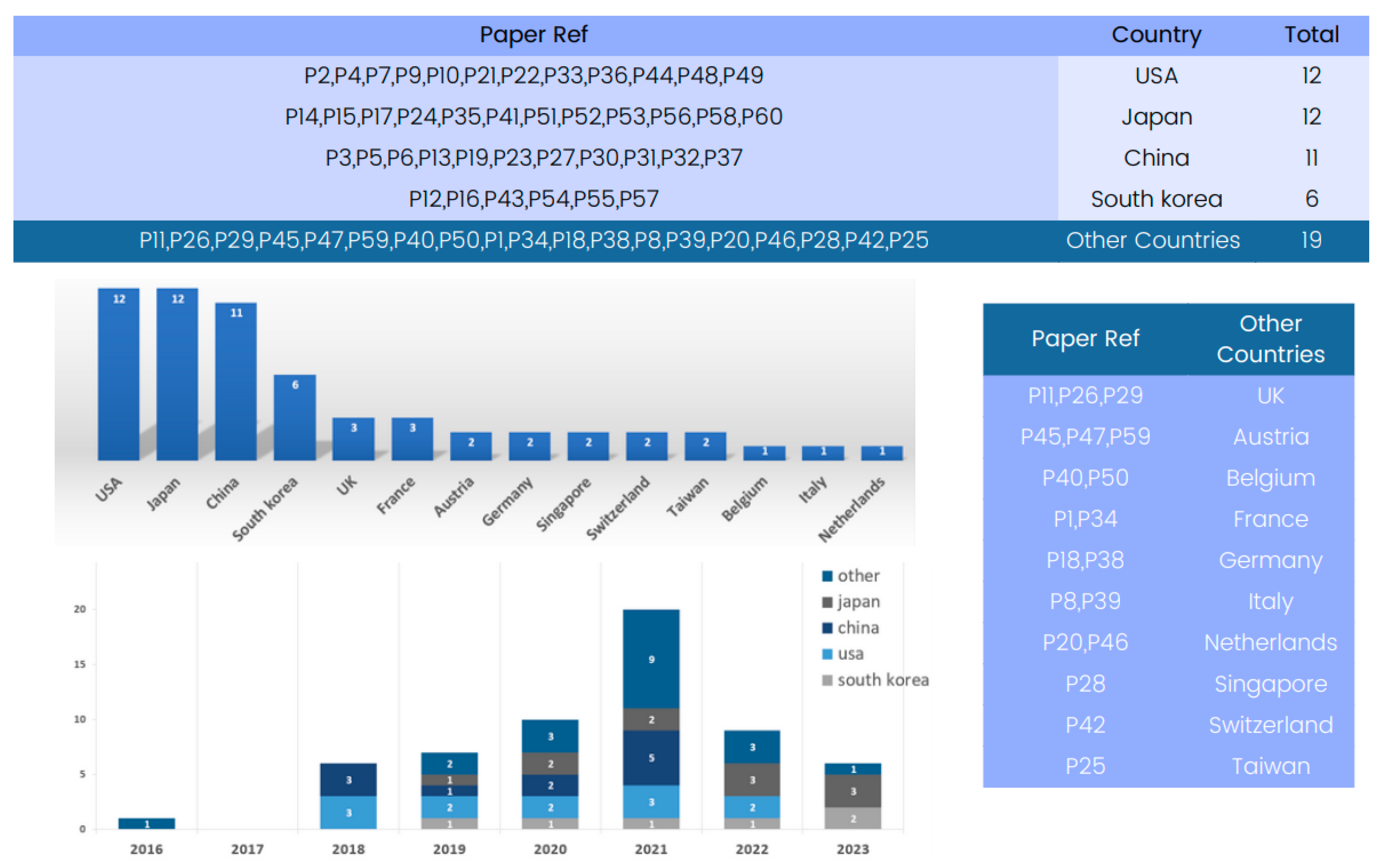
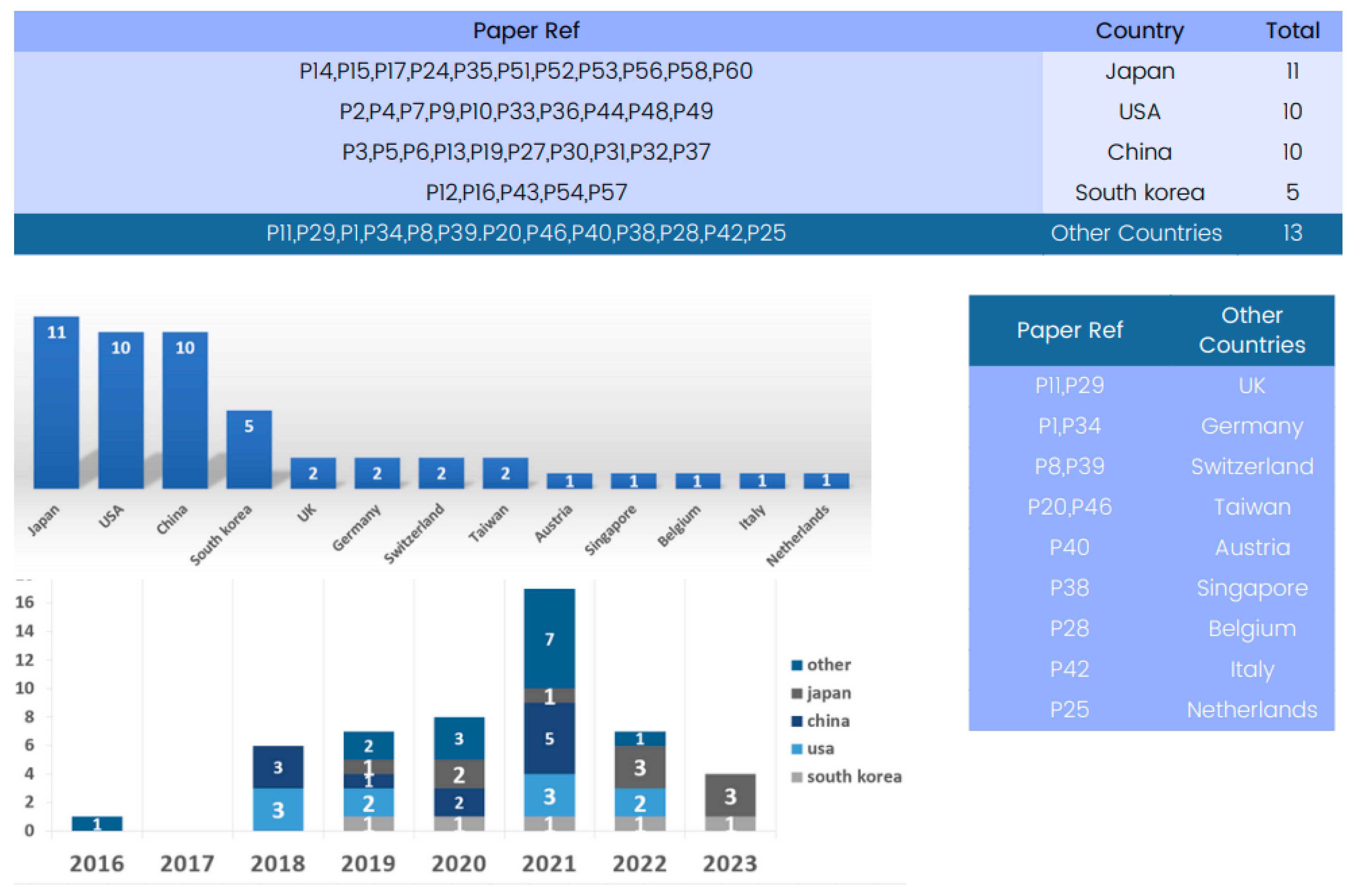
Figure 3 provides a country-based analysis of the state-of-the-art 3D deep learning methods applied to computed tomography reconstruction. A review of the “Country” column reveals a global view of innovation in this field.
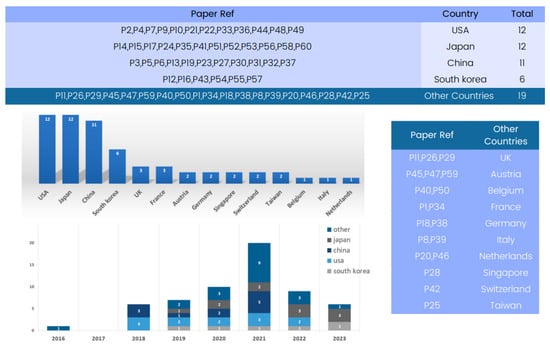
Figure 3.
Global contribution to 3D deep learning research in computed tomography reconstruction over the years.
Several countries have emerged as significant contributors to the advancement of 3D deep learning in computed tomography reconstruction. Notably, the United States and Japan have multiple entries, reflecting their foundational role in developing deep learning methods for CT reconstruction, particularly in lung nodule classification and pneumothorax detection. China and Republic of Korea also appear strongly, with numerous submissions indicating significant contributions in areas such as CT segmentation and image denoising.
4.2.4. Database-Driven
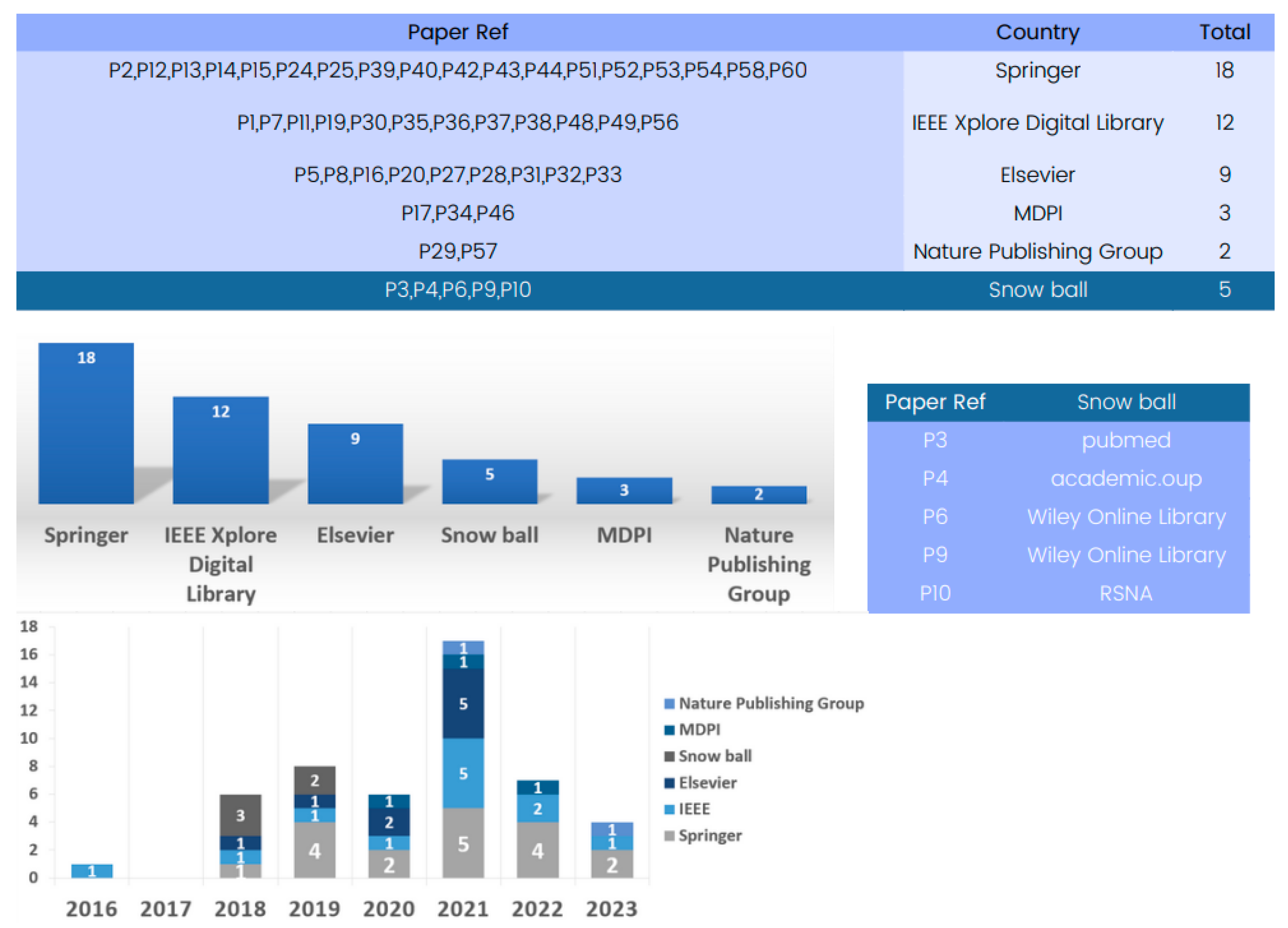
The state-of-the-art methods for 3D deep learning in computed tomography (CT) reconstruction have significantly improved patient care and diagnosis, as evidenced by the comprehensive database analysis presented in Figure 4.
