A UV/Vis Spectroscopy-Based Assay for Monitoring of Transformations Between Nucleosides and Nucleobases
Abstract
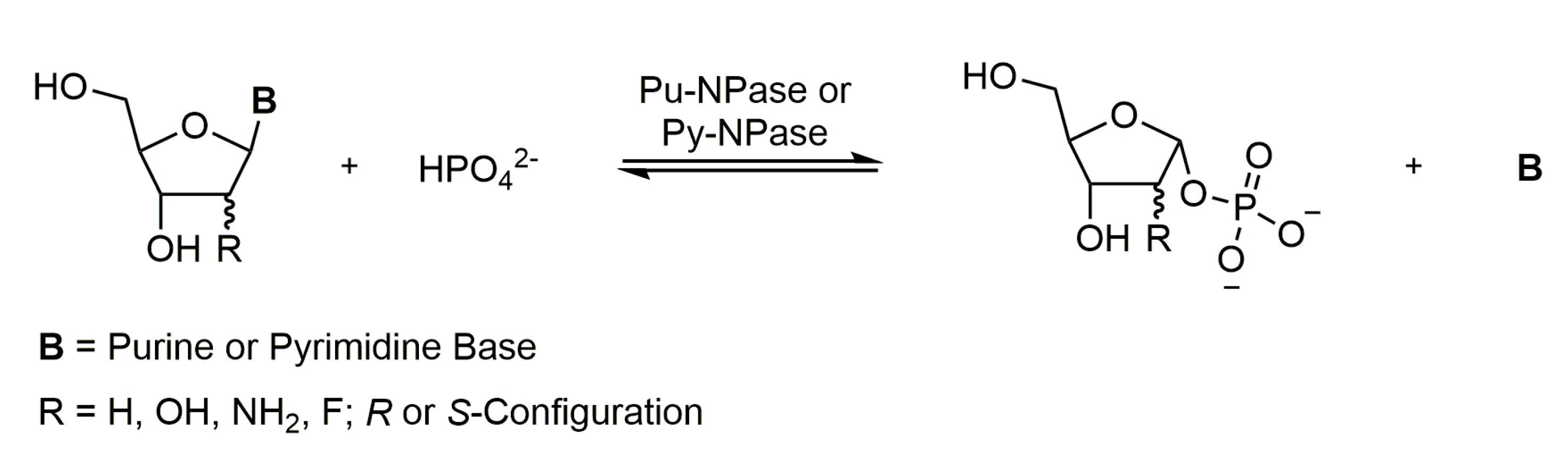
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Enzymes
2.3. Recording of Spectra
2.4. Investigating the pH Dependence of Spectra
2.5. Enzymatic Reactions
2.6. Enzyme Reaction Sampling
2.7. Spectral Unmixing
- = representation of the molar fractions of compounds for sample k in a vector form (in this study: )
- = experimentally determined spectrum for sample ,
- = number of compounds (in this study: ),
- = molar fraction of compound , and
- = spectra of pure compound , normalized to the corresponding isosbestic point of the mixture (see below),
2.8. Normalization of Spectra to Accommodate Concentration Differences
- = the normalized spectrum of compound ,
- = raw spectrum of compound ,
- = raw blank spectrum of NaOH solution,
- = the absorption of the blank-corrected spectrum at wavelength .
