Sequential Isolation of Microglia and Astrocytes from Young and Aged Adult Mouse Brains for Downstream Transcriptomic Analysis
Abstract
1. Introduction
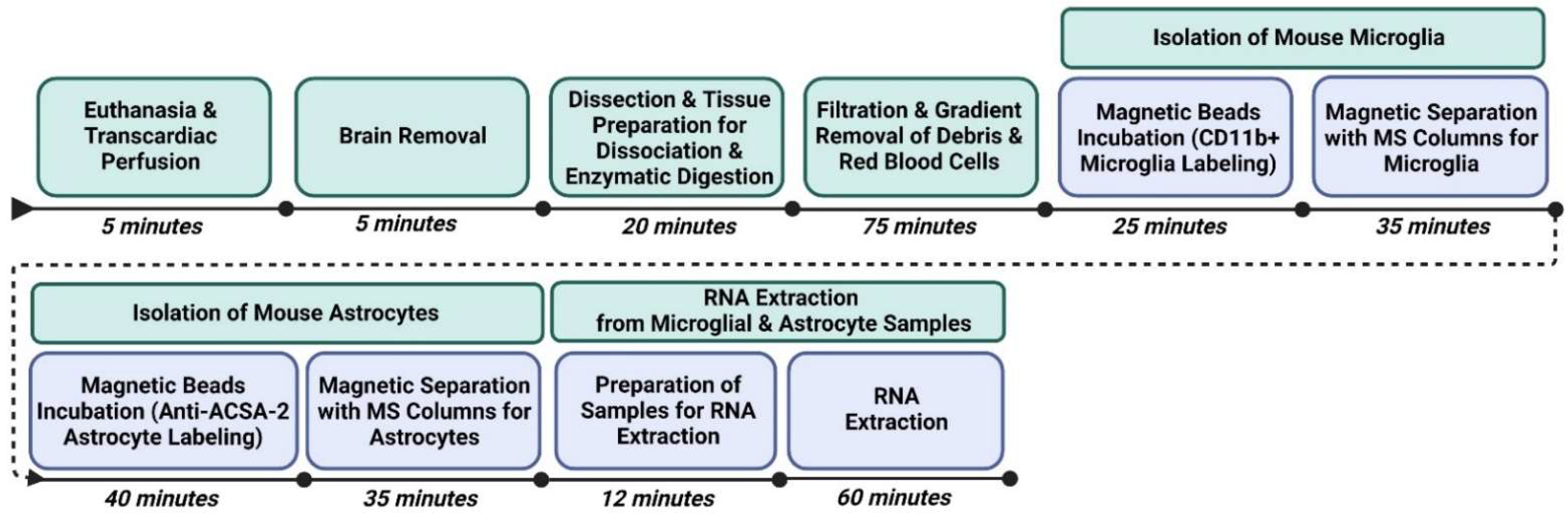
2. Experimental Design
3. Procedure
3.1. Euthanasia and Transcardiac Perfusion
- Euthanize mouse (C57Bl/6J) via CO2 chamber at a flow rate of 4 L/min for approximately 3 min;
- Place mouse on ice and to lift the upper ventral abdomen using Graefe forceps;
- While holding Graefe forceps, make cutaneal incisions with light operating scissors, 3 cm horizontally and 3 cm vertically from the abdominal midline to expose the peritoneal cavity;
- After cutaneal incisions have been made, confirm secondary euthanasia by lifting the xyphoid process with Graefe forceps and creating lateral incisions of the diaphragm along the upper thoracic line;
- Make bilateral incisions of the thoracic vertebra and using Halsted mosquito forceps hold the xyphoid process above the chest cavity;
- With the heart now exposed, insert a 21-gauge needle 5 mm into the left ventricle;
- Turn on peristaltic pump to initiate transcardiac flush of ice-cold 1X PBS at 5 mL/min (see Table 3);
- After turning on the pump, immediately incise the right atrium by 3 mm using micro dissecting scissors (see Figure 2);
- Continue perfusion for approximately 2 min or until visible color change of liver indicating successful perfusion.