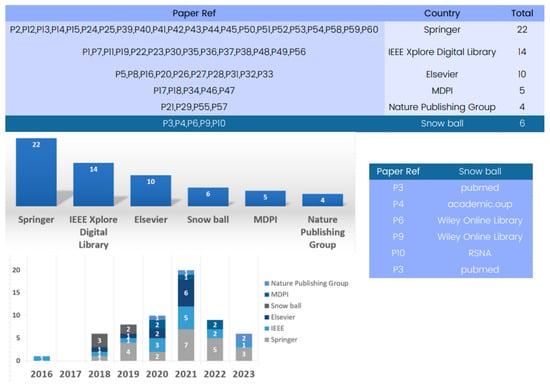
Figure 4.
Evolution of databases utilized in 3D deep learning for computed tomography reconstruction over the years.
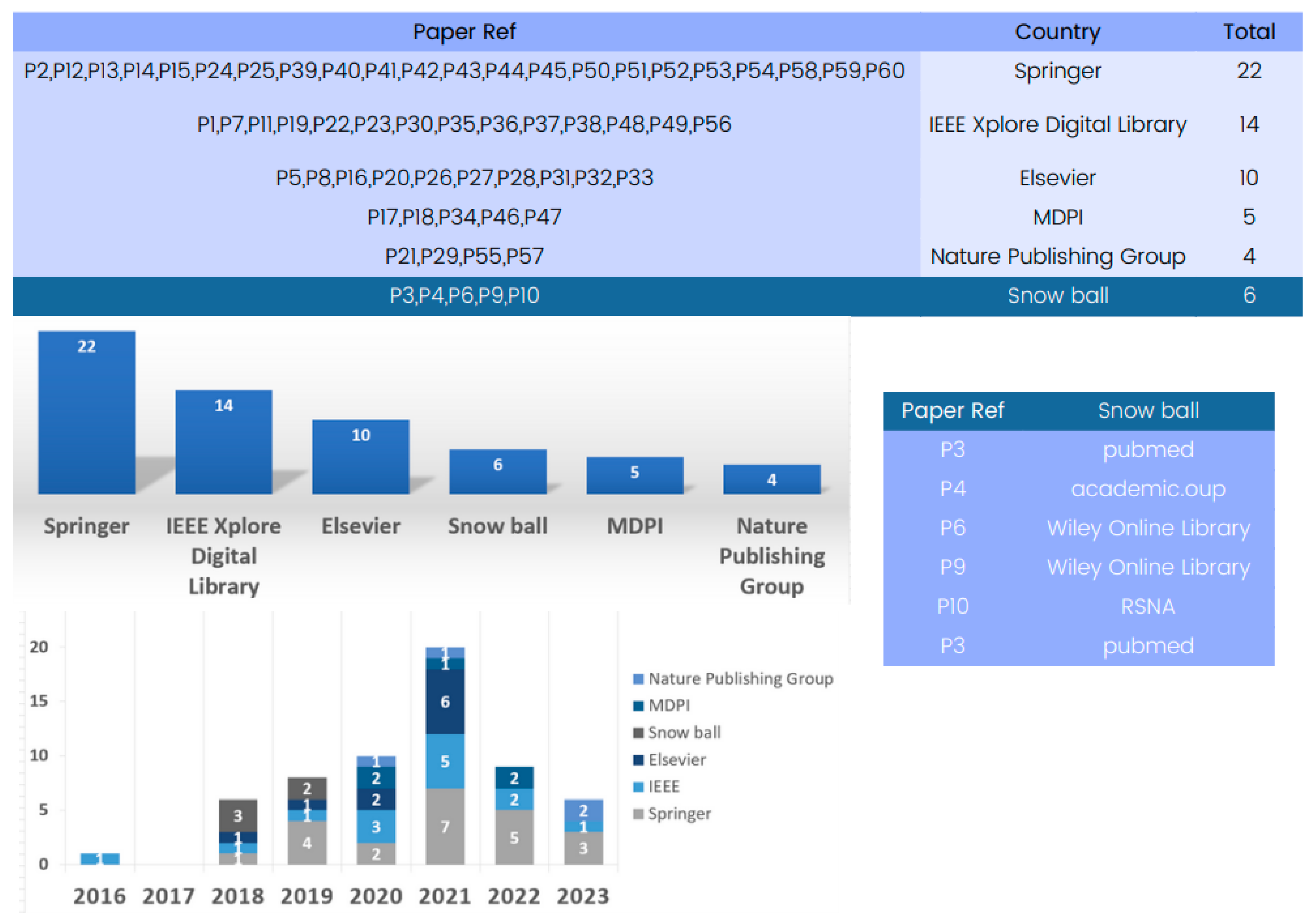
The database analysis reveals numerous research articles on 3D deep learning for CT reconstruction, with Springer being a key platform with 22 papers for showcasing advancements. IEEE and Elsevier have also contributed significantly to the field, with numerous articles highlighting the impressive performance of deep learning models in CT reconstruction.
The database analysis emphasizes the crucial role played by prominent publishers in advancing research on 3D deep learning methods in CT reconstruction, thereby shaping the future landscape of medical imaging.
4.2.5. Methodology Review
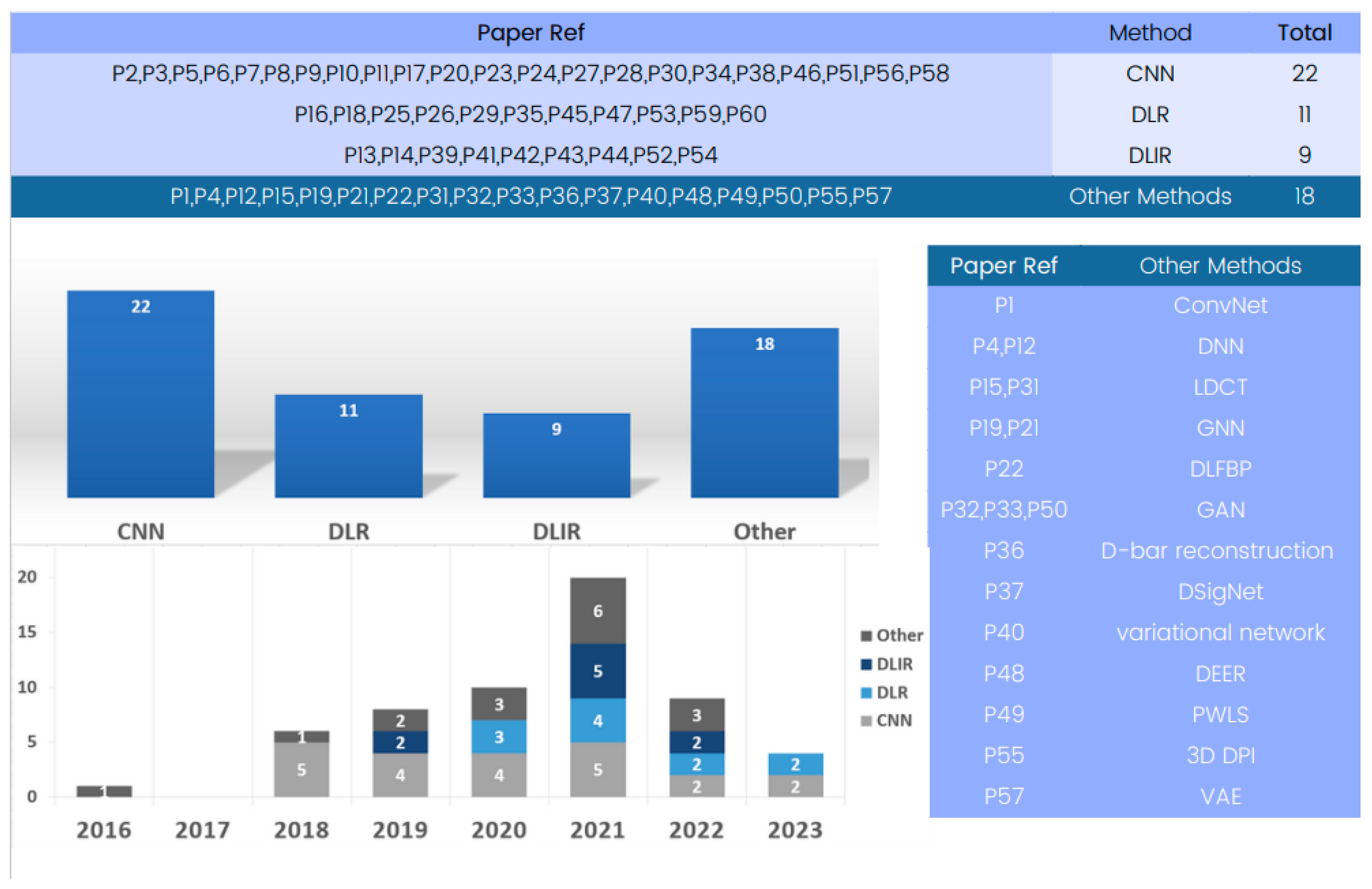
The included papers’ analyses showed a variety of models and methods used in 3D deep learning in computed tomography reconstruction. An examination of Figure 5 reveals methodologies employed in the field of 3D deep learning in CT reconstruction. Among these methodologies, convolutional neural networks (CNNs) emerge as an important method for research and are also prominently featured in numerous entries.
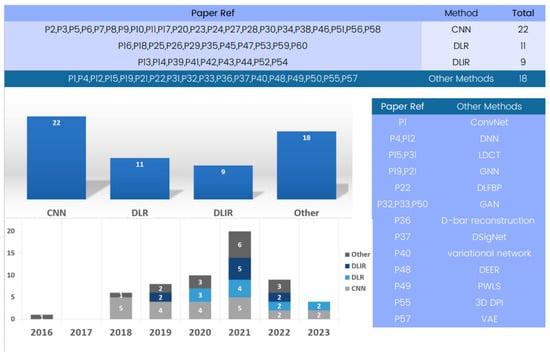
Figure 5.
Evolution of 3D deep learning methods in computed tomography reconstruction over the years.