2.9. Statistical Analyses
- = residual sum of squares of all data points;
- = total sum of squares of all data points;
- = predicted molar fraction of the data point of the data set (i.e., all data points for one compounds, considering both replicates), in percent,
- = average of all actual molar fraction values of the data set (equal to 50%), in percent,
- = actual molar fraction value for the data point in percent,
- = total number of data points in the data set,
2.10. HPLC Analysis of Samples
3. Results
3.1. Nucleoside–Nucleobase Pairs Show Discriminable UV/Vis Spectra under Alkaline Conditions
3.2. Spectral Unmixing Can Resolve the Composition of Nucleoside–Nucleobase Mixtures
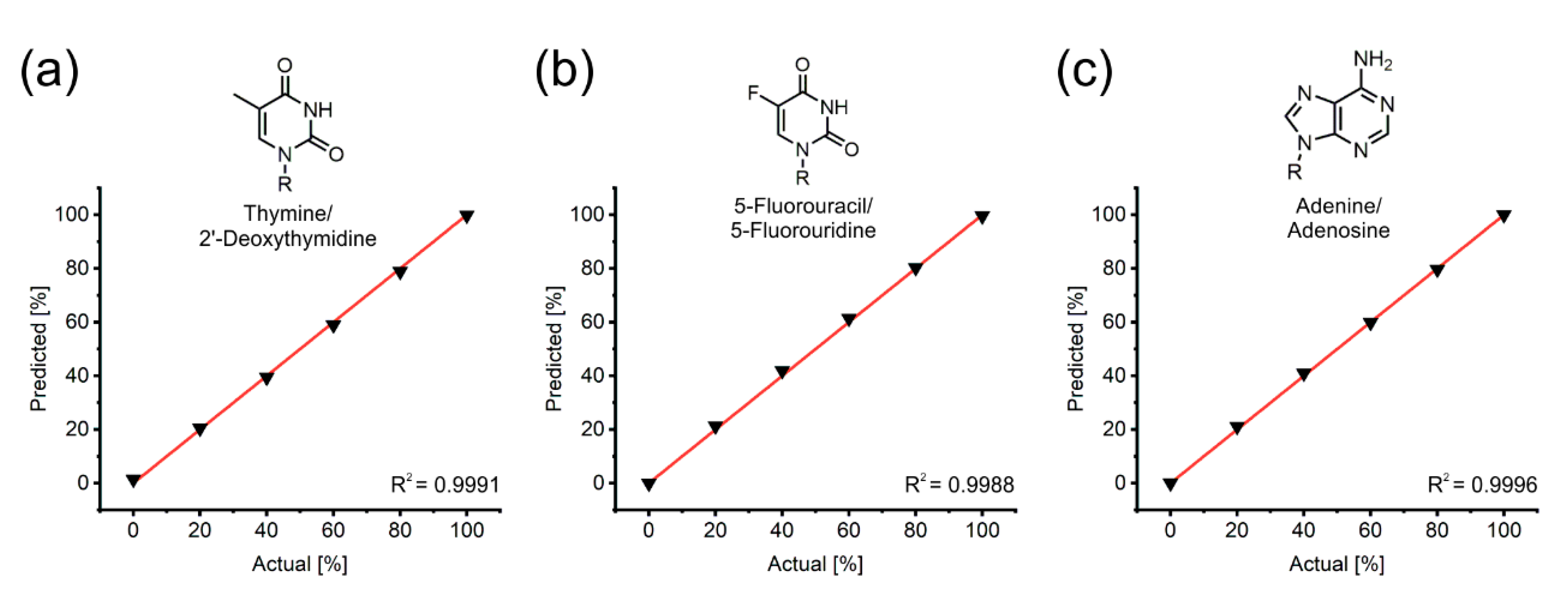
3.3. The Accuracy of Spectral Unmixing Holds up Excellently across All Possible Mixture Compositions
3.4. Spectral Unmixing Empowers Practical Applications in Enzymatic Conversion Research
3.5. The Spectral Unmixing Algorithm Can be Fine-Tuned to Specific Areas of Interest
4. Discussion
4.1. The Prerequisites of Spectral Unmixing are Met by All Nucleoside–Nucleobase Pairs
4.2. Previous Assays in the Field
4.3. Improvements and Adaptations of the Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Kamel, S.; Yehia, H.; Neubauer, P.; Wagner, A. Enzymatic synthesis of nucleoside analogues by nucleoside phosphorylases. In Enzymatic and Chemical Synthesis of Nucleic Acid Derivatives; Wiley-VCH Verlag GmbH & Co.: Weinheim, Germany, 2019; pp. 1–28. [Google Scholar]
- Yehia, H.; Kamel, S.; Paulick, K.; Wagner, A.; Neubauer, P. Substrate spectra of nucleoside phosphorylases and their potential in the production of pharmaceutically active compounds. Curr. Pharm. Des. 2017, 23, 6913–6935. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Lucas, J.; Arroyo, M. Enzymatic synthesis of nucleic acid derivatives by immobilized enzymes. In Enzymatic and Chemical Synthesis of Nucleic Acid Derivatives; Wiley-VCH Verlag GmbH & Co.: Weinheim, Germany, 2019; pp. 107–128. [Google Scholar]
- Yamada, E.W. Uridine phosphorylase from rat liver. Methods Enzymol. 1978, 51, 423–431. [Google Scholar] [CrossRef] [PubMed]
- Ubiali, D.; Rocchietti, S.; Scaramozzino, F.; Terreni, M.; Albertini, A.M.; Fernández-Lafuente, R.; Guisán, J.M.; Pregnolato, M. Synthesis of 2′-deoxynucleosides by transglycosylation with new immobilized and stabilized uridine phosphorylase and purine nucleoside phosphorylase. Adv. Synth. Catal. 2004, 346, 1361–1366. [Google Scholar] [CrossRef]