3.2. Brain Removal
- Immediately post perfusion, decapitate the mouse with 6.5” operating scissors;
- Cut the skin above the midline with micro dissecting scissors, to expose the skull and externally rotate the skin above the head;
- Make bilateral incisions between the posterior midline and the parietal bone (approximately 10 mm) and between the posterior midline and the occipital bone (approximately 5 mm), creating a t-formation;
- Using closed micro dissecting scissors, insert tip into the junction of the interfrontal suture and frontonasal sutures approximately 2 mm into the skull, then gently open the scissors to crack the calvaria midline down the interfrontal and sagittal suture, opening the skull into two hemispheres;
- Using Graefe forceps, carefully separate both calvariae hemispheres away from brain;
- Remove connective tissue from skull, and remove brain using curved forceps or micro spatula;
- Place brain on a cold plate for immediate dissection of brain regions.
3.3. Dissection and Preparation of Tissue for Dissociation and Enzymatic Digestion
- Position the brain on ice cold plate so that the cerebral cortices are facing upwards.
- 2.
- Using one brain hemisphere, turn over with cortex facing the cold block. Using small curved forceps, pinch out striatal/thalamus region leaving only cortical and hippocampal tissue;
- 3.
- Take a micro punch through cortical and hippocampal tissue (20–50 mg);
- 4.
- Using a sterile scalpel, slice/mince selected micro punch into small pieces;
- 5.
- Place segments of minced micro punch tissue around the cap of gentleMACS C Tube in a circular arrangement;
- 6.
- Prepare both enzyme mix 1 and 2 (see Table 3);
- 7.
- Add 1950 µL of enzyme mix 1 and 30 µL of enzyme mix 2 into gentleMACS C Tube;
- 8.
- Place cap with circular arranged tissue on gentleMACS C tube and close;
- 9.
- Tip C tube upside down, ensuring enzyme mix 1 and 2 solution is covering tissue;
- 10.
- Place closed C tube with tissue and enzymes on gentleMACS Octo Dissociator to proceed with gentleMACS Program 37C_ABDK_02;
CRITICAL STEP Samples should be prepared directly after perfusion for optimal cell suspension performance. Keep samples on ice, especially when preparing larger sample sizes, to maintain optimal tissue integrity.
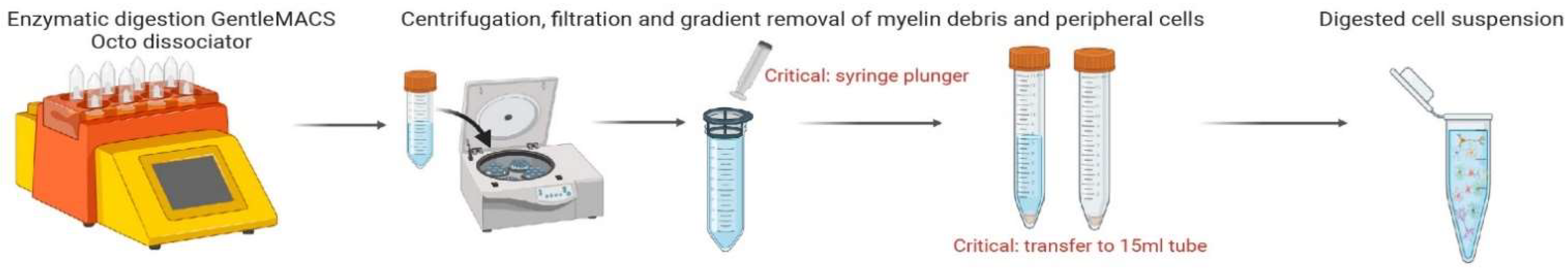
3.4. Filtration and Gradient Removal of Debris and Red Blood Cells (Figure 3)
- Prepare MACS SmartStrainers (70 µm) by lightly prewetting surface with cold D-PBS;
- After termination of the ABDK_02 program, detach C Tube from the gentleMACS Octo Dissociator with Heaters;
- Resuspend sample by adding 10 mL of cold D-PBS to the gentleMACS C Tube. Close the tube and gently shake to collect samples at the bottom. Gentle inversions may be necessary to retrieve any remaining tissue on the lid;
- Add components of C Tube through MACS SmartStrainers (70 µm) into a 50 mL conical tube;
CRITICAL STEP Use the top end of a syringe plunger to mash and fully dissolve sample through strainer;
CRITICAL STEP Transfer samples from 50 mL conical tubes to new 15 mL conical tubes after filtration;
- 5.
- Centrifuge samples at 300× g for 10 min at 4 °C. Remove supernatant completely;
- 6.
- Resