In Situ DNA/Protein Interaction Assay to Visualize Transcriptional Factor Activation
Abstract
1. Introduction
2. Experimental Design
2.1. Materials
2.1.1. Probes
- Biotin or Cy5-labeled CRE oligo-probe forward 5′-AGAGATTGCCTGACGTCAGAGAGCTAG -3′ (Sigma, Merck group).
- Biotin or Cy5-labeled CRE oligo-probe reverse 5′-CTAGCTCTCTGACGTCAGGCAATCTCT-3′ (Sigma, Merck group).
- Biotin or Cy5-labeled CRE scramble oligo-probe sense 5′-AGACATTGCCTGGATAGGGAGAGTTAG-3′ (Sigma, Merck group).
- Biotin or Cy5-labeled CRE scramble oligo-probe nonsense 5′-CTAACTCTCCCTATCCAGGCAATGTCT-3′ (Sigma, Merck group).
2.1.2. Reagents
- Alginic Acid Sodium Salt powder (Sigma, Merck Group–cat n° A2158).
- Calcium Chloride dihydrate (Sigma, Merck Group–cat n° C3306).
- PBS (Dulbecco’s Phosphate-buffered saline w/o Ca and Mg, 10×; Lonza–cat n° BE17–517Q). Dilute 1:10 for working solution.
- Triton X-100 (Sigma, Merck Group–cat n° T8787). ! CAUTION Triton X-100 is hazardous. Avoid contact with skin and eyes.
- Killik (Optimal cutting temperature compound (OCT); Bio-Optica–cat n° 05-9801).
- Salmon Sperm DNA (ThermoFisher-cat n° AM9680).
- Avidin Blocking Reagent (Vector Laboratories Inc.–cat n° SP-2001).
- Methanol (Sigma, Merck Group–cat n° 32213). ! CAUTION Methanol is toxic. Manipulate in a fume hood.
- Streptavidin AlexaFluor 488 or 594 (Molecular Probes–cat n° S11223–S11227).
- DAPI (4′,6-diamidin-2-fenilindolo) (Sigma, Merck Group–cat n° D8412). Dilute 1:15000 for working solution.
- Dako Fluorescence Mounting Medium (Dako Cytomation–cat n° S3023).
2.1.3. Eggs
- Fertilized white leghorn eggs.
2.2. Equipment
- Static incubator for eggs (FIEM snc, Buttigliera, Italy).
- Surgery straight forceps (2Biological Instrument–cat n° 11255-20).
- Surgery straight scissors (2Biological Instrument–cat n° 14094-11).
- Surgery curved scissors (2Biological Instrument–cat n° 14095-11).
- 5 ml syringe (Terumo–cat n° SS*05SE1).
- Leukosilk Silk Tape 2.5 cm × 5 m (BSN Medical–cat n° 01022-00).
- Transparent tape.
- Cryostats.
- Fluorescence Microscope and/or Confocal Microscope.
3. Procedure
3.1. Egg Preparation
- Gently wash the eggs in 25–28 °C water, and incubate sagittally at 37 °C in a humidified incubator.
