Comparison of Different Fixation Methods for Combined Histological and Biomolecular Analysis of Fixed and Decalcified Bone Samples
Abstract
:1. Introduction
2. Material and Methods
2.1. Sample Collection and Reparation
2.2. Histological Analysis
2.3. Biomolecular Analysis
2.3.1. Isolation of Total RNA from Unfixed Frozen Bone Samples
2.3.2. Isolation of Total RNA from Fixed and Paraffin Embedded Bone Samples
2.3.3. Quantification of RNA and Quality Control
2.3.4. Gene Expression
2.4. Statistical Evaluation
3. Experiment Results
3.1. Histological and Immunohistological Evaluation
3.2. Biomolecular Evaluation
3.2.1. Extracted RNA from Differently Treated Samples
3.2.2. Gene Expression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Marsell, R.; Einhorn, T.A. The biology of fracture healing. Injury 2011, 42, 551–555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghanaati, S.; Kirkpatrick, C.J.; Kovács, A.F.; Landes, C.; Lorenz, J.; Sader, R.A.; Seitz, O.; Stuebinger, S.; Barbeck, M. Synthetic bone substitute material comparable with xenogeneic material for bone tissue regeneration in oral cancer patients: First and preliminary histological, histomorphometrical and clinical results. Ann. Maxillofac. Surg. 2013, 3, 126–138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Succo, G.; Berrone, M.; Battiston, B.; Tos, P.; Goia, F.; Appendino, P.; Crosetti, E. Step-by-step surgical technique for mandibular reconstruction with fibular free flap: Application of digital technology in virtual surgical planning. Eur. Arch. Oto-Rhino-Laryngol. 2015, 272, 1491–1501. [Google Scholar] [CrossRef] [PubMed]
- Younger, E.M.; Chapman, M.W. Morbidity at Bone Graft Donor Sites. J. Orthop. Trauma 1989, 3, 192–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Omar, O.; Dahlin, A.; Gasser, A.; Dahlin, C. Tissue dynamics and regenerative outcome in two resorbable non-cross-linked collagen membranes for guided bone regeneration: A preclinical molecular and histological study in vivo. Clin. Oral Implant. Res. 2018, 29, 7–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dohle, E.; Singh, S.; Nishiguchi, A.; Fischer, T.; Wessling, M.; Möller, M.; Sader, R.; Kasper, J.Y.; Ghanaati, S.; Kirkpatrick, C.J. Human Co- and Triple-Culture Model of the Alveolar-Capillary Barrier on a Basement Membrane Mimic. Tissue Eng. Part C Methods 2018, 24, 495–503. [Google Scholar] [CrossRef]