We include 3D CNNs in the category of CNNs, which are designed to operate on volumetric data and offer improved spatial awareness and accuracy compared to their 2D counterparts. Their three-dimensional convolutional layers allow for more in-depth understanding of anatomical components, making them appropriate for tasks such as segmentation. CNNs are widely accepted in medical imaging due to their consistent usage and effectiveness. They consistently achieve high sensitivity, specificity, and accuracy in tasks like lung nodule classification and pneumothorax detection, demonstrating their enduring relevance and relevance.
Another noteworthy methodology is deep learning reconstruction (DLR) [94], a widely used method that significantly improves image quality and reduces radiation dose. It offers enhanced performance and shorter reconstruction times compared to traditional methods, like filtered back projection (FBP).
Additionally, Figure 5 presents various methods, like deep learning iterative reconstruction (DLIR), variational autoencoders (VAEs), and generative adversarial networks (GANs) [95,96], to tackle medical imaging challenges, enhancing the accuracy, efficiency, and quality of medical image analysis, showcasing the field’s dynamic and innovative nature.
Deep learning reconstruction (DLR) serves as a versatile approach that integrates various methods, including convolutional neural networks (CNNs), generative adversarial networks (GANs), variational autoencoders (VAEs), and recurrent neural networks (RNNs). Many of the reviewed papers utilize DLR as a comprehensive framework for 3D deep learning in computed tomography reconstruction, unifying multiple methods to enhance image quality, reduce radiation dose, and improve overall performance. It is worth noting that some papers mention DLR without specifying individual methods.
4.2.6. Convolutional Neural Networks (CNN)
For a wide range of computer vision applications [97], convolutional neural networks (CNNs) [98,99,100] have become a dominant deep learning model. Given their efficiency in learning hierarchical features from CT images, CNNs play a crucial role in computed tomography reconstruction. CNNs [101] analyze the input data in the context of computed tomography reconstruction by applying several trainable filters that convolve across the input’s structural dimensions. Local patterns and characteristics may be extracted from the computed tomography data using this convolution procedure [102,103]. CNNs may learn representations that are especially suited to added tomography reconstruction tasks by stacking many convolutional layers, which allows them to capture increasingly complicated and abstract aspects from the input data. The hierarchical structure of CNNs enables them to learn features at various abstraction levels. Higher layers of the network learn more sophisticated and meaningful representations, whereas lower levels tend to capture simpler elements, like edges and textures. Due to their capacity to extract hierarchical features, CNNs are highly suited for computed tomography reconstruction [104,105,106] because they can identify important structures and patterns in volumetric CT images.
In this research article on 3D deep learning in computed tomography reconstruction, the use of CNNs is constantly emphasized as a critical strategy. These studies demonstrate how CNNs are widely used for computed tomography reconstruction tasks in a variety of variants and topologies. The extensive use of CNNs underscores their effectiveness in enhancing reconstruction accuracy [107] by leveraging learned characteristics from CT images.
These models may learn to recognize common patterns and structures present in the data by training CNNs on massive datasets of CT images. By assuming missing information from the input scans, CNNs can produce high-quality reconstructions [108,109,110,111]. In comparison to conventional techniques, CNN-based computed tomography reconstruction models have demonstrated significant advancements [112], yielding reconstructions that exhibit higher precision and aesthetic appeal. Furthermore, enhancements in regularization methods can prove beneficial for the application of CNNs in computed tomography reconstruction. Computed tomography reconstruction has been made more effective and efficient by using deep learning regularization (DLR), which typically refers to the use of regularization in machine learning or neural network models to prevent overfitting and enhance model generalization [113,114] using techniques such as the efficient dense learning framework (EDLF). These regularization techniques make use of techniques to improve deep learning models, producing better reconstruction outcomes. Their incorporation also enhances the effectiveness of regularization-based computed tomography reconstruction models. The use of CNNs in computed tomography reconstruction has a lot of potential to advance the science and to advance medical imaging applications.
4.2.7. 3D Convolutional Neural Networks (3DCNN)
Operating on volumetric data like 3D computed tomography scans [115], 3D convolutional neural networks (3D CNNs) [116] have become a potent extension of conventional CNNs. 3D CNNs expand convolutional procedures to capture spatial relationships and complicated patterns throughout the entire volume, in contrast to typical CNNs that analyze 2D pictures individually. A significant advantage of 3D CNNs in computed tomography reconstruction lies in extending convolutional processes to the third dimension. 3D CNNs [117,118,119] can efficiently capture the spatial interdependence and connections that exist within computed tomography scans by taking into account the entire 3D environment of the volumetric data. The precision and robustness of reconstruction approaches are increased as a result of 3D CNNs’ enhanced ability to grasp the complex interactions between various areas and structures.
Research papers on 3D deep learning in computed tomography reconstruction frequently examine the usage of 3D CNNs as a cutting-edge model architecture. These studies show the benefits of employing 3D CNNs for reconstruction as well as their ability to recognize complex patterns and features in CT images. 3D CNNs improve the accuracy of reconstruction by using the volumetric part of the data to include contextual connections and spatial information. Both of these are needed for correct reconstruction. An ongoing project is to apply cutting-edge architectural designs to improve reconstruction results using 3D CNNs in computed tomography reconstruction. By augmenting CNNs’ ability to analyze volumetric data, researchers have overcome the limitations of traditional 2D-based methods. Computed tomography reconstruction using 3D CNNs can capture the intricate anatomical details visible in volumetric CT images. Due to the data’s multidimensionality, 3D CNNs can recognize and comprehend the connections between neighboring slices, which improves the quality of the complete volume reconstruction. This capacity is especially important when the structures of interest span several slices or have complex spatial arrangements.
3D CNNs gain from improvements in regularization methods in addition to their capacity to capture spatial relationships. To further improve the accuracy and robustness of reconstruction approaches, deep neural networks (DNNs) with more complicated architectural designs have been used.
The extensive adoption of 3D CNNs in academic publications attests to their effectiveness in enhancing reconstruction accuracy and their role in advancing the field of computed tomography reconstruction. By using the whole 3D context of computed tomography scans, 3D CNNs offer insights about the intricate structures seen in medical imaging, enabling higher-quality reconstructions and more accurate medical diagnosis and treatment.
4.3. RQ2 What Datasets Are Available for Training and Validating 3D Deep Learning in Computed Tomography Reconstruction?
Understanding the available datasets for training and validating 3D deep learning in computed tomography reconstruction is essential for researchers and practitioners alike. The primary aim of this systematic literature review is to provide a review of available datasets for training and validation in 3D deep learning in computed tomography reconstruction.
The review was conducted through a systematic search of relevant journals and selected studies based on predefined inclusion criteria, ensuring comprehensive coverage of recent developments.
The presented Table 4 provides an overview of 3D deep learning methods for computed tomography reconstruction, encompassing various methodologies and their outcomes in CT images, serving as a valuable resource for researchers, clinicians, and anyone interested in the intersection of 3D deep learning and medical imaging. It showcases the diverse range of available datasets and their potential to impact various CT reconstructions.

Table 4.
Overview of datasets available for training and validation in 3D deep learning for computed tomography reconstruction.
4.3.1. Country-Based Analysis
Figure 6 provides a country-based analysis of available datasets for training and validation in 3D deep learning methods applied to computed tomography reconstruction.
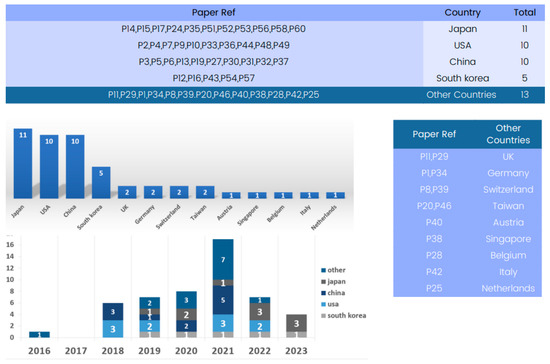
Figure 6.
Global contribution available datasets for training and validation in 3D deep learning research in computed tomography reconstruction over the years.
A complete review of the “Country” column reveals a global view of innovation in this field. Several countries stand out as significant contributors to the advancement of 3D deep learning in computed tomography reconstruction. Based on the review, Japan, the United States, China, and Republic of Korea have made substantial contributions to this field, providing datasets from various medical imaging modalities. These datasets vary in size, content, and application, catering to different aspects of CT reconstruction research. Researchers worldwide have the opportunity to access and utilize these datasets to advance 3D deep learning in CT reconstruction.
4.3.2. Database-Driven Evaluation
Available datasets for training and validating 3D deep learning in computed tomography reconstruction have significantly improved patient care and diagnosis, as evidenced by the comprehensive database analysis presented in Figure 7.
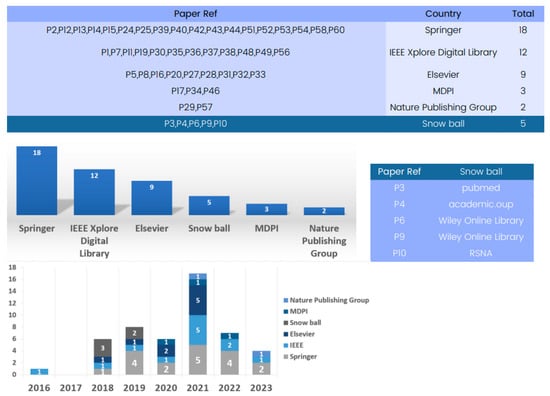
Figure 7.
Evolution of databases utilized for available datasets for training and validation in 3D deep learning for computed tomography reconstruction over the years.
These databases are sourced from various reputable publishers, such as IEEE, Springer, and Elsevier, encompassing datasets primarily focused on CT imaging. Springer serves as a key platform with 18 papers showcasing advancements. Additionally, IEEE and Elsevier have significantly contributed to the field, featuring numerous articles that underscore the impressive performance of available datasets for training and validating 3D deep learning in computed tomography reconstruction.