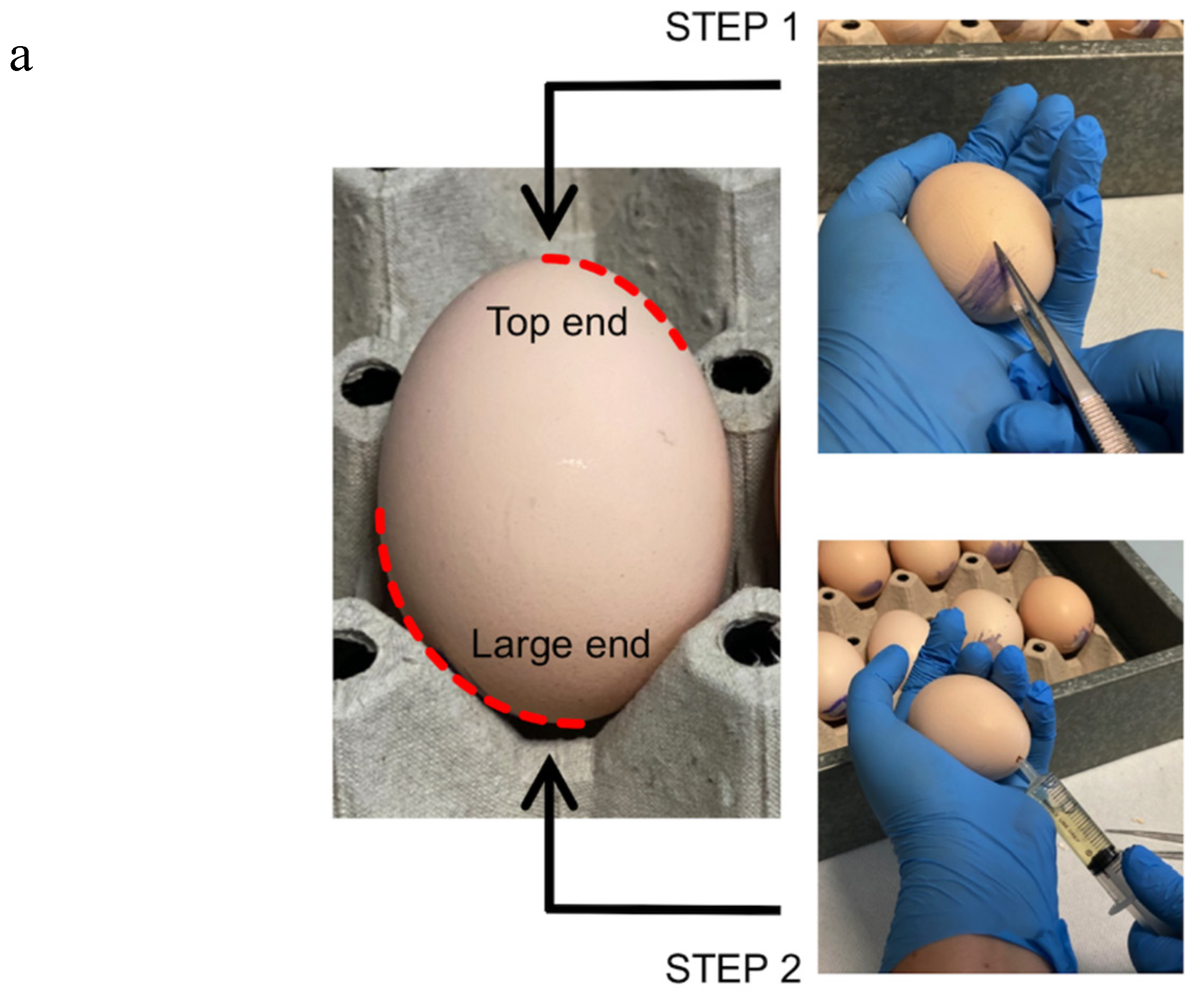
- Four days post-incubation, prick the large end of the shell in correspondence with the air space with a forceps (Figure 1–STEP 1). This procedure allows us to break the inner shell membrane and detach the embryo from the eggshell.
- Drill a hole on the opposite side to the first one, and aspirate 3 to 5 mL of egg albumen with a syringe without a needle (Figure 1–STEP 2).
- Promptly close the holes with silk tape. Incubate the eggs at 37 °C for 24 h.