- Giessmann, R.T.; Krausch, N.; Kaspar, F.; Bournazou, M.N.C.; Wagner, A.; Neubauer, P.; Gimpel, M. Dynamic modelling of enzymatic deoxyribose phosphorylation reactions. Processes 2019, 7, 380. [Google Scholar] [CrossRef]
- Goetz, A.F.H.; Vane, G.; Solomon, J.E.; Rock, B.N. Imaging spectrometry for earth remote sensing. Science 1985, 228, 1147–1153. [Google Scholar] [CrossRef] [PubMed]
- Plaza, A.; Benediktsson, J.A.; Boardman, J.W.; Brazile, J.; Bruzzone, L.; Camps-Valls, G.; Chanussot, J.; Fauvel, M.; Gamba, P.; Gualtieri, A.; et al. Recent advances in techniques for hyperspectral image processing. Remote Sens. Environ. 2009, 113, 110–122. [Google Scholar] [CrossRef]
- Lorente, D.; Aleixos, N.; Gómez-Sanchis, J.; Cubero, S.; García-Navarrete, O.L.; Blasco, J. Recent advances and applications of hyperspectral imaging for fruit and vegetable quality assessment. Food Bioprocess Technol. 2012, 5, 1121–1142. [Google Scholar] [CrossRef]
- Dickinson, M.E.; Bearman, G.; Tille, S.; Lansford, R.; Fraser, S.E. Multi-spectral imaging and linear unmixing add a whole new dimension to laser scanning fluorescence microscopy. Biotechniques 2001, 31, 1272–1278. [Google Scholar] [CrossRef]
- Quintano, C.; Fernández-Manso, A.; Shimabukuro, Y.E.; Pereira, G. Spectral unmixing. Int. J. Remote Sens. 2012, 33, 5307–5340. [Google Scholar] [CrossRef]
- Giessmann, R.T. Python code [Software]. Available online: https://doi:10.5281/zenodo.3243376 (accessed on 12 June 2019).
- Szeker, K.; Zhou, X.; Schwab, T.; Casanueva, A.; Cowan, D.; Mikhailopulo, I.A.; Neubauer, P. Comparative investigations on thermostable pyrimidine nucleoside phosphorylases from Geobacillus thermoglucosidasius and Thermus thermophilus. J. Mol. Catal. B Enzym. 2012, 84, 27–34. [Google Scholar] [CrossRef]
- Zhou, X.; Szeker, K.; Jiao, L.-Y.; Oestreich, M.; Mikhailopulo, I.A.; Neubauer, P. Synthesis of 2,6-dihalogenated purine nucleosides by thermostable nucleoside phosphorylases. Adv. Synth. Catal. 2015, 357, 1237–1244. [Google Scholar] [CrossRef]
- Newville, M.; Stensitzki, T.; Allen, D.B.; Ingargiola, A. LMFIT: Non-linear least-square minimization and curve-fitting for Python. Astrophys. Source Code Library 2016. [Google Scholar] [CrossRef]
- Giessmann, R.T. Supplementary Material. Available online: https://doi:10.5281/zenodo.3245012 (accessed on 12 June 2019).