4.3.3. Dataset Review
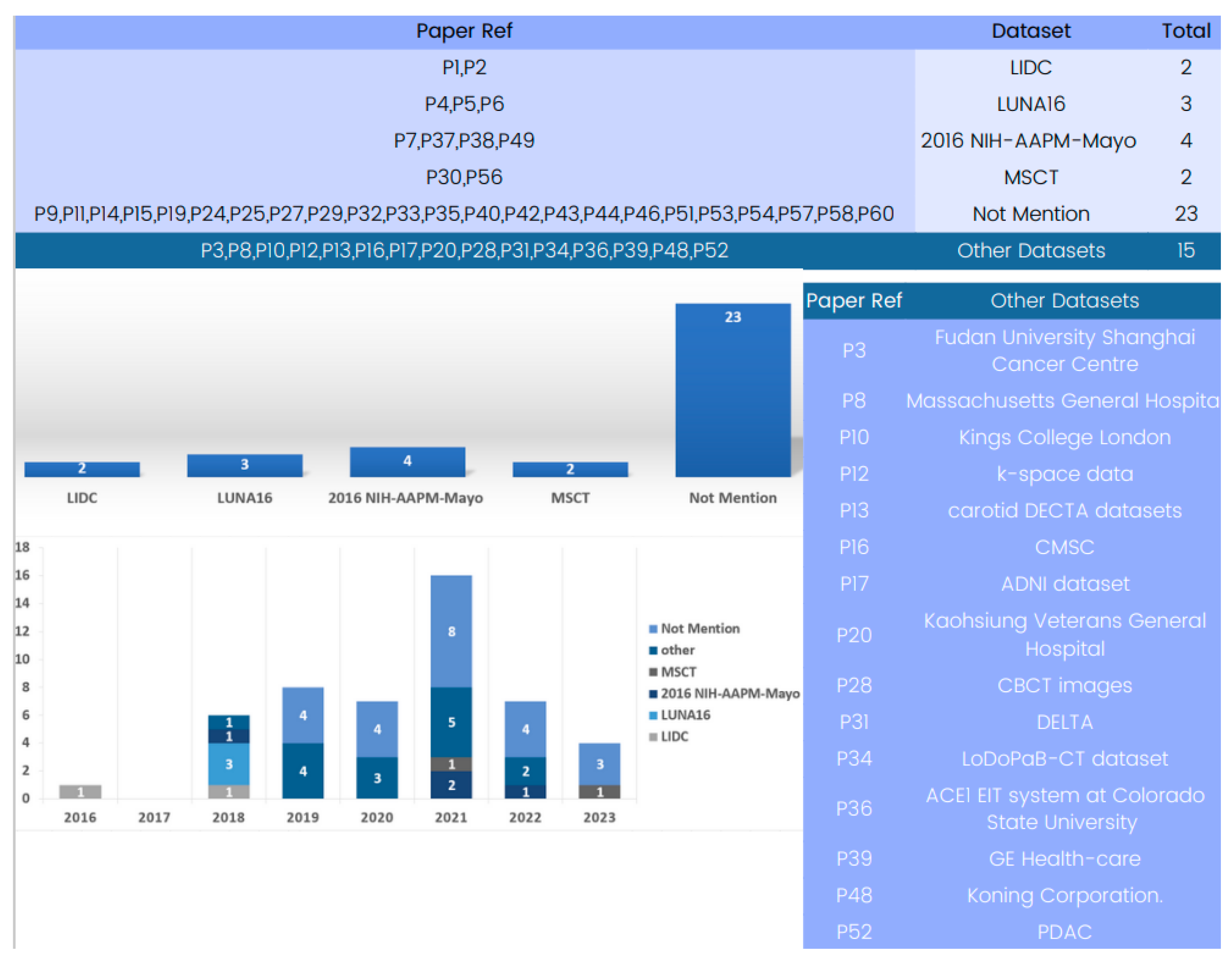
Several datasets have been employed in 3D deep learning for computed tomography (CT) reconstruction, as indicated by the analysis of the included papers. An examination of Figure 8 reveals the available datasets for training and validating 3D deep learning in computed tomography reconstruction. Some studies utilized the LIDC-IDRI dataset [122], which offers an extensive collection of lung nodule CT scans, providing crucial data for the development and evaluation of CT reconstruction models. However, this dataset has not been used in research after 2017. Similarly, LUNA16 [123], employed in three different studies, is a frequently cited dataset but was primarily used in 2018 for testing nodule detection algorithms and has seen limited usage since then.
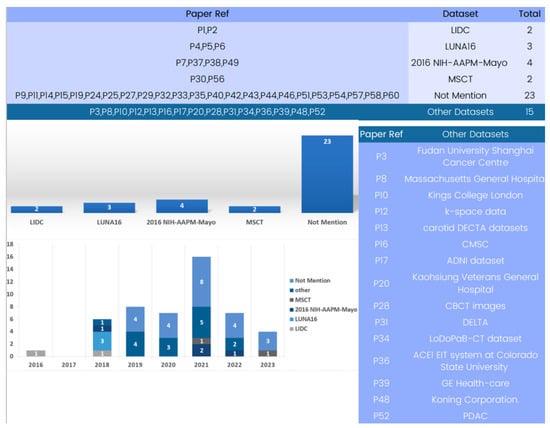
Figure 8.
Evolution of 3D deep learning datasets in computed tomography reconstruction over the years.
In the past few years, the 2016 NIH-AAPM-Mayo and MSCT datasets have become advanced and useful tools for 3D deep learning in CT reconstruction. Notably, several papers use only patient datasets which do not explicitly define the dataset source due to the use of patient data.
The study of datasets used in computed tomography reconstruction with 3D deep learning shows how varied and specific the data used in this field is. For training and evaluating computed tomography reconstruction models in a variety of settings, from lung nodule identification to HRCT and X-ray-based reconstruction, datasets like 2016 NIH-AAPM-Mayo and MSCT are accessible and offer helpful resources. Researchers should carefully consider the characteristics, size, and clinical value of the datasets when selecting the most suitable data for their specific applications. By using a range of datasets, researchers may enhance the robustness, generalizability, and clinical usefulness of 3D deep learning models in computed tomography reconstruction.
4.3.4. Lung Nodule Analysis 2016 (LUNA16)
The study and development of 3D deep learning for computed tomography reconstruction has greatly benefited from the dataset LUNA1651 [124,125] (Lung Nodule Analysis 2016). Since LUNA16 was developed primarily for evaluating nodule detection algorithms, it has gained recognition as a useful tool for creating and validating computed tomography reconstruction models. One of LUNA16’s key benefits is the vast array of lung nodule pictures it offers. A total of 1186 CT pictures make up the collection, which offers a wide range of examples and annotations pertaining to lung nodules. Due to this variability, deep learning models may be created and tested on a large range of instances, accurately representing the heterogeneity and complexity of lung nodules seen in actual computed tomography scans.
Due to the significance of LUNA16 as a benchmark dataset, researchers frequently use it. The performance and generalizability of deep learning models in the context of lung nodule recognition and CT reconstruction are evaluated using this standardized assessment approach. Researchers may evaluate various strategies, algorithms, and methods using LUNA16, making it easier to pinpoint the benefits and drawbacks of computed tomography reconstruction models as well as potential areas for development.
For researchers, the annotations offered by LUNA16 are of great use. These annotations contain ground truth labels and nodule delineations, providing deep learning algorithms with trustworthy benchmarks for training and assessment. The presence of such annotations greatly improves the efficacy and accuracy of the created models.
Many research papers talk about LUNA16 and how important it is for testing and measuring how accurate and useful computed tomography reconstruction models are. The dataset has evolved into a benchmark for comparing and assessing the effectiveness of various algorithms, giving academics a uniform yardstick for judging their strategies. The frequent use of LUNA16 in academic works emphasizes how important it is as a trustworthy and extensively used dataset in the area.
Furthermore, improvements in lung nodule identification and computed tomography reconstruction have been made because of the use of LUNA16. Researchers have improved the detection and reconstruction of lung nodules by using the dataset to build novel deep-learning architectures, feature extraction methods, and post-processing approaches. Researchers can improve the precision and effectiveness of computed tomography reconstruction and, hence, aid in the early identification and diagnosis of lung illnesses by training and evaluating their models using LUNA16.
The fact that LUNA16 is used so often in research papers shows how useful this framework is as a standard evaluation tool and a reliable source for checking the accuracy and usefulness of CT reconstruction models.
4.3.5. Lung Image Database Consortium–Image Database Resource Initiative (LIDC-IDRI)
The study of 3D deep learning in computed tomography reconstruction has greatly benefited from the dataset LIDC-IDRI [126] (Lung Image Database Consortium–Image Database Resource Initiative). It is commonly used and extensively cited in research works in this field. An important resource for the creation and assessment of computed tomography reconstruction models, LIDC-IDRI has a comprehensive collection of about 1186 CT images.
The extensive and varied collection of lung nodule scans that LIDC-IDRI includes is one of its main advantages. Researchers may use real-world computed tomography scans to train and test deep learning algorithms since the dataset provides a wide variety of examples and annotations pertaining to lung nodules.
Deep learning methods for computed tomography reconstruction have advanced and have been improved greatly as a result of the use of LIDC-IDRI. The use of this dataset has allowed researchers to refine their models and explore new avenues for increasing the precision, robustness, and generalizability of computed tomography reconstruction methods. Due to the availability of LIDC-IDRI, it is now easier to evaluate the efficiency and performance of various algorithms, allowing academics to compare and verify their models against industry standards.
The prominence of LIDC-IDRI in the area is demonstrated by the volume of references to and citations of its work in various research articles. Researchers frequently use LIDC-IDRI to confirm the effectiveness of their suggested methods, highlighting the significance and dependability of this dataset.
Radiology as a whole and computed tomography reconstruction methods have advanced as a result of the use of LIDC-IDRI in research. Researchers have utilized the LIDC-IDRI dataset to develop deep learning architectures, feature extraction strategies, and post-processing approaches for CT scan reconstruction. This has led to more accurate and effective models, and evaluation of their effectiveness, robustness, and generalizability. The dataset’s extensive use in academic publications highlights its crucial role in advancing 3D deep learning in computed tomography reconstruction.
4.3.6. 2016 NIH-AAPM-Mayo
The 2016 NIH-AAPM-Mayo [127] dataset is a collection of CT images used in the “2016 Low-Dose CT Grand Challenge [128]”, sponsored by the National Institutes of Health, the American Association of Physicists in Medicine, and the Mayo Clinic. The dataset focused on CT reconstruction and denoising techniques and was made publicly available to the scientific community. Researchers worldwide have used this dataset to improve CT image reconstruction and noise reduction techniques, facilitating further research and development in the field of medical imaging.
The 2016 NIH-AAPM-Mayo dataset [129], used in research papers P07, P37, P38, and P49, has significantly aided in the development of deep learning techniques for CT reconstruction. The dataset, which comprises 500 CT images with 1493 pixels in the view direction and 720 views, has been instrumental in improving image quality and enhancing CT reconstruction methods. In P37 and P38, the dataset was used to train deep learning models, allowing for the exploration of advanced CT reconstruction techniques.
The dataset’s large number of slices and epochs has facilitated robust model training, leading to improved CT reconstruction outcomes. In P49, the dataset was used to obtain LDCT and SDCT images with a size of 512 × 512. This made it possible to test different CT image reconstruction methods, especially those used in low-dose CT imaging. This allowed researchers to assess and compare the performance of other reconstruction methods, leading to improvements in image quality and dose-reduction strategies. The 2016 NIH-AAPM-Mayo dataset has proven to be a valuable resource for researchers, resulting in advancements in CT reconstruction techniques and enhanced deep learning approaches in CT imaging.
4.3.7. MSCT
Multi-slice computed tomography (MSCT) [130] is a non-invasive medical imaging technique that uses X-ray beams and liquid dye to create 3D images of the heart and blood vessels. Unlike traditional coronary angiography, which injects dye into a vein, MSCT injects the dye into a superficial vein. The dye travels through the bloodstream to the coronary arteries, where a CT scanner scans it. The procedure is quick, safe, and requires minimal radiation exposure, making it a safer option for cardiac health assessment.
MSCT datasets [131] have significantly improved CT reconstruction techniques, resulting in sharper and more detailed images as reported in research papers P30 and P56. They have also enhanced the capabilities of deep learning models for CT image reconstruction, and their widespread application across various clinical scenarios highlights their significance in medical imaging.
5. Discussion
In this systematic literature review, we explored and analyzed contemporary state-of-the-art methods for 3D deep learning in CT reconstruction. In addition, we also worked on the availability of datasets to train and validate these methods. By conducting this review, the authors aimed to identify the techniques and approaches used in CT reconstruction and to assess the datasets available for training and validating these models. This paper chiefly focuses on the challenges associated with conventional computed tomography image reconstruction techniques, such as filtered back-projection (FBP), which often results in artifacts and noise that affect the image quality and diagnostic accuracy. The study of deep learning algorithms offered an auspicious solution for the improvement of the quality and competence of CT image reconstruction.
The authors recognized the ability of 3D deep learning to produce more accurate and detailed 3D pictures, hence boosting the diagnostic capabilities of CT scans. The purpose of this study was to enhance the knowledge and uptake of these novel techniques in the field of medical imaging by examining the state of research and developments in 3D deep learning for CT reconstruction.
This systematic literature review has accomplished all its goals. The researchers conducted a comprehensive search on five major databases, identified 60 relevant research articles, and followed the Preferred Reporting Items for Systematic Review and Meta-Analysis (PRISMA) statement to ensure the quality and rigor of the review process. Convolutional neural networks and 3D convolutional neural networks were shown to be the most utilized 3D deep learning approaches for CT reconstruction through this investigation.
In addition, two significant datasets, the 2016 NIH-AAPM-Mayo and MSCT, were recognized as acceptable sources for training and evaluating 3D deep learning systems in CT reconstruction. The primary conclusions of the systematic analysis of the literature underlined the widespread use of CNNs and 3D CNNs in the field of deep-learning-based CT image reconstruction. These cutting-edge algorithms have the potential to increase CT image reconstruction’s efficacy and efficiency, resulting in better picture quality and perhaps even less radiation exposure for patients.
Consideration of the 2016 NIH-AAPM-Mayo and MSCT datasets revealed important information on the availability of pertinent data for developing and analyzing deep learning models in the context of CT reconstruction. The study has important ramifications for diagnostic radiology and the field of medical imaging. The research illustrates the use of 3D deep learning in CT reconstruction, highlighting the potential to improve patient outcomes and diagnostic precision. Adoption of these deep learning techniques has the potential to transform CT reconstruction, making it more accurate and effective, which would be advantageous to patients and healthcare systems alike.
The article concludes that 3D deep learning, especially when using CNNs and 3D CNNs, has enhanced CT reconstruction. These methods may be able to overcome the limitations of traditional imaging techniques, improving image quality, improving diagnostic accuracy, and possibly reducing radiation exposure. In addition, the identification of the datasets that the 2016 NIH-AAPM-Mayo and MSCT approaches provide offers valuable resources for researchers and practitioners interested in developing and validating deep learning models in CT reconstruction.
6. Limitaion of the Literature
There are several limitations to the literature on 3D deep learning in computed tomography reconstruction. Firstly, there is a lack of variety in the test datasets, with an emphasis on particular datasets, which cannot accurately reflect the larger difficulties in computed tomography reconstruction across different bodily systems and disorders. Small sample sizes may limit the generalizability of the results in some research, necessitating larger and more varied datasets for validation. Furthermore, the ability to compare the effectiveness of 3D deep learning models across various populations and imaging techniques is hampered by the lack of external validation. Additionally, it is not easy to fully comprehend the relative performance of multiple methodologies or procedures without comparative analysis. Furthermore, there has been little investigation into imaging techniques outside of computed tomography. Finally, there is a need to close the gap between research results and clinical application in the real world, taking into account elements like clinical workflow integration and validation in sizable clinical trials. By overcoming these restrictions, 3D deep learning in computed tomography reconstruction will become more reliable, generalizable, and clinically applicable, opening the door for its effective use in clinical settings.
7. Conclusions
The systematic literature review explores the current state-of-the-art methods and datasets for training and validation in 3D deep learning for computed tomography (CT) reconstruction. To achieve our goals, we searched major databases and identified relevant research papers. The study has revealed that convolutional neural networks (CNNs), 3D convolutional neural networks (3D CNNs), and deep learning reconstruction (DLR) are the most frequently utilized approaches in this specific field. Moreover, the paper also focuses on the accessibility of datasets for training and validating 3D deep learning models in CT reconstruction. The 2016 NIH-AAPM-Mayo and MSCT datasets were recognized as valuable sources suitable for the purpose of research, highlighting the transformative impact of adopting 3D deep learning techniques in medical imaging, offering the promise of improved patient outcomes and diagnostic precision. By providing a perspicuous insight into both research questions, the paper offers a comprehensive understanding of the current advancements and resources in 3D deep learning in CT reconstruction.
3D deep learning in CT reconstruction faces challenges due to limited test datasets, small sample sizes, and limited external validation. Comparative analysis and real-world application are needed to improve reliability, generalizability, and clinical applicability. Future studies should focus on real-time reconstruction techniques, integrating imaging modalities, and enhancing interpretability. Collaborations between scientists, doctors, and data scientists can create standardized datasets, assessment measures, and benchmarking frameworks for improved precision and effectiveness in medical imaging applications.
Author Contributions
H.R. Conceptualisation, Validation, Methodology, Visualisation, Supervision, Formal analysis, Investigation, Data curation, Writing—original draft. A.R.K. Conceptualisation, Writing—original draft, Formal analysis, Project administration, Resources, Validation. T.S. Formal analysis, review, resources. A.H.F. Formal analysis, review, resources, review and editing. I.U.K. Formal analysis, review, resources. W.H.L. review, Formal analysis. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Jonas, D.E.; Reuland, D.S.; Reddy, S.M.; Nagle, M.; Clark, S.D.; Weber, R.P.; Enyioha, C.; Malo, T.L.; Brenner, A.T.; Armstrong, C.; et al. Screening for lung cancer with low-dose computed tomography: Updated evidence report and systematic review for the US Preventive Services Task Force. JAMA 2021, 325, 971–987. [Google Scholar] [CrossRef] [PubMed]
- Awan, D.; Rehman, H.U. Grid Load Balancing Using Parallel Genetic Algorithm. Int. J. Electron. Electr. Eng. 2015, 3, 451–456. [Google Scholar]
- Greenwood, J.P.; Maredia, N.; Younger, J.F.; Brown, J.M.; Nixon, J.; Everett, C.C.; Bijsterveld, P.; Ridgway, J.P.; Radjenovic, A.; Dickinson, C.J.; et al. Cardiovascular magnetic resonance and single-photon emission computed tomography for diagnosis of coronary heart disease (CE-MARC): A prospective trial. Lancet 2012, 379, 453–460. [Google Scholar] [CrossRef] [PubMed]
- McKeon, A.; Apiwattanakul, M.; Lachance, D.H.; Lennon, V.A.; Mandrekar, J.N.; Boeve, B.F.; Mullan, B.; Mokri, B.; Britton, J.W.; Drubach, D.A.; et al. Positron emission tomography–computed tomography in paraneoplastic neurologic disorders: Systematic analysis and review. Arch. Neurol. 2010, 67, 322–329. [Google Scholar] [CrossRef] [PubMed]
- Abdalla, Y.; Brown, L.; Sonnesen, L. Effects of rapid maxillary expansion on upper airway volume: A three-dimensional cone-beam computed tomography study. Angle Orthod. 2019, 89, 917–923. [Google Scholar] [CrossRef] [PubMed]
- Rahman, H.; Arshad, H.; Mahmud, R.; Mahayuddin, Z.R.; Obeidy, W.K. A framework to visualize 3d breast tumor using x-ray vision technique in mobile augmented reality. J. Telecommun. Electron. Comput. Eng. (JTEC) 2017, 9, 145–149. [Google Scholar]
- Zheng, W.; Zhang, H.; Huang, C.; McQuillan, K.; Li, H.; Xu, W.; Xia, J. Deep-E Enhanced Photoacoustic Tomography Using Three-Dimensional Reconstruction for High-Quality Vascular Imaging. Sensors 2022, 22, 7725. [Google Scholar] [CrossRef]
- Lagerwerf, M.J.; Pelt, D.M.; Palenstijn, W.J.; Batenburg, K.J. A computationally efficient reconstruction algorithm for circular cone-beam computed tomography using shallow neural networks. J. Imaging 2020, 6, 135. [Google Scholar] [CrossRef]
- Obeidy, W.K.; Arshad, H.; Yee Tan, S.; Rahman, H. Developmental analysis of a markerless hybrid tracking technique for mobile augmented reality systems. In Proceedings of the Advances in Visual Informatics: 4th International Visual Informatics Conference, IVIC 2015, Bangi, Malaysia, 17–19 November 2015; Proceedings 4. Springer International Publishing: Berlin/Heidelberg, Germany, 2015; pp. 99–110. [Google Scholar]
- Naresh, K.; Khan, K.; Umer, R.; Cantwell, W.J. The use of X-ray computed tomography for design and process modeling of aerospace composites: A review. Mater. Des. 2020, 190, 108553. [Google Scholar] [CrossRef]
- Nishimoto, S.; Saito, T.; Ishise, H.; Fujiwara, T.; Kawai, K.; Kakibuchi, M. Three-Dimensional Craniofacial Landmark Detection in Series of CT Slices Using Multi-Phased Regression Networks. Diagnostics 2023, 13, 1930. [Google Scholar] [CrossRef]
- Shen, L.; Zhao, W.; Xing, L. Patient-specific reconstruction of volumetric computed tomography images from a single projection view via deep learning. Nat. Biomed. Eng. 2019, 3, 880–888. [Google Scholar] [CrossRef] [PubMed]
- Ang, K.M.; Lim, W.H.; Tiang, S.S.; Rahman, H.; Ang, C.K.; Natarajan, E.; Khan, M.K.A.A.; Pan, L. Training Feedforward Neural Networks Using Arithmetic Optimization Algorithm for Medical Classification. In Proceedings of the Advances in Intelligent Manufacturing and Mechatronics: Selected Articles from the Innovative Manufacturing, Mechatronics & Materials Forum (iM3F 2022), Pahang, Malaysia; Springer Nature: Singapore, 2023; pp. 313–323. [Google Scholar]
- Xie, H.; Shan, H.; Wang, G. Deep encoder-decoder adversarial reconstruction (DEAR) network for 3D CT from few-view data. Bioengineering 2019, 6, 111. [Google Scholar] [CrossRef] [PubMed]
- Yasmeen, B.; Arshad, H.; Hameedur, R. Suspicious Activity Detection Using CCTV Surveillance Video. J. Inf. Syst. Technol. Manag. (JISTM) 2021, 6, 60–70. [Google Scholar] [CrossRef]
- Mahesh, B. Machine learning algorithms-a review. Int. J. Sci. Res. (IJSR) 2020, 9, 381–386. [Google Scholar]
- Junaid, T.; Ayman, A.; Ijaz1, A.; Ali, H.; Imran, A.; Hameedur, R.; Ammar, A.; Inzamam, M.; Saad, R. Fast Intra Mode Selection in HEVC Using Statistical Model. Comput. Mater. Contin. 2022, 70, 3903–3918. [Google Scholar]
- Bano, R.; Toqir, S.M.; Hameedur, R. Detection of Anthracnose Disease in Chili Using IOT and Field Data. LC Int. J. STEM 2020, 1, 75–82. [Google Scholar]
- Kamaruddin, N.S.; Kamsin, A.; Por, L.Y.; Rahman, H. A review of text watermarking: Theory, methods, and applications. IEEE Access 2018, 6, 8011–8028. [Google Scholar] [CrossRef]
- Singh, S.P.; Wang, L.; Gupta, S.; Goli, H.; Padmanabhan, P.; Gulyás, B. 3D deep learning on medical images: A review. Sensors 2020, 20, 5097. [Google Scholar] [CrossRef]
- Apivanichkul, K.; Phasukkit, P.; Dankulchai, P.; Sittiwong, W.; Jitwatcharakomol, T. Enhanced Deep-Learning-Based Automatic Left-Femur Segmentation Scheme with Attribute Augmentation. Sensors 2023, 23, 5720. [Google Scholar] [CrossRef]
- Salleh, S.; Mahmud, R.; Rahman, H.; Yasiran, S. Speed up Robust Features (SURF) with Principal Component Analysis-Support Vector Machine (PCA-SVM) for benign and malignant classifications. J. Fundam. Appl. Sci. 2017, 9, 624–643. [Google Scholar] [CrossRef]
- Rahman, H.; Lashkari, A.H.; Weng, E.N.G.; Parhizkar, B. Appliance Mobile Positioning System (AMPS) (An Advanced mobile Application). Int. J. Comput. Sci. Inf. Secur. 2010, 8, 207–215. [Google Scholar]
- Ang, K.M.; Natarajan, E.; Isa, N.A.M.; Sharma, A.; Rahman, H.; Then, R.Y.S.; Alrifaey, M.; Tiang, S.S.; Lim, W.H. Modified teaching-learning-based optimization and applications in multi-response machining processes. Comput. Ind. Eng. 2022, 174, 108719. [Google Scholar] [CrossRef]
- MacCormick, I.J.; Williams, B.M.; Zheng, Y.; Li, K.; Al-Bander, B.; Czanner, S.; Cheeseman, R.; Willoughby, C.E.; Brown, E.N.; Spaeth, G.L.; et al. Accurate, fast, data efficient and interpretable glaucoma diagnosis with automated spatial analysis of the whole cup to disc profile. PLoS ONE 2019, 14, e0209409. [Google Scholar]
- Sameen, D.; Faryal, I.; Hameedur, R. Skin Cancer Disease Detection Using Image Processing Techniques. LC Int. J. STEM 2020, 1, 50–58. [Google Scholar]
- Ahmad, R.; Khalid, A.; Hameedur, R. Brain Tumor Detection Using Image Segmentation and Classification. LC Int. J. STEM 2020, 1, 59–65. [Google Scholar]
- Keele, S. Guidelines for Performing Systematic Literature Reviews in Software Engineering; 2007. Technical Report, Ver. 2.3 Ebse Technical Report. Ebse. Available online: https://www.researchgate.net/profile/Barbara-Kitchenham/publication/302924724_Guidelines_for_performing_Systematic_Literature_Reviews_in_Software_Engineering/links/61712932766c4a211c03a6f7/Guidelines-for-performing-Systematic-Literature-Reviews-in-Software-Engineering.pdf (accessed on 26 November 2023).
- Bukht, T.F.N.; Rahman, H.; Jalal, A. A Novel Framework for Human Action Recognition Based on Features Fusion and Decision Tree. In Proceedings of the 2023 4th International Conference on Advancements in Computational Sciences (ICACS), Lahore, Pakistan, 20–22 February 2023; pp. 1–6. [Google Scholar]
- Aromataris, E.; Pearson, A. The systematic review: An overview. AJN Am. J. Nurs. 2014, 114, 53–58. [Google Scholar] [CrossRef]
- Improving tomographic reconstruction from limited data using mixed-scale dense convolutional neural networks. LC Int. J. STEM 2018, 4, 128.
- Devaney, A.J. A filtered backpropagation algorithm for diffraction tomography. Ultrason. Imaging 1982, 4, 336–350. [Google Scholar] [CrossRef]
- Tang, X.; Hsieh, J.; Nilsen, R.A.; Dutta, S.; Samsonov, D.; Hagiwara, A. A three-dimensional-weighted cone beam filtered backprojection (CB-FBP) algorithm for image reconstruction in volumetric CT—Helical scanning. Phys. Med. Biol. 2006, 51, 855. [Google Scholar] [CrossRef]
- Tang, X.; Hsieh, J.; Hagiwara, A.; Nilsen, R.A.; Thibault, J.B.; Drapkin, E. A three-dimensional weighted cone beam filtered backprojection (CB-FBP) algorithm for image reconstruction in volumetric CT under a circular source trajectory. Phys. Med. Biol. 2005, 50, 3889. [Google Scholar] [CrossRef]
- Beister, M.; Kolditz, D.; Kalender, W.A. Iterative reconstruction methods in X-ray CT. Phys. Medica 2012, 28, 94–108. [Google Scholar] [CrossRef] [PubMed]
- Willemink, M.J.; de Jong, P.A.; Leiner, T.; de Heer, L.M.; Nievelstein, R.A.; Budde, R.P.; Schilham, A.M. Iterative reconstruction techniques for computed tomography Part 1: Technical principles. Eur. Radiol. 2013, 23, 1623–1631. [Google Scholar] [CrossRef] [PubMed]
- Geyer, L.L.; Schoepf, U.J.; Meinel, F.G.; Nance, J.W., Jr.; Bastarrika, G.; Leipsic, J.A.; Paul, N.S.; Rengo, M.; Laghi, A.; De Cecco, C.N. State of the art: Iterative CT reconstruction techniques. Radiology 2015, 276, 339–357. [Google Scholar] [CrossRef] [PubMed]
- Higaki, T.; Nakamura, Y.; Zhou, J.; Yu, Z.; Nemoto, T.; Tatsugami, F.; Awai, K. Deep learning reconstruction at CT: Phantom study of the image characteristics. Acad. Radiol. 2020, 27, 82–87. [Google Scholar] [CrossRef] [PubMed]
- McLeavy, C.; Chunara, M.; Gravell, R.; Rauf, A.; Cushnie, A.; Talbot, C.S.; Hawkins, R. The future of CT: Deep learning reconstruction. Clin. Radiol. 2021, 76, 407–415. [Google Scholar] [CrossRef]
- Wohlin, C. Guidelines for snowballing in systematic literature studies and a replication in software engineering. In Proceedings of the 18th International Conference on Evaluation and Assessment in Software Engineering, London, UK, 13–14 May 2014; pp. 1–10. [Google Scholar]
- Setio, A.A.A.; Ciompi, F.; Litjens, G.; Gerke, P.; Jacobs, C.; Van Riel, S.J.; Wille, M.M.W.; Naqibullah, M.; Sánchez, C.I.; Van Ginneken, B. Pulmonary nodule detection in CT images: False positive reduction using multi-view convolutional networks. IEEE Trans. Med. Imaging 2016, 35, 1160–1169. [Google Scholar] [CrossRef]
- Li, M.; Shen, S.; Gao, W.; Hsu, W.; Cong, J. Computed tomography image enhancement using 3D convolutional neural network. In Proceedings of the Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support: 4th International Workshop, DLMIA 2018, and 8th International Workshop, ML-CDS 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, 20 September 2018; Springer: Berlin/Heidelberg, Germany, 2018; pp. 291–299. [Google Scholar]
- Wang, S.; Wang, R.; Zhang, S.; Li, R.; Fu, Y.; Sun, X.; Li, Y.; Sun, X.; Jiang, X.; Guo, X.; et al. 3D convolutional neural network for differentiating pre-invasive lesions from invasive adenocarcinomas appearing as ground-glass nodules with diameters ≤ 3 cm using HRCT. Quant. Imaging Med. Surg. 2018, 8, 491. [Google Scholar] [CrossRef]
- Gruetzemacher, R.; Gupta, A.; Paradice, D. 3D deep learning for detecting pulmonary nodules in CT scans. J. Am. Med. Informatics Assoc. 2018, 25, 1301–1310. [Google Scholar] [CrossRef]
- Gu, Y.; Lu, X.; Yang, L.; Zhang, B.; Yu, D.; Zhao, Y.; Gao, L.; Wu, L.; Zhou, T. Automatic lung nodule detection using a 3D deep convolutional neural network combined with a multi-scale prediction strategy in chest CTs. Comput. Biol. Med. 2018, 103, 220–231. [Google Scholar] [CrossRef]
- Ren, X.; Xiang, L.; Nie, D.; Shao, Y.; Zhang, H.; Shen, D.; Wang, Q. Interleaved 3D-CNN s for joint segmentation of small-volume structures in head and neck CT images. Med. Phys. 2018, 45, 2063–2075. [Google Scholar] [CrossRef]
- Gupta, H.; Jin, K.H.; Nguyen, H.Q.; McCann, M.T.; Unser, M. CNN-based projected gradient descent for consistent CT image reconstruction. IEEE Trans. Med. Imaging 2018, 37, 1440–1453. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Thrall, J.H.; Digumarthy, S.R.; Kalra, M.K.; Pandharipande, P.V.; Zhang, B.; Nitiwarangkul, C.; Singh, R.; Khera, R.D.; Li, Q. Deep learning-enabled system for rapid pneumothorax screening on chest CT. Eur. J. Radiol. 2019, 120, 108692. [Google Scholar] [CrossRef] [PubMed]
- Uthoff, J.; Stephens, M.J.; Newell, J.D., Jr.; Hoffman, E.A.; Larson, J.; Koehn, N.; De Stefano, F.A.; Lusk, C.M.; Wenzlaff, A.S.; Watza, D.; et al. Machine learning approach for distinguishing malignant and benign lung nodules utilizing standardized perinodular parenchymal features from CT. Med. Phys. 2019, 46, 3207–3216. [Google Scholar] [CrossRef] [PubMed]
- Annarumma, M.; Withey, S.J.; Bakewell, R.J.; Pesce, E.; Goh, V.; Montana, G. Automated triaging of adult chest radiographs with deep artificial neural networks. Radiology 2019, 291, 196–202. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Lee, J.; Kim, H.; Cho, B.; Cho, S. Deep-neural-network-based sinogram synthesis for sparse-view CT image reconstruction. IEEE Trans. Radiat. Plasma Med. Sci. 2018, 3, 109–119. [Google Scholar] [CrossRef]
- Jung, W.; Kim, J.; Ko, J.; Jeong, G.; Kim, H.G. Highly accelerated 3D MPRAGE using deep neural network–based reconstruction for brain imaging in children and young adults. Eur. Radiol. 2022, 32, 5468–5479. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Jin, D.; Liu, Z.; Zhang, Y.; Ni, M.; Yuan, H. Deep learning image reconstruction algorithm for carotid dual-energy computed tomography angiography: Evaluation of image quality and diagnostic performance. Insights Imaging 2022, 13, 182. [Google Scholar] [CrossRef]
- Sato, M.; Ichikawa, Y.; Domae, K.; Yoshikawa, K.; Kanii, Y.; Yamazaki, A.; Nagasawa, N.; Nagata, M.; Ishida, M.; Sakuma, H. Deep learning image reconstruction for improving image quality of contrast-enhanced dual-energy CT in abdomen. Eur. Radiol. 2022, 32, 5499–5507. [Google Scholar] [CrossRef]
- Park, S.; Yoon, J.H.; Joo, I.; Yu, M.H.; Kim, J.H.; Park, J.; Kim, S.W.; Han, S.; Ahn, C.; Kim, J.H.; et al. Image quality in liver CT: Low-dose deep learning vs standard-dose model-based iterative reconstructions. Eur. Radiol. 2022, 32, 2865–2874. [Google Scholar] [CrossRef]
- Zhang, J.; Gong, L.R.; Yu, K.; Qi, X.; Wen, Z.; Hua, Q.; Myint, S.H. 3D reconstruction for super-resolution CT images in the Internet of health things using deep learning. IEEE Access 2020, 8, 121513–121525. [Google Scholar] [CrossRef]
- Liang, C.H.; Liu, Y.C.; Wu, M.T.; Garcia-Castro, F.; Alberich-Bayarri, A.; Wu, F.Z. Identifying pulmonary nodules or masses on chest radiography using deep learning: External validation and strategies to improve clinical practice. Clin. Radiol. 2020, 75, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Ye, J.C.; De Man, B. Deep learning for tomographic image reconstruction. Nat. Mach. Intell. 2020, 2, 737–748. [Google Scholar] [CrossRef]
- Fu, J.; Dong, J.; Zhao, F. A deep learning reconstruction framework for differential phase-contrast computed tomography with incomplete data. IEEE Trans. Image Process. 2019, 29, 2190–2202. [Google Scholar] [CrossRef] [PubMed]
- Jiao, F.; Gui, Z.; Li, K.; Shangguang, H.; Wang, Y.; Liu, Y.; Zhang, P. A dual-domain CNN-based network for CT reconstruction. IEEE Access 2021, 9, 71091–71103. [Google Scholar] [CrossRef]
- Ichikawa, S.; Hamada, M.; Sugimori, H. A deep-learning method using computed tomography scout images for estimating patient body weight. Sci. Rep. 2021, 11, 15627. [Google Scholar] [CrossRef]
- Oostveen, L.J.; Meijer, F.J.; de Lange, F.; Smit, E.J.; Pegge, S.A.; Steens, S.C.; van Amerongen, M.J.; Prokop, M.; Sechopoulos, I. Deep learning–based reconstruction may improve non-contrast cerebral CT imaging compared to other current reconstruction algorithms. Eur. Radiol. 2021, 31, 5498–5506. [Google Scholar] [CrossRef]
- Zeng, L.; Xu, X.; Zeng, W.; Peng, W.; Zhang, J.; Sixian, H.; Liu, K.; Xia, C.; Li, Z. Deep learning trained algorithm maintains the quality of half-dose contrast-enhanced liver computed tomography images: Comparison with hybrid iterative reconstruction: Study for the application of deep learning noise reduction technology in low dose. Eur. J. Radiol. 2021, 135, 109487. [Google Scholar] [CrossRef]