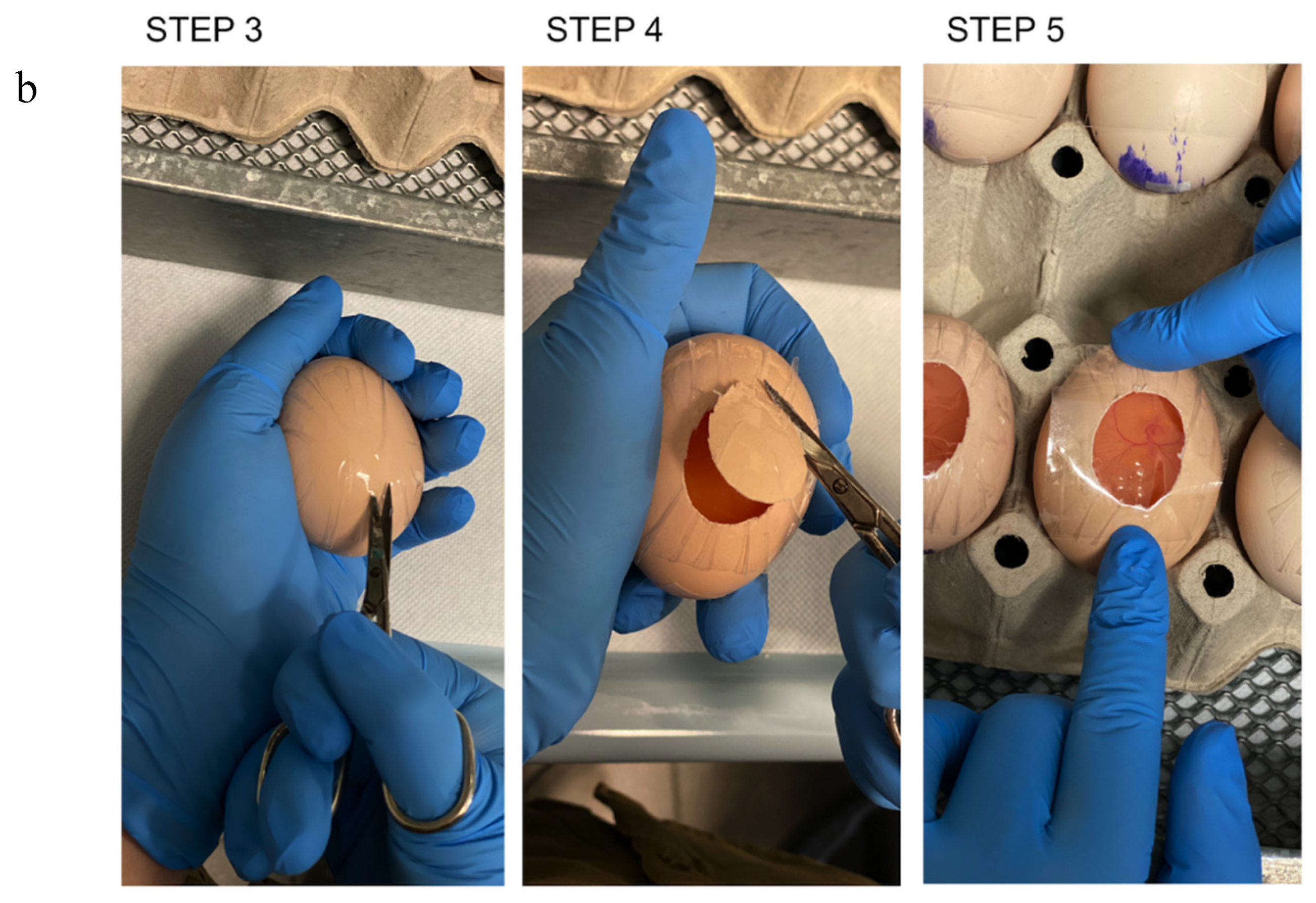
- Place a piece of transparent tape on the eggshell, and make it adhere perfectly.
- Cut a window in the eggshell approximately 1.5 × 2.5 cm wide with a curve surgical scissor (Figure 1–STEP 3 and STEP 4).
- Close the egg with transparent tape (Figure 1–STEP 5). CRITICAL STEP: ensure that the tape is well tightened to prevent the embryo from drying out.
- Incubate the eggs at 37 °C for 7 d.
3.1.1. Egg Treatment
- Prepare a solution of Alginic acid 6% (w/v) in sterile milliQ H2O at least 16–18 h before starting.
- Spot a drop (3–4 μL) of Alginic acid solution into the lid of a culture dish and immediately add 3 uL of the molecule under testing.