- Al-Maawi, S.; Mota, C.; Kubesch, A.; James Kirkpatrick, C.; Moroni, L.; Ghanaati, S. Multiwell three-dimensional systems enable in vivo screening of immune reactions to biomaterials: A new strategy toward translational biomaterial research. J. Mater. Sci. Mater. Med. 2019, 30, 61. [Google Scholar] [CrossRef]
- Mourão, C.F.; Machado, R.; Javid, K.; Moraschini, V. The use of leukocyte- and platelet-rich fibrin in the management of soft tissue healing and pain in post-extraction sockets: A randomized clinical trial. J. Cranio-Maxillofac. Surg. 2020, 48, 452–457. [Google Scholar] [CrossRef]
- Lorenz, J.; Korzinskas, T.; Chia, P.; Al Maawi, S.; Eichler, K.; Sader, R.A.; Ghanaati, S. Do Clinical and Radiological Assessments Contribute to the Understanding of Biomaterials? Results from a Prospective Randomized Sinus Augmentation Split-Mouth Trial. J. Oral Implant. 2018, 44, 62–69. [Google Scholar] [CrossRef]
- Lorenz, J.; Kubesch, A.; Korzinskas, T.; Barbeck, M.; Landes, C.; Sader, R.A.; Kirkpatrick, C.J.; Ghanaati, S. TRAP-Positive Multinucleated Giant Cells Are Foreign Body Giant Cells Rather Than Osteoclasts: Results from a Split-Mouth Study in Humans. J. Oral Implant. 2015, 41, e257–e266. [Google Scholar] [CrossRef] [Green Version]
- Canellas, J.V.D.S.; da Costa, R.C.; Breves, R.C.; de Oliveira, G.P.; Figueredo, C.M.D.S.; Fischer, R.G.; Thole, A.A.; Medeiros, P.J.D.; Ritto, F.G. Tomographic and histomorphometric evaluation of socket healing after tooth extraction using leukocyte- and platelet-rich fibrin: A randomized, single-blind, controlled clinical trial. J. Cranio-Maxillofac. Surg. 2020, 48, 24–32. [Google Scholar] [CrossRef]
- Halloran, D.; Durbano, H.W.; Nohe, A. Bone Morphogenetic Protein-2 in Development and Bone Homeostasis. J. Dev. Biol. 2020, 8, 19. [Google Scholar] [CrossRef]
- Stübinger, S.; Ghanaati, S.; Orth, C.; Hilbig, U.; Saldamli, B.; Biesterfeld, S.; Kirkpatrick, C.; Sader, R. Maxillary Sinus Grafting with a Nano-Structured Biomaterial: Preliminary Clinical and Histological Results. Rech. Chir. Eur. 2009, 42, 143–149. [Google Scholar] [CrossRef]
- Castro, A.B.; Van Dessel, J.; Temmerman, A.; Jacobs, R.; Quirynen, M. Effect of different platelet-rich fibrin matrices for ridge preservation in multiple tooth extractions: A split-mouth randomized controlled clinical trial. J. Clin. Periodontol. 2021, 48, 984–995. [Google Scholar] [CrossRef]
- Kilic, S.C.; Güngörmüş, M.; Parlak, S.N. Histologic and histomorphometric assessment of sinus-floor augmentation with beta-tricalcium phosphate alone or in combination with pure-platelet-rich plasma or platelet-rich fibrin: A randomized clinical trial. Clin. Implant Dent. Relat. Res. 2017, 19, 959–967. [Google Scholar] [CrossRef]