- Verhelst, P.J.; Smolders, A.; Beznik, T.; Meewis, J.; Vandemeulebroucke, A.; Shaheen, E.; Van Gerven, A.; Willems, H.; Politis, C.; Jacobs, R. Layered deep learning for automatic mandibular segmentation in cone-beam computed tomography. J. Dent. 2021, 114, 103786. [Google Scholar] [CrossRef]
- Aggarwal, R.; Sounderajah, V.; Martin, G.; Ting, D.S.; Karthikesalingam, A.; King, D.; Ashrafian, H.; Darzi, A. Diagnostic accuracy of deep learning in medical imaging: A systematic review and meta-analysis. NPJ Digit. Med. 2021, 4, 65. [Google Scholar] [CrossRef]
- Chung, Y.W.; Choi, I.Y. Detection of abnormal extraocular muscles in small datasets of computed tomography images using a three-dimensional variational autoencoder. Sci. Rep. 2023, 13, 1765. [Google Scholar] [CrossRef]
- Jiang, H.; Tang, S.; Liu, W.; Zhang, Y. Deep learning for COVID-19 chest CT (computed tomography) image analysis: A lesson from lung cancer. Comput. Struct. Biotechnol. J. 2021, 19, 1391–1399. [Google Scholar] [CrossRef] [PubMed]
- Hsu, K.T.; Guan, S.; Chitnis, P.V. Comparing deep learning frameworks for photoacoustic tomography image reconstruction. Photoacoustics 2021, 23, 100271. [Google Scholar] [CrossRef] [PubMed]
- Leuschner, J.; Schmidt, M.; Ganguly, P.S.; Andriiashen, V.; Coban, S.B.; Denker, A.; Bauer, D.; Hadjifaradji, A.; Batenburg, K.J.; Maass, P.; et al. Quantitative comparison of deep learning-based image reconstruction methods for low-dose and sparse-angle CT applications. J. Imaging 2021, 7, 44. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, M.; Zhou, J.; Akino, N.; Yu, Z. Feature-aware deep-learning reconstruction for context-sensitive X-ray computed tomography. IEEE Trans. Radiat. Plasma Med. Sci. 2020, 5, 99–107. [Google Scholar] [CrossRef]
- Capps, M.; Mueller, J.L. Reconstruction of organ boundaries with deep learning in the D-bar method for electrical impedance tomography. IEEE Trans. Biomed. Eng. 2020, 68, 826–833. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Chen, S.; Zhang, H.; Tao, X.; Lin, W.; Zhang, S.; Zeng, D.; Ma, J. Downsampled imaging geometric modeling for accurate CT reconstruction via deep learning. IEEE Trans. Med. Imaging 2021, 40, 2976–2985. [Google Scholar] [CrossRef] [PubMed]
- Ding, Q.; Nan, Y.; Gao, H.; Ji, H. Deep learning with adaptive hyper-parameters for low-dose CT image reconstruction. IEEE Trans. Comput. Imaging 2021, 7, 648–660. [Google Scholar] [CrossRef]
- Benz, D.C.; Ersözlü, S.; Mojon, F.L.; Messerli, M.; Mitulla, A.K.; Ciancone, D.; Kenkel, D.; Schaab, J.A.; Gebhard, C.; Pazhenkottil, A.P.; et al. Radiation dose reduction with deep-learning image reconstruction for coronary computed tomography angiography. Eur. Radiol. 2022, 32, 2620–2628. [Google Scholar] [CrossRef]
- Hammernik, K.; Würfl, T.; Pock, T.; Maier, A. A deep learning architecture for limited-angle computed tomography reconstruction. In Proceedings of the Bildverarbeitung für die Medizin 2017: Algorithmen-Systeme-Anwendungen. Proceedings des Workshops vom 12. bis 14. März 2017 in Heidelberg; Springer: Berlin/Heidelberg, Germany, 2017; pp. 92–97. [Google Scholar]
- Noda, Y.; Kawai, N.; Nagata, S.; Nakamura, F.; Mori, T.; Miyoshi, T.; Suzuki, R.; Kitahara, F.; Kato, H.; Hyodo, F.; et al. Deep learning image reconstruction algorithm for pancreatic protocol dual-energy computed tomography: Image quality and quantification of iodine concentration. Eur. Radiol. 2022, 32, 384–394. [Google Scholar] [CrossRef]
- De Santis, D.; Polidori, T.; Tremamunno, G.; Rucci, C.; Piccinni, G.; Zerunian, M.; Pugliese, L.; Del Gaudio, A.; Guido, G.; Barbato, L.; et al. Deep learning image reconstruction algorithm: Impact on image quality in coronary computed tomography angiography. La Radiol. Medica 2023, 128, 434–444. [Google Scholar] [CrossRef]
- Kim, I.; Kang, H.; Yoon, H.J.; Chung, B.M.; Shin, N.Y. Deep learning–based image reconstruction for brain CT: Improved image quality compared with adaptive statistical iterative reconstruction-Veo (ASIR-V). Neuroradiology 2021, 63, 905–912. [Google Scholar] [CrossRef] [PubMed]
- Thapaliya, S.; Brady, S.L.; Somasundaram, E.; Anton, C.G.; Coley, B.D.; Towbin, A.J.; Zhang, B.; Dillman, J.R.; Trout, A.T. Detection of urinary tract calculi on CT images reconstructed with deep learning algorithms. Abdom. Radiol. 2022, 47, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Greffier, J.; Durand, Q.; Frandon, J.; Si-Mohamed, S.; Loisy, M.; de Oliveira, F.; Beregi, J.P.; Dabli, D. Improved image quality and dose reduction in abdominal CT with deep-learning reconstruction algorithm: A phantom study. Eur. Radiol. 2023, 33, 699–710. [Google Scholar] [CrossRef] [PubMed]
- Kuo, C.F.J.; Liao, Y.S.; Barman, J.; Liu, S.C. Semi-supervised deep learning semantic segmentation for 3D volumetric computed tomographic scoring of chronic rhinosinusitis: Clinical correlations and comparison with Lund-Mackay scoring. Tomography 2022, 8, 718–729. [Google Scholar] [CrossRef]
- Lenfant, M.; Comby, P.O.; Guillen, K.; Galissot, F.; Haioun, K.; Thay, A.; Chevallier, O.; Ricolfi, F.; Loffroy, R. Deep Learning-Based Reconstruction vs. Iterative Reconstruction for Quality of Low-Dose Head-and-Neck CT Angiography with Different Tube-Voltage Protocols in Emergency-Department Patients. Diagnostics 2022, 12, 1287. [Google Scholar] [CrossRef]
- Xie, H.; Shan, H.; Cong, W.; Liu, C.; Zhang, X.; Liu, S.; Ning, R.; Wang, G. Deep efficient end-to-end reconstruction (DEER) network for few-view breast CT image reconstruction. IEEE Access 2020, 8, 196633–196646. [Google Scholar] [CrossRef]
- Park, H.S.; Kim, K.; Jeon, K. Low-dose CT image reconstruction with a deep learning prior. IEEE Access 2020, 8, 158647–158655. [Google Scholar] [CrossRef]
- Thaler, F.; Hammernik, K.; Payer, C.; Urschler, M.; Štern, D. Sparse-view CT reconstruction using wasserstein GANs. In Proceedings of the International Workshop on Machine Learning for Medical Image Reconstruction, Granada, Spain, 16 September 2018; Springer: Berlin/Heidelberg, Germany, 2018; pp. 75–82. [Google Scholar]
- Koike, Y.; Ohira, S.; Teraoka, Y.; Matsumi, A.; Imai, Y.; Akino, Y.; Miyazaki, M.; Nakamura, S.; Konishi, K.; Tanigawa, N.; et al. Pseudo low-energy monochromatic imaging of head and neck cancers: Deep learning image reconstruction with dual-energy CT. Int. J. Comput. Assist. Radiol. Surg. 2022, 17, 1271–1279. [Google Scholar] [CrossRef]
- Noda, Y.; Iritani, Y.; Kawai, N.; Miyoshi, T.; Ishihara, T.; Hyodo, F.; Matsuo, M. Deep learning image reconstruction for pancreatic low-dose computed tomography: Comparison with hybrid iterative reconstruction. Abdom. Radiol. 2021, 46, 4238–4244. [Google Scholar] [CrossRef]
- Nakamura, Y.; Narita, K.; Higaki, T.; Akagi, M.; Honda, Y.; Awai, K. Diagnostic value of deep learning reconstruction for radiation dose reduction at abdominal ultra-high-resolution CT. Eur. Radiol. 2021, 31, 4700–4709. [Google Scholar] [CrossRef]
- Nam, J.G.; Hong, J.H.; Kim, D.S.; Oh, J.; Goo, J.M. Deep learning reconstruction for contrast-enhanced CT of the upper abdomen: Similar image quality with lower radiation dose in direct comparison with iterative reconstruction. Eur. Radiol. 2021, 31, 5533–5543. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.; Kim, M.W.; Jin, K.H.; Yi, K.M.; Kohmura, Y.; Ishikawa, T.; Je, J.H.; Park, J. Deep 3D reconstruction of synchrotron X-ray computed tomography for intact lungs. Sci. Rep. 2023, 13, 1738. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Zhang, X.; Kawasumi, Y.; Usui, A.; Ichiji, K.; Funayama, M.; Homma, N. A 2.5 D deep learning-based method for drowning diagnosis using post-mortem computed tomography. IEEE J. Biomed. Health Informatics 2022, 27, 1026–1035. [Google Scholar] [CrossRef] [PubMed]
- Shiode, R.; Kabashima, M.; Hiasa, Y.; Oka, K.; Murase, T.; Sato, Y.; Otake, Y. 2D–3D reconstruction of distal forearm bone from actual X-ray images of the wrist using convolutional neural networks. Sci. Rep. 2021, 11, 15249. [Google Scholar] [CrossRef]
- Bornet, P.A.; Villani, N.; Gillet, R.; Germain, E.; Lombard, C.; Blum, A.; Gondim Teixeira, P.A. Clinical acceptance of deep learning reconstruction for abdominal CT imaging: Objective and subjective image quality and low-contrast detectability assessment. Eur. Radiol. 2022, 32, 3161–3172. [Google Scholar] [CrossRef]
- Kroll, N.; Abu-Zurayk, M.; Dimitrov, D.; Franz, T.; Führer, T.; Gerhold, T.; Görtz, S.; Heinrich, R.; Ilic, C.; Jepsen, J.; et al. DLR project Digital-X: Towards virtual aircraft design and flight testing based on high-fidelity methods. CEAS Aeronaut. J. 2016, 7, 3–27. [Google Scholar] [CrossRef]
- Pan, Z.; Yu, W.; Yi, X.; Khan, A.; Yuan, F.; Zheng, Y. Recent progress on generative adversarial networks (GANs): A survey. IEEE Access 2019, 7, 36322–36333. [Google Scholar] [CrossRef]
- Naeem, A.; Rizwan, M.; Alsubai, S.; Almadhor, A.; Akhtaruzzaman, M.; Islam, S.; Rahman, H. Enhanced clustering based routing protocol in vehicular ad-hoc networks. IET Electr. Syst. Transp. 2023, 13, e12069. [Google Scholar] [CrossRef]
- Arshad, M.Z.; Rahman, H.; Tariq, J.; Riaz, A.; Imran, A.; Yasin, A.; Ihsan, I. Digital Forensics Analysis of IoT Nodes using Machine Learning. J. Comput. Biomed. Inform. 2022, 4, 1–12. [Google Scholar]
- Kattenborn, T.; Leitloff, J.; Schiefer, F.; Hinz, S. Review on Convolutional Neural Networks (CNN) in vegetation remote sensing. ISPRS J. Photogramm. Remote Sens. 2021, 173, 24–49. [Google Scholar] [CrossRef]
- Jubeen, M.; Rahman, H.; Rahman, A.U.; Wahid, S.A.; Imran, A.; Yasin, A.; Ihsan, I. An Automatic Breast Cancer Diagnostic System Based on Mammographic Images Using Convolutional Neural Network Classifier. J. Comput. Biomed. Inform. 2022, 4, 77–86. [Google Scholar]
- Tariq, T.; Hassan, M.; Hameedur, R.; Shah, A. Predictive Model for Lung Cancer Detection. LC Int. J. STEM 2020, 1, 61–74. [Google Scholar]
- Bonechi, S.; Andreini, P.; Mecocci, A.; Giannelli, N.; Scarselli, F.; Neri, E.; Bianchini, M.; Dimitri, G.M. Segmentation of aorta 3D CT images based on 2D convolutional neural networks. Electronics 2021, 10, 2559. [Google Scholar] [CrossRef]
- Tariq, J.; Javed, M.; Ayub, B.; Armghan, A.; Ijaz, A.; Alenezi, F.; Rahman, H.; Zulfiqar, A. Nature inspired algorithm based fast intra mode decision in HEVC. Multimed. Tools Appl. 2023, 82, 29789–29804. [Google Scholar] [CrossRef]
- Rahman, H.; Naik Bukht, T.F.; Ahmad, R.; Almadhor, A.; Javed, A.R. Efficient Breast Cancer Diagnosis from Complex Mammographic Images Using Deep Convolutional Neural Network. Comput. Intell. Neurosci. 2023, 2023, 7717712. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yu, H. Convolutional neural network based metal artifact reduction in x-ray computed tomography. IEEE Trans. Med. Imaging 2018, 37, 1370–1381. [Google Scholar] [CrossRef] [PubMed]
- Rahman, H.; Tariq, J.; Masood, M.A.; Subahi, A.F.; Khalaf, O.I.; Alotaibi, Y. Multi-tier sentiment analysis of social media text using supervised machine learning. Comput. Mater. Contin 2023, 74, 5527–5543. [Google Scholar] [CrossRef]
- Tasleem, S.; Bano, P.; Hameedur, R. Students Attendance Management System Based On Face Recognition. LC Int. J. STEM 2020, 1, 66–74. [Google Scholar]
- Jackson, A.S.; Bulat, A.; Argyriou, V.; Tzimiropoulos, G. Large pose 3D face reconstruction from a single image via direct volumetric CNN regression. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 1031–1039. [Google Scholar]
- Rahman, H.; Bukht, T.F.N.; Imran, A.; Tariq, J.; Tu, S.; Alzahrani, A. A Deep Learning Approach for Liver and Tumor Segmentation in CT Images Using ResUNet. Bioengineering 2022, 9, 368. [Google Scholar] [CrossRef]
- Eilertsen, G.; Kronander, J.; Denes, G.; Mantiuk, R.K.; Unger, J. HDR image reconstruction from a single exposure using deep CNNs. ACM Trans. Graph. (TOG) 2017, 36, 1–15. [Google Scholar] [CrossRef]
- Abbas, M.; Arshad, M.; Hameedur, R. Detection of Breast Cancer Using Neural Networks. LC Int. J. STEM 2020, 1, 75–88. [Google Scholar]
- Abid, I.; Almakdi, S.; Rahman, H.; Almulihi, A.; Alqahtani, A.; Rajab, K.; Alqhatani, A.; Shaikh, A. A Convolutional Neural Network for Skin Lesion Segmentation Using Double U-Net Architecture. Intell. Autom. Soft Comput. 2022, 33, 1407–1421. [Google Scholar] [CrossRef]
- Shi, Z.; Chen, C.; Xiong, Z.; Liu, D.; Wu, F. Hscnn+: Advanced cnn-based hyperspectral recovery from rgb images. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops, Salt Lake City, UT, USA, 18–22 June 2018; pp. 939–947. [Google Scholar]
- Riaz, A.; Rahman, H.; Arshad, M.A.; Nabeel, M.; Yasin, A.; Al-Adhaileh, M.H.; Eldin, E.T.; Ghamry, N.A. Augmentation of Deep Learning Models for Multistep Traffic Speed Prediction. Appl. Sci. 2022, 12, 9723. [Google Scholar] [CrossRef]
- Sulaiman, A.; Rahman, H.; Ali, N.; Shaikh, A.; Akram, M.; Lim, W.H. An augmented reality PQRST based method to improve self-learning skills for preschool autistic children. Evol. Syst. 2022, 14, 859–872. [Google Scholar] [CrossRef]
- Krumm, M.; Kasperl, S.; Franz, M. Reducing non-linear artifacts of multi-material objects in industrial 3D computed tomography. Ndt E Int. 2008, 41, 242–251. [Google Scholar] [CrossRef]
- Torng, W.; Altman, R.B. 3D deep convolutional neural networks for amino acid environment similarity analysis. BMC Bioinform. 2017, 18, 302. [Google Scholar] [CrossRef] [PubMed]
- Seol, Y.J.; Kim, Y.J.; Kim, Y.S.; Cheon, Y.W.; Kim, K.G. A study on 3D deep learning-based automatic diagnosis of nasal fractures. Sensors 2022, 22, 506. [Google Scholar] [CrossRef] [PubMed]
- Zia, Z.U.R.; Hameedur, R.; Malik, M.H.; Jahngir, A. Technical Challenges in Achieving Ultra-Reliable & Low Latency Communication in 5G Cellular-V2X Systems. LC Int. J. STEM 2020, 1, 89–95. [Google Scholar]
- Lim, K.S.; Ang, K.M.; Isa, N.A.M.; Tiang, S.S.; Rahman, H.; Chandrasekar, B.; Hussin, E.E.; Lim, W.H. Optimized Machine Learning Model with Modified Particle Swarm Optimization for Data Classification. In Proceedings of the Advances in Intelligent Manufacturing and Mechatronics: Selected Articles from the Innovative Manufacturing, Mechatronics & Materials Forum (iM3F 2022), Pahang, Malaysia; Springer Nature: Singapore, 2023; pp. 211–223. [Google Scholar]
- Image-based 3D object reconstruction: State-of-the-art and trends in the deep learning era. LC Int. J. STEM 2019, 43, 1578–1604.
- Lenfant, M.; Chevallier, O.; Comby, P.O.; Secco, G.; Haioun, K.; Ricolfi, F.; Lemogne, B.; Loffroy, R. Deep learning versus iterative reconstruction for CT pulmonary angiography in the emergency setting: Improved image quality and reduced radiation dose. Diagnostics 2020, 10, 558. [Google Scholar] [CrossRef]
- Jacobs, C.; van Rikxoort, E.M.; Murphy, K.; Prokop, M.; Schaefer-Prokop, C.M.; van Ginneken, B. Computer-aided detection of pulmonary nodules: A comparative study using the public LIDC/IDRI database. Eur. Radiol. 2016, 26, 2139–2147. [Google Scholar] [CrossRef] [PubMed]
- Setio, A.A.A.; Traverso, A.; De Bel, T.; Berens, M.S.; Van Den Bogaard, C.; Cerello, P.; Chen, H.; Dou, Q.; Fantacci, M.E.; Geurts, B.; et al. Validation, comparison, and combination of algorithms for automatic detection of pulmonary nodules in computed tomography images: The LUNA16 challenge. Med. Image Anal. 2017, 42, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Li, A.; Hu, Z.; Wang, L. Accurate pulmonary nodule detection in computed tomography images using deep convolutional neural networks. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2017: 20th International Conference, Quebec City, QC, Canada, 11–13 September 2017; Proceedings, Part III 20. Springer: Berlin/Heidelberg, Germany, 2017; pp. 559–567. [Google Scholar]
- Asif, S.; Ambreen, M.; Muhammad, Z.; ur Rahman, H.; Iqbal, S. Cloud Computing in Healthcare-Investigation of Threats, Vulnerabilities, Future Challenges and Counter Measure. LC Int. J. STEM 2022, 3, 63–74. [Google Scholar]
- Armato III, S.G.; McLennan, G.; Bidaut, L.; McNitt-Gray, M.F.; Meyer, C.R.; Reeves, A.P.; Zhao, B.; Aberle, D.R.; Henschke, C.I.; Hoffman, E.A.; et al. The lung image database consortium (LIDC) and image database resource initiative (IDRI): A completed reference database of lung nodules on CT scans. Med. Phys. 2011, 38, 915–931. [Google Scholar] [CrossRef] [PubMed]
- McCollough, C. TU-FG-207A-04: Overview of the low dose CT grand challenge. Med. Phys. 2016, 43, 3759–3760. [Google Scholar] [CrossRef]
- McCollough, C.H.; Bartley, A.C.; Carter, R.E.; Chen, B.; Drees, T.A.; Edwards, P.; Holmes III, D.R.; Huang, A.E.; Khan, F.; Leng, S.; et al. Low-dose CT for the detection and classification of metastatic liver lesions: Results of the 2016 low dose CT grand challenge. Med. Phys. 2017, 44, e339–e352. [Google Scholar] [CrossRef]
- Bera, S.; Biswas, P.K. Noise conscious training of non local neural network powered by self attentive spectral normalized Markovian patch GAN for low dose CT denoising. IEEE Trans. Med. Imaging 2021, 40, 3663–3673. [Google Scholar] [CrossRef]
- Latsios, G.; Spyridopoulos, T.N.; Toutouzas, K.; Synetos, A.; Trantalis, G.; Stathogiannis, K.; Penesopoulou, V.; Oikonomou, G.; Brountzos, E.; Tousoulis, D. Multi-slice CT (MSCT) imaging in pretrans-catheter aortic valve implantation (TAVI) screening. How to perform and how to interpret. Hell. J. Cardiol. 2018, 59, 3–7. [Google Scholar] [CrossRef]
- Michiels, K.; Heffinck, E.; Astudillo, P.; Wong, I.; Mortier, P.; Bavo, A.M. Automated MSCT analysis for planning left atrial appendage occlusion using artificial intelligence. J. Interv. Cardiol. 2022, 2022, 5797431. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).