- Add a drop of CaCl2 0.1 M to allow the polymerization of Alginic acid.
- Gently transfer the alginate pellets on top of the CAM using a forcep.
3.1.2. Sample Harvest and Inclusion
- Following the timescale provided by your protocol, collect the samples without fixing them.
- Gently wash the CAM with cold PBS for 2–3 min.
- Include the CAM into tissue embedding resin OCT for cryosections and immediately submerge in liquid nitrogen for snap freezing at least 30 sec. ! CAUTION Extremely cold liquid (−196 °C) can cause severe frostbite and cold burns. Gloves and face shield required.
- Cut 4–5 μm thick sections, and store the samples at −80 °C.
3.1.3. Hybridization and Staining
- Wash tissue sections in phosphate buffer saline (PBS) 1% Triton X-100 for 2–3 min.
- Rinse them in PBS for 5 min.
- Incubate with Avidin blocking solution 1:20 in PBS for 1 h at room temperature (RT).
- Rinse in PBS for 5 min.
- Incubate with Salmon sperm DNA 100 ug/mL in PBS for 1h at RT.
- Add biotinylated CREB oligos 600 pmol/reaction and incubate 45 min at RT.
- Rinse twice with PBS for 5 min.
- Fix with cold methanol for 10 min.
- Rinse twice with PBS for 5 min.
- Add AlexaFluor 488-conjugated Streptavidin or AlexaFluor 594-conjugated Streptavidin 10 μg/mL in PBS for 1 h at RT in a dark humidified chamber.
- Rinse twice with PBS for 5 min.
- In the dark, counterstain nuclei with DAPI 50 ng/mL in PBS for 15 min at RT.
- Mount the slices with Dako Mounting Medium and an appropriate coverslip.
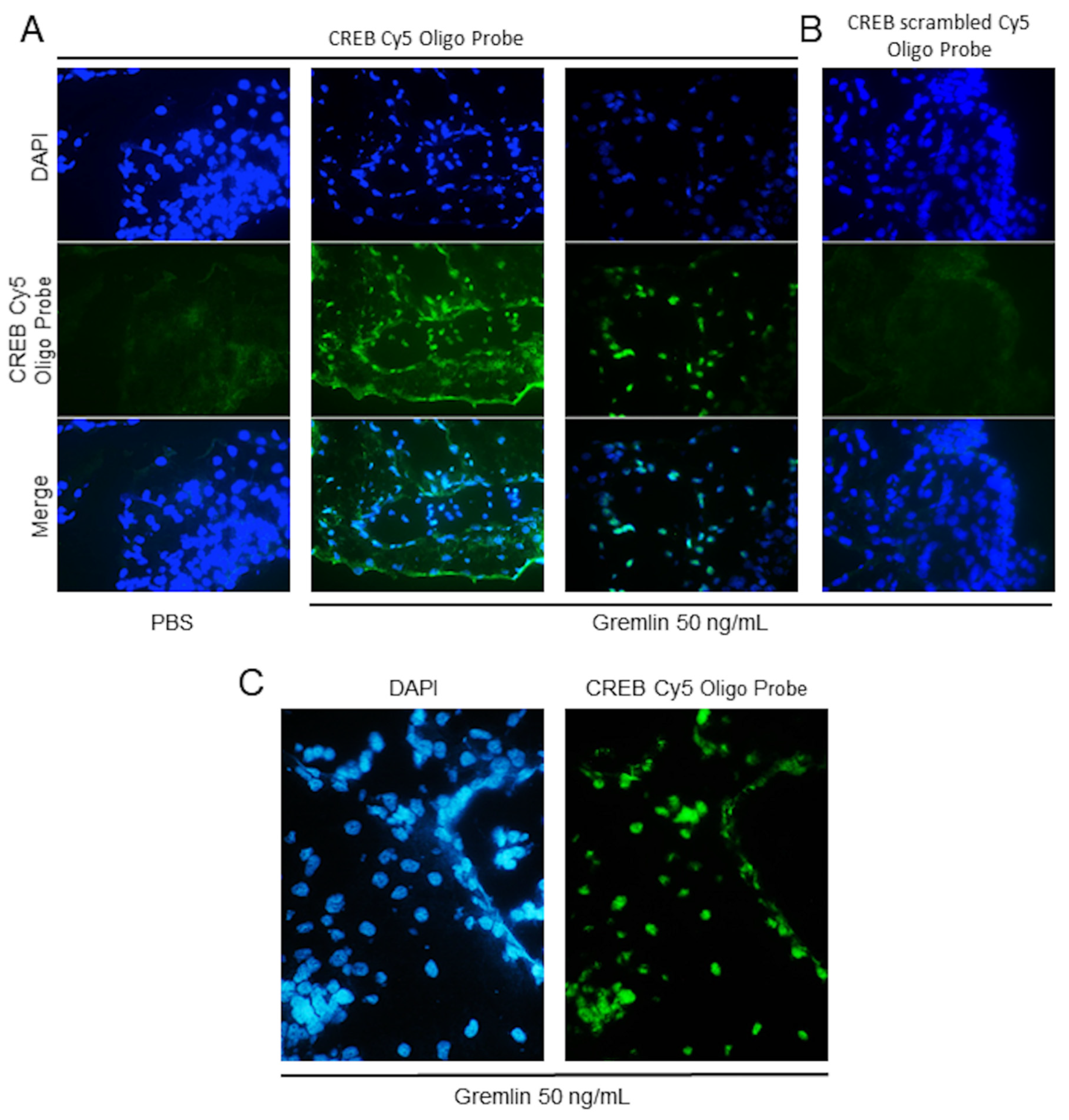
4. Expected Results
Author Contributions
Funding
Conflicts of Interest
References
- Gabrielli, M.G.; Accili, D. The Chick Chorioallantoic Membrane: A Model of Molecular, Structural, and Functional Adaptation to Transepithelial Ion Transport and Barrier Function during Embryonic Development. J. Biomed. Biotechnol. 2010, 2010, 1–12. [Google Scholar] [CrossRef]
- Ribatti, D. The chick embryo chorioallantoic membrane (CAM). A multifaceted experimental model. Mech. Dev. 2016, 141, 70–77. [Google Scholar] [CrossRef]
- Ribatti, D. The chick embryo chorioallantoic membrane as a model for tumor biology. Exp. Cell Res. 2014, 328, 314–324. [Google Scholar] [CrossRef] [PubMed]
- Gnutti, A.; Signoroni, A.; Leonardi, R.; Corsini, M.; Presta, M.; Mitola, S. A tool for the quantification of radial neo-vessels in chick chorioallantoic membrane angiogenic assays. In Proceedings of the 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Milano, Italy, 25–29 August 2015; pp. 763–766. [Google Scholar]