- Kamel, S.; Weiß, M.; Klare, H.F.T.; Mikhailopulo, I.A.; Neubauer, P.; Wagner, A. Chemo-enzymatic synthesis of α-D-pentofuranose-1-phosphates using thermostable pyrimidine nucleoside phosphorylases. Mol. Catal. 2018, 458, 52–59. [Google Scholar] [CrossRef]
- Wittenburg, E. Untersuchung der tautomeren Struktur von Thymin und seinen Alkylderivaten mit Hilfe von UV-Spektren. Chem. Ber. 1966, 99, 2391–2398. [Google Scholar] [CrossRef]
- Ganguly, S.; Kundu, K.K. Deprotonation energetics of some nucleosides in water from EMF measurements. Can. J. Chem. 1995, 73, 70–78. [Google Scholar] [CrossRef]
- Fox, J.J.; Yung, N.; Wempen, I. Spectrophotometric studies of nucleic acid derivatives and related compounds as a function of pH: IV. On the structure of orotidine. A study of N-methylated orotic acids. Biochim. Biophys. Acta 1957, 23, 295–305. [Google Scholar] [CrossRef]
- Fox, J.J.; Shugar, D. Spectrophotometric studies on nucleic acid derivatives and related compounds as a function of pH: II. Natural and synthetic pyrimidine nucleosides. Biochim. Biophys. Acta 1952, 9, 369–384. [Google Scholar] [CrossRef]
- Calabrese, I.; Merli, M.; Turco Liveri, M.L. Deconvolution procedure of the UV-vis spectra. A powerful tool for the estimation of the binding of a model drug to specific solubilisation loci of bio-compatible aqueous surfactant-forming micelle. Spectrochim. Acta. A. Mol. Biomol. Spectrosc. 2015, 142, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Novo, D.; Gregori, G.; Rajwa, B. Generalized unmixing model for multispectral flow cytometry utilizing nonsquare compensation matrices. Cytometry A 2013, 83, 508–520. [Google Scholar] [CrossRef] [PubMed]
- De Luca, M.; Ioele, G.; Spatari, C.; Ragno, G. Optimization of wavelength range and data interval in chemometric analysis of complex pharmaceutical mixtures. J. Pharm. Anal. 2016, 6, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Lacey, R.F. Deconvolution of overlapping chromatographic peaks. Anal. Chem. 1986, 58, 1404–1410. [Google Scholar] [CrossRef]
- Stachelska-Wierzchowska, A.; Wierzchowski, J.; Górka, M.; Bzowska, A.; Wielgus-Kutrowska, B. Tri-Cyclic Nucleobase Analogs and Their Ribosides as Substrates of Purine-Nucleoside Phosphorylases. II Guanine and Isoguanine Derivatives. Molecules 2019, 24, 1493. [Google Scholar] [CrossRef] [PubMed]
- Bzowska, A.; Kulikowska, E.; Shugar, D. Purine nucleoside phosphorylases: properties, functions, and clinical aspects. Pharmacol. Ther. 2000, 88, 349–425. [Google Scholar] [CrossRef]
- Panova, N.G.; Shcheveleva, E.V.; Alexeev, C.S.; Mukhortov, V.G.; Zuev, A.N.; Mikhailov, S.N.; Esipov, R.S.; Chuvikovsky, D.V.; Miroshnikov, A.I. Use of 4-Thiouridine and 4-Thiothymidine in Studies on Pyrimidine Nucleoside Phosphorylases. Mol. Biol. 2004, 38, 770–776. [Google Scholar] [CrossRef]
- Wierzchowski, J.; Stachelska-Wierzchowska, A.; Wielgus-Kutrowska, B.; Bzowska, A. 1,N6-ethenoadenine and other fluorescent nucleobase analogs as substrates for purine-nucleoside phosphorylases: Spectroscopic and kinetic studies. Curr. Pharm. Des. 2017, 23, 6948–6966. [Google Scholar] [CrossRef] [PubMed]
- Wierzchowski, J.; Ogiela, M.; Iwańska, B.; Shugar, D. Selective fluorescent and fluorogenic substrates for purine-nucleoside phosphorylases from various sources, and direct fluorimetric determination of enzyme levels in human and animal blood. Anal. Chim. Acta 2002, 472, 63–74. [Google Scholar] [CrossRef]
- Wierzchowski, J.; Stachelska-Wierzchowska, A.; Wielgus-Kutrowska, B.; Mikleušević, G. Two fluorogenic substrates for purine nucleoside phosphorylase, selective for mammalian and bacterial forms of the enzyme. Anal. Biochem. 2014, 446, 25–27. [Google Scholar] [CrossRef]
- Stachelska-Wierzchowska, A.; Wierzchowski, J.; Bzowska, A.; Wielgus-Kutrowska, B. Tricyclic nitrogen base 1,N6-ethenoadenine and its ribosides as substrates for purine-nucleoside phosphorylases: Spectroscopic and kinetic studies. Nucleosides Nucleotides Nucleic Acids 2018, 37, 89–101. [Google Scholar] [CrossRef]
- Porter, D.J. Purine nucleoside phosphorylase. Kinetic mechanism of the enzyme from calf spleen. J. Biol. Chem. 1992, 267, 7342–7351. [Google Scholar]
- Singh, D.; Schaaper, R.M.; Hochkoeppler, A. A continuous spectrophotometric enzyme-coupled assay for deoxynucleoside triphosphate triphosphohydrolases. Anal. Biochem. 2016, 496, 43–49. [Google Scholar] [CrossRef][Green Version]
- Seamon, K.J.; Stivers, J.T. A High-Throughput Enzyme-Coupled Assay for SAMHD1 dNTPase. J. Biomol. Screen. 2015, 20, 801–809. [Google Scholar] [CrossRef] [PubMed]
- Surette, M.; Gill, T.; MacLean, S. Purification and characterization of purine nucleoside phosphorylase from Proteus vulgaris. Appl. Environ. Microbiol. 1990, 56, 1435–1439. [Google Scholar] [PubMed]



| Compound | Nucleoside [nm] | Base [nm] | Isosbestic Point for Base Cleavage [nm] | Upper Limit of Range for Fitting [nm] | |
|---|---|---|---|---|---|
| Pyrimidines | Uridine | 262 | 281 | 271 | 310 |
| 2’-Deoxyuridine | 262 | 272 | 310 | ||
| 5-Methyluridine + | 267 | 290 | 277 | 320 | |
| 2’-Deoxythymidine $ | 266 | 278 | 320 | ||
| 5-Fluorouridine * | 269 | 281 | 282 | 325 | |
| 2’-Deoxy-5-fluorouridine * | 268 | 280 | 325 | ||
| 5-Bromouridine | 276 | 290 | 283 | 330 | |
| 2’-Deoxy-5-bromouridine | 275 | 282 | 330 | ||
| 5-Iodouridine * | 281 | 291 | 283 | 340 | |
| 2’-Deoxy-5-Iodouridine * | 279 | 282 | 340 | ||
| 5-Ethynyluridine * | 285 | 298 | 262, 288 | 340 | |
| 2’-Deoxy-5-ethynyluridine * | 284 | 262, 288 | 340 | ||
| Cytidine # | 271 | 281 | 271 | 310 | |
| 2’-Deoxycytidine # | 271 | 271 | 310 | ||
| Purines | Adenosine | 259 | 268 | 267 | 310 |
| 2’-Deoxyadenosine | 259 | 267 | 310 | ||
| Guanosine | 264 | 273 | 279 | 310 | |
| 2’-Deoxyguanosine | 264 | 279 | 310 | ||
| Inosine | 252 | 262 | 263 | 320 | |
| 2’-Deoxyinosine | 252 | 263 | 320 |
| Actual [%] | Predicted [%] | Difference [%] (UV/Vis-Assay - HPLC) | |
|---|---|---|---|
| HPLC | UV/Vis Assay | ||
| 0 | 0 ± 0 | 1.47 ± 1.02 | +1.47 |
| 20 | 21.08 ± 0.02 | 20.47 ± 0.25 | −0.60 |
| 40 | 40.89 ± 0.15 | 39.46 ± 0.09 | −1.43 |
| 60 | 60.03 ± 0.05 | 58.99 ± 0.38 | −1.04 |
| 80 | 79.38 ± 0.19 | 78.91 ± 0.13 | −0.46 |
| 100 | 99.67 ± 0.36 | 99.85 ± 0.15 | +0.18 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaspar, F.; Giessmann, R.T.; Krausch, N.; Neubauer, P.; Wagner, A.; Gimpel, M. A UV/Vis Spectroscopy-Based Assay for Monitoring of Transformations Between Nucleosides and Nucleobases. Methods Protoc. 2019, 2, 60. https://doi.org/10.3390/mps2030060
Kaspar F, Giessmann RT, Krausch N, Neubauer P, Wagner A, Gimpel M. A UV/Vis Spectroscopy-Based Assay for Monitoring of Transformations Between Nucleosides and Nucleobases. Methods and Protocols. 2019; 2(3):60. https://doi.org/10.3390/mps2030060
Chicago/Turabian StyleKaspar, Felix, Robert T. Giessmann, Niels Krausch, Peter Neubauer, Anke Wagner, and Matthias Gimpel. 2019. "A UV/Vis Spectroscopy-Based Assay for Monitoring of Transformations Between Nucleosides and Nucleobases" Methods and Protocols 2, no. 3: 60. https://doi.org/10.3390/mps2030060
APA StyleKaspar, F., Giessmann, R. T., Krausch, N., Neubauer, P., Wagner, A., & Gimpel, M. (2019). A UV/Vis Spectroscopy-Based Assay for Monitoring of Transformations Between Nucleosides and Nucleobases. Methods and Protocols, 2(3), 60. https://doi.org/10.3390/mps2030060