- Zhang, Y.; Al-Maawi, S.; Wang, X.; Sader, R.; James Kirkpatrick, C.; Ghanaati, S. Biomaterial-induced multinucleated giant cells express proinflammatory signaling molecules: A histological study in humans. J. Biomed. Mater. Res. Part A 2019, 107, 780–790. [Google Scholar] [CrossRef]
- Hughes, A.; Stewart, T.L.; Mann, V. Extraction of Nucleic Acids from Bone. Methods Mol. Biol. 2012, 816, 249–259. [Google Scholar] [CrossRef]
- Farragher, S.M.; Tanney, A.; Kennedy, R.D.; Harkin, D.P. RNA expression analysis from formalin fixed paraffin embedded tissues. Histochem. Cell Biol. 2008, 130, 435–445. [Google Scholar] [CrossRef]
- Masuda, N.; Ohnishi, T.; Kawamoto, S.; Monden, M.; Okubo, K. Analysis of chemical modification of RNA from formalin-fixed samples and optimization of molecular biology applications for such samples. Nucleic Acids Res. 1999, 27, 4436–4443. [Google Scholar] [CrossRef]
- Groelz, D.; Sobin, L.; Branton, P.; Compton, C.; Wyrich, R.; Rainen, L. Non-formalin fixative versus formalin-fixed tissue: A comparison of histology and RNA quality. Exp. Mol. Pathol. 2013, 94, 188–194. [Google Scholar] [CrossRef] [Green Version]
- Pedersen, K.B.; Williams, A.; Watt, J.; Ronis, M.J. Improved method for isolating high-quality RNA from mouse bone with RNAlater at room temperature. Bone Rep. 2019, 11, 100211. [Google Scholar] [CrossRef]
- Suhovskih, A.V.; Kazanskaya, G.M.; Volkov, A.M.; Tsidulko, A.Y.; Aidagulova, S.V.; Grigorieva, E.V. Suitability of RNALater solution as a tissue-preserving reagent for immunohistochemical analysis. Histochem. Cell Biol. 2019, 152, 239–247. [Google Scholar] [CrossRef]
- Shao, Y.Y.; Wang, L.; Hicks, D.G.; Ballock, R.T. Analysis of gene expression in mineralized skeletal tissues by laser capture microdissection and RT-PCR. Lab. Investig. 2006, 86, 1089–1095. [Google Scholar] [CrossRef] [Green Version]
- Miquelestorena-Standley, E.; Jourdan, M.-L.; Collin, C.; Bouvier, C.; Larousserie, F.; Aubert, S.; Gomez-Brouchet, A.; Guinebretière, J.-M.; Tallegas, M.; Brulin, B.; et al. Effect of decalcification protocols on immunohistochemistry and molecular analyses of bone samples. Mod. Pathol. 2020, 33, 1505–1517. [Google Scholar] [CrossRef]
- Waissbluth, S.; Chan, S.W.; Chen, J.Z.; McIntosh, M.; Daniel, S.J. RNA Preservation in Decalcified Cochlear Samples. Otol. Neurotol. 2013, 34, 331–337. [Google Scholar] [CrossRef]
- Ghanaati, S.; Schlee, M.; Webber, M.J.; Willershausen, I.; Barbeck, M.; Balic, E.; Görlach, C.; Stupp, S.I.; Sader, R.A.; Kirkpatrick, C.J. Evaluation of the tissue reaction to a new bilayered collagen matrix in vivo and its translation to the clinic. Biomed. Mater. 2011, 6, 015010. [Google Scholar] [CrossRef]
- Al-Maawi, S.; Rother, S.; Halfter, N.; Fiebig, K.M.; Moritz, J.; Moeller, S.; Schnabelrauch, M.; Kirkpatrick, C.J.; Sader, R.; Wiesmann, H.-P.; et al. Covalent linkage of sulfated hyaluronan to the collagen scaffold Mucograft® enhances scaffold stability and reduces proinflammatory macrophage activation in vivo. Bioact. Mater. 2021, 8, 420–434. [Google Scholar] [CrossRef]
- Cepollaro, S.; Della Bella, E.; De Biase, D.; Visani, M.; Fini, M. Evaluation of RNA from human trabecular bone and identification of stable reference genes. J. Cell. Physiol. 2018, 233, 4401–4407. [Google Scholar] [CrossRef] [PubMed]
- Fleige, S.; Pfaffl, M.W. RNA integrity and the effect on the real-time qRT-PCR performance. Mol. Asp. Med. 2006, 27, 126–139. [Google Scholar] [CrossRef] [PubMed]
- Kuang, J.; Yan, X.; Genders, A.J.; Granata, C.; Bishop, D.J. An overview of technical considerations when using quantitative real-time PCR analysis of gene expression in human exercise research. PLoS ONE 2018, 13, e0196438. [Google Scholar] [CrossRef] [PubMed]
- Mandarino, D.; Luz, D.; Moraschini, V.; Rodrigues, D.; Barboza, E. Alveolar ridge preservation using a non-resorbable membrane: Randomized clinical trial with biomolecular analysis. Int. J. Oral Maxillofac. Surg. 2018, 47, 1465–1473. [Google Scholar] [CrossRef]
- Poundarik, A.A.; Boskey, A.; Gundberg, C.; Vashishth, D. Biomolecular regulation, composition and nanoarchitecture of bone mineral. Sci. Rep. 2018, 8, 1191. [Google Scholar] [CrossRef] [Green Version]
- Miron, R.J.; Bosshardt, D.D. OsteoMacs: Key players around bone biomaterials. Biomaterials 2016, 82, 1–19. [Google Scholar] [CrossRef]
- Cox, M.L.; Schray, C.L.; Luster, C.N.; Stewart, Z.S.; Korytko, P.J.; Khan, K.N.M.; Paulauskis, J.D.; Dunstan, R.W. Assessment of fixatives, fixation, and tissue processing on morphology and RNA integrity. Exp. Mol. Pathol. 2006, 80, 183–191. [Google Scholar] [CrossRef]
- Milcheva, R.; Janega, P.; Celec, P.; Russev, R.; Babál, P. Alcohol based fixatives provide excellent tissue morphology, protein immunoreactivity and RNA integrity in paraffin embedded tissue specimens. Acta Histochem. 2013, 115, 279–289. [Google Scholar] [CrossRef]
- Dotti, I.; Bonin, S.; Basili, G.; Nardon, E.; Balani, A.; Siracusano, S.; Zanconati, F.; Palmisano, S.; De Manzini, N.; Stanta, G. Effects of Formalin, Methacarn, and FineFIX Fixatives on RNA Preservation. Diagn. Mol. Pathol. 2010, 19, 112–122. [Google Scholar] [CrossRef]
- Takagi, H.; Shibutani, M.; Kato, N.; Fujita, H.; Lee, K.-Y.; Takigami, S.; Mitsumori, K.; Hirose, M. Microdissected Region-specific Gene Expression Analysis with Methacarn-fixed, Paraffin-embedded Tissues by Real-time RT-PCR. J. Histochem. Cytochem. 2004, 52, 903–913. [Google Scholar] [CrossRef]
- Salmon, C.R.; Silvério, K.G.; Giorgetti, A.P.D.O.; Sallum, E.A.; Casati, M.Z.; Nociti, F.H. Gene Expression Analysis in Microdissected Samples from Decalcified Tissues. Diagn. Mol. Pathol. 2012, 21, 120–126. [Google Scholar] [CrossRef]
- Skrypina, N.A.; Timofeeva, A.V.; Khaspekov, G.L.; Savochkina, L.P.; Beabealashvilli, R.S. Total RNA suitable for molecular biology analysis. J. Biotechnol. 2003, 105, 1–9. [Google Scholar] [CrossRef]
- Soheilypour, M.; Mofrad, M.R.K. Quality control of mRNAs at the entry of the nuclear pore: Cooperation in a complex molecular system. Nucleus 2018, 9, 202–211. [Google Scholar] [CrossRef] [Green Version]
- Bell, D.A.; DeMarini, D.M. Excessive cycling converts PCR products to randomlength higher molecular weight fragments. Nucleic Acids Res. 1991, 19, 5079. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.-O.; Kim, H.-N.; Hwang, M.-H.; Shin, H.-I.; Kim, S.-Y.; Park, R.-W.; Park, E.-Y.; Kim, I.-S.; Van Wijnen, A.J.; Stein, J.L.; et al. Differential gene expression analysis using paraffin-embedded tissues after laser microdissection. J. Cell. Biochem. 2003, 90, 998–1006. [Google Scholar] [CrossRef]
- Shibutani, M.; Uneyama, C.; Miyazaki, K.; Toyoda, K.; Hirose, M. Methacarn Fixation: A Novel Tool for Analysis of Gene Expressions in Paraffin-Embedded Tissue Specimens. Lab. Investig. 2000, 80, 199–208. [Google Scholar] [CrossRef] [Green Version]
- Srinivasan, M.; Sedmak, D.; Jewell, S. Effect of Fixatives and Tissue Processing on the Content and Integrity of Nucleic Acids. Am. J. Pathol. 2002, 161, 1961–1971. [Google Scholar] [CrossRef] [Green Version]
- Puchtler, H.; Waldrop, F.S.; Meloan, S.N.; Terry, M.S.; Conner, H.M. Methacarn (methanol-Carnoy) fixation. Histochemie 1970, 21, 97–116. [Google Scholar] [CrossRef]