- Dohle, D.S.; Pasa, S.D.; Gustmann, S.; Laub, M.; Wissler, J.H.; Jennissen, H.P.; Dünker, N. Chick ex ovo Culture and ex ovo CAM Assay: How it Really Works. J. Vis. Exp. 2009, 2009, e1620. [Google Scholar] [CrossRef]
- Hamburger, V.; Hamilton, H.L. A series of normal stages in the development of the chick embryo. J. Morphol. 1951, 88, 49–92. [Google Scholar] [CrossRef] [PubMed]
- Ravelli, C.; Mitola, S.; Corsini, M.; Presta, M. Involvement of alphavbeta3 integrin in gremlin-induced angiogenesis. Angiogenesis 2013, 16, 235–243. [Google Scholar] [CrossRef]
- Maule, F.; Bresolin, S.; Rampazzo, E.; Boso, D.; Della Puppa, A.; Esposito, G.; Porcù, E.; Mitola, S.; Lombardi, G.; Accordi, B.; et al. Annexin 2A sustains glioblastoma cell dissemination and proliferation. Oncotarget 2016, 7, 54632–54649. [Google Scholar] [CrossRef][Green Version]
- González-González, A.; Rueda, N.; Alonso-González, C.; Menéndez, J.M.; Martínez-Campa, C.; Mitola, S.; Cos, S. Usefulness of melatonin as complementary to chemotherapeutic agents at different stages of the angiogenic process. Sci. Rep. 2020, 10, 1–20. [Google Scholar] [CrossRef]
- Mohammed, R.; Cavallaro, G.; Kessels, C.G.; Villamor, E. Functional differences between the arteries perfusing gas exchange and nutritional membranes in the late chicken embryo. J. Comp. Physiol. B 2015, 185, 783–796. [Google Scholar] [CrossRef]
- Valdes, T.I.; Klueh, U.; Kreutzer, D.; Moussy, F. Ex ova chick chorioallantoic membrane as a novelin vivo model for testing biosensors. J. Biomed. Mater. Res. 2003, 67, 215–223. [Google Scholar] [CrossRef]
- Di Somma, M.; Schaafsma, W.; Grillo, E.; Vliora, M.; Dakou, E.; Corsini, M.; Ravelli, C.; Ronca, R.; Sakellariou, P.; Vanparijs, J.; et al. Natural Histogel-Based Bio-Scaffolds for Sustaining Angiogenesis in Beige Adipose Tissue. Cells 2019, 8, 1457. [Google Scholar] [CrossRef] [PubMed]
- Samkoe, K.S.; Clancy, A.A.; Karotki, A.; Wilson, B.C.; Cramb, D.T. Complete blood vessel occlusion in the chick chorioallantoic membrane using two-photon excitation photodynamic therapy: Implications for treatment of wet age-related macular degeneration. J. Biomed. Opt. 2007, 12, 034025. [Google Scholar] [CrossRef] [PubMed]
- Schneider-Stock, R.; Ribatti, D. The CAM Assay as an Alternative In Vivo Model for Drug Testing. Handb. Exp. Pharmacol. 2020, 1–21. [Google Scholar] [CrossRef]
- Pink, D.B.S.; Schulte, W.; Parseghian, M.H.; Zijlstra, A.; Lewis, J.D. Real-Time Visualization and Quantitation of Vascular Permeability In Vivo: Implications for Drug Delivery. PLoS ONE 2012, 7, e33760. [Google Scholar] [CrossRef] [PubMed]
- Lokman, N.A.; Elder, A.S.F.; Ricciardelli, C.; Oehler, M.K. Chick Chorioallantoic Membrane (CAM) Assay as an In Vivo Model to Study the Effect of Newly Identified Molecules on Ovarian Cancer Invasion and Metastasis. Int. J. Mol. Sci. 2012, 13, 9959–9970. [Google Scholar] [CrossRef]
- Nowak-Sliwinska, P.; Segura, T.; Iruela-Arispe, M.L. The chicken chorioallantoic membrane model in biology, medicine and bioengineering. Angiogenesis 2014, 17, 779–804. [Google Scholar] [CrossRef]
- Steven, A.; Friedrich, M.; Jank, P.; Heimer, N.; Budczies, J.; Denkert, C.; Seliger, B. What turns CREB on? And off? And why does it matter? Cell. Mol. Life Sci. 2020, 77, 4049–4067. [Google Scholar] [CrossRef]
- Smith, S.A.; Newby, A.C.; Bond, M. Ending Restenosis: Inhibition of Vascular Smooth Muscle Cell Proliferation by cAMP. Cells 2019, 8, 1447. [Google Scholar] [CrossRef]
- Bekesi, A.; Abdellaoui, S.; Holroyd, N.; Van Delm, W.; Pardon, E.; Pauwels, J.; Gevaert, K.; Steyaert, J.; Derveaux, S.; Borysik, A.; et al. Challenges in the Structural–Functional Characterization of Multidomain, Partially Disordered Proteins CBP and p300: Preparing Native Proteins and Developing Nanobody Tools. Methods Enzym. 2018, 611, 607–675. [Google Scholar] [CrossRef]
- Comerford, K.M.; Leonard, M.O.; Karhausen, J.; Carey, R.; Colgan, S.P.; Taylor, C.T. Small ubiquitin-related modifier-1 modification mediates resolution of CREB-dependent responses to hypoxia. Proc. Natl. Acad. Sci. USA 2003, 100, 986–991. [Google Scholar] [CrossRef]
- Naqvi, S.; Martin, K.J.; Arthur, J.S.C. CREB phosphorylation at Ser133 regulates transcription via distinct mechanisms downstream of cAMP and MAPK signalling. Biochem. J. 2014, 458, 469–479. [Google Scholar] [CrossRef] [PubMed]
- Briand, L.A.; Lee, B.G.; Lelay, J.; Kaestner, K.H.; Blendy, J.A. Serine 133 phosphorylation is not required for hippocampal CREB-mediated transcription and behavior. Learn. Mem. 2015, 22, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Craig, J.C.; Schumacher, M.A.; Mansoor, S.E.; Farrens, D.L.; Brennan, R.G.; Goodman, R.H. Consensus and Variant cAMP-regulated Enhancers Have Distinct CREB-binding Properties. J. Biol. Chem. 2000, 276, 11719–11728. [Google Scholar] [CrossRef] [PubMed]
- Underwood, K.F.; Mochin, M.T.; Brusgard, J.L.; Choe, M.; Gnatt, A.; Passaniti, A. A Quantitative Assay to Study Protein:DNA Interactions, Discover Transcriptional Regulators of Gene Expression, and Identify Novel Anti-tumor Agents. J. Vis. Exp. 2013, 2013. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Odom, D.T.; Koo, S.-H.; Conkright, M.D.; Canettieri, G.; Best, J.; Chen, H.; Jenner, R.; Herbolsheimer, E.; Jacobsen, E.; et al. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc. Natl. Acad. Sci. USA 2005, 102, 4459–4464. [Google Scholar] [CrossRef] [PubMed]
- Corsini, M.; Moroni, E.; Ravelli, C.; Andrés, G.; Grillo, E.; Ali, I.H.; Brazil, D.P.; Presta, M.; Mitola, S. Cyclic Adenosine Monophosphate-Response Element–Binding Protein Mediates the Proangiogenic or Proinflammatory Activity of Gremlin. Arter. Thromb. Vasc. Biol. 2014, 34, 136–145. [Google Scholar] [CrossRef] [PubMed]
- Mitola, S.; Ravelli, C.; Moroni, E.; Salvi, V.; Leali, D.; Ballmer-Hofer, K.; Zammataro, L.; Presta, M. Gremlin is a novel agonist of the major proangiogenic receptor VEGFR2. Blood 2010, 116, 3677–3680. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Corsini, M.; Moroni, E.; Ravelli, C.; Grillo, E.; Presta, M.; Mitola, S. In Situ DNA/Protein Interaction Assay to Visualize Transcriptional Factor Activation. Methods Protoc. 2020, 3, 80. https://doi.org/10.3390/mps3040080
Corsini M, Moroni E, Ravelli C, Grillo E, Presta M, Mitola S. In Situ DNA/Protein Interaction Assay to Visualize Transcriptional Factor Activation. Methods and Protocols. 2020; 3(4):80. https://doi.org/10.3390/mps3040080
Chicago/Turabian StyleCorsini, Michela, Emanuela Moroni, Cosetta Ravelli, Elisabetta Grillo, Marco Presta, and Stefania Mitola. 2020. "In Situ DNA/Protein Interaction Assay to Visualize Transcriptional Factor Activation" Methods and Protocols 3, no. 4: 80. https://doi.org/10.3390/mps3040080
APA StyleCorsini, M., Moroni, E., Ravelli, C., Grillo, E., Presta, M., & Mitola, S. (2020). In Situ DNA/Protein Interaction Assay to Visualize Transcriptional Factor Activation. Methods and Protocols, 3(4), 80. https://doi.org/10.3390/mps3040080