- Beckstead, J.H. A simple technique for preservation of fixation-sensitive antigens in paraffin-embedded tissues. J. Histochem. Cytochem. 1994, 42, 1127–1134. [Google Scholar] [CrossRef] [Green Version]

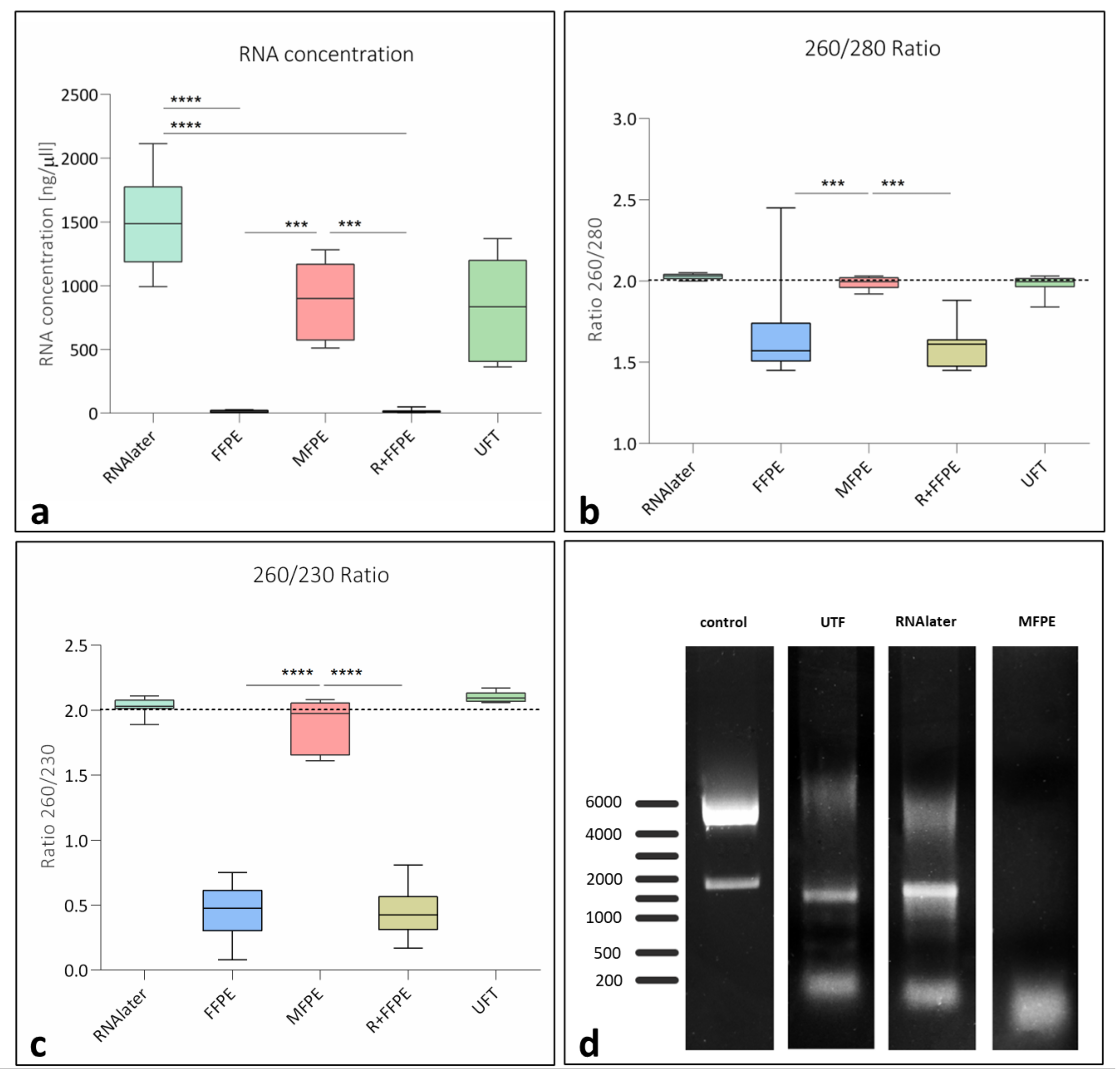
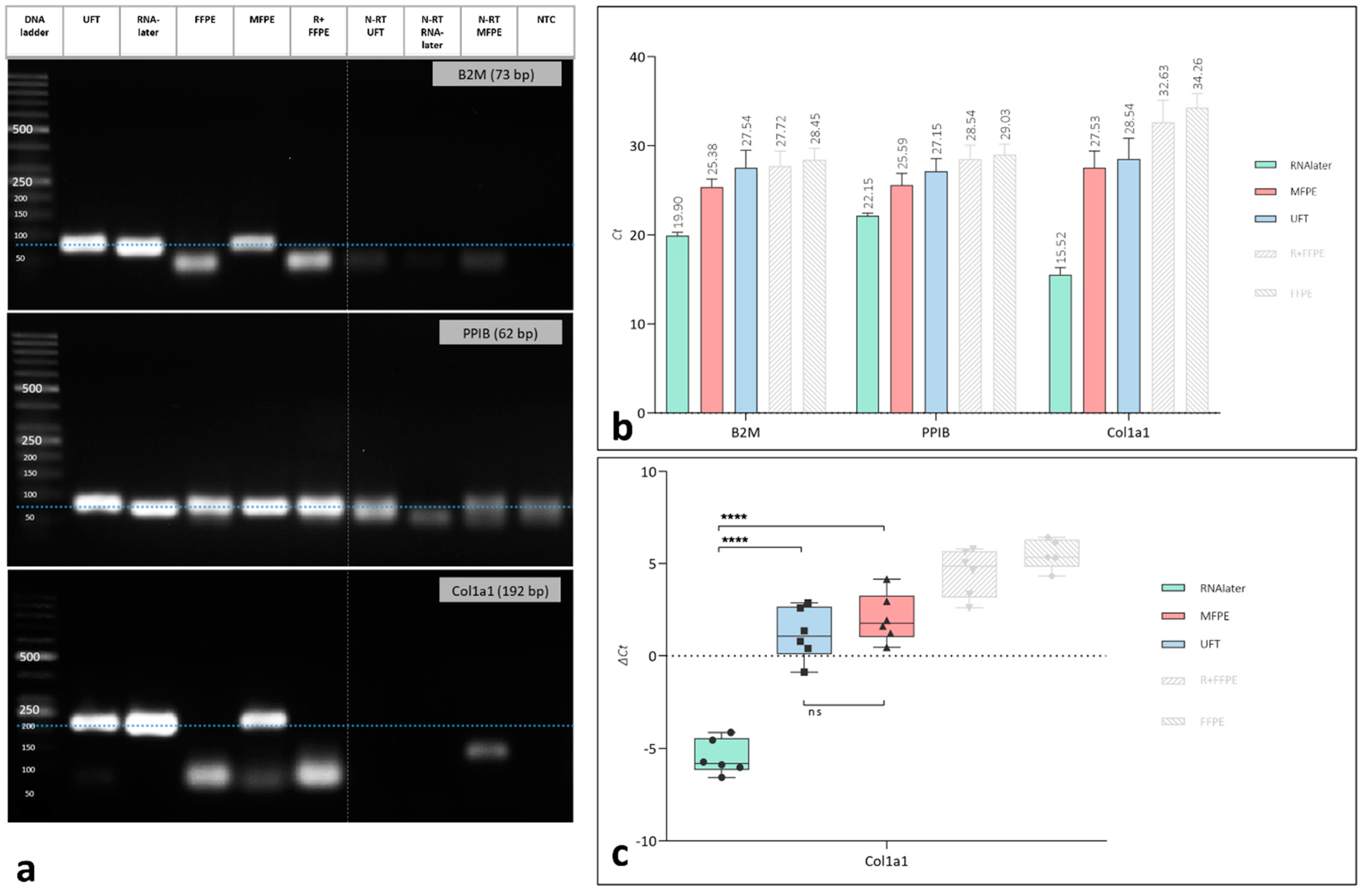
| Group | Category | Fixation Medium | Incubation Time | P + PE |
|---|---|---|---|---|
| UFT | control | snap-frozen (in liquid nitrogen) | 15 min | - |
| RNAlater | control | RNAlater | 1 week | - |
| FFPE | test | Formaldehyde (Roti®-Histofix) | 1 week | + |
| MFPE | test | Methacarn | 1 week | + |
| R + FFPE | test | RNAlater (6 days) + formaldehyde (Roti®-Histofix) (24 h) | 1 week | + |
| Gene | NCBI Accession Number (mRNA) | mRNA Length (bp) | 5′-Forward-Primer-3′ 5′-Reverse-Primer-3′ | Primer Length (bp) |
|---|---|---|---|---|
| Reference Genes | ||||
| B2M | NM_012512.2 | 1845 | TCTCTCTGGCCGTCGTGCTT | 20 |
| TTCTCCGGTGGATGGCGAGA | 20 | |||
| PPIB | NM_022536.2 | 838 | ACG TGG TTT TCG GCA AAG T | 19 |
| CTT GGT GTT CTC CAC CTT CC | 20 | |||
| Target Gene | ||||
| Col1a1 | NM_053304.1 | 5843 | CCTGACGCATGGCCAAGAAG | 20 |
| CACTCGCCCTCCCGTTTTTG | 20 | |||
| Criteria | FFPE | MFPE | R + FFPE |
|---|---|---|---|
| Staining characteristics | 3 | 3 | 3 |
| Tissue architecture | 3 | 3 | 3 |
| Cell morphology and sharpness | 3 | 3 | 2 |
| Total | 9 | 9 | 8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Maawi, S.; Valenzuela, P.; Dohle, E.; Heselich, A.; Sader, R.; Ghanaati, S. Comparison of Different Fixation Methods for Combined Histological and Biomolecular Analysis of Fixed and Decalcified Bone Samples. Methods Protoc. 2022, 5, 64. https://doi.org/10.3390/mps5040064
Al-Maawi S, Valenzuela P, Dohle E, Heselich A, Sader R, Ghanaati S. Comparison of Different Fixation Methods for Combined Histological and Biomolecular Analysis of Fixed and Decalcified Bone Samples. Methods and Protocols. 2022; 5(4):64. https://doi.org/10.3390/mps5040064
Chicago/Turabian StyleAl-Maawi, Sarah, Priscilia Valenzuela, Eva Dohle, Anja Heselich, Robert Sader, and Shahram Ghanaati. 2022. "Comparison of Different Fixation Methods for Combined Histological and Biomolecular Analysis of Fixed and Decalcified Bone Samples" Methods and Protocols 5, no. 4: 64. https://doi.org/10.3390/mps5040064
APA StyleAl-Maawi, S., Valenzuela, P., Dohle, E., Heselich, A., Sader, R., & Ghanaati, S. (2022). Comparison of Different Fixation Methods for Combined Histological and Biomolecular Analysis of Fixed and Decalcified Bone Samples. Methods and Protocols, 5(4), 64. https://doi.org/10.3390/mps5040064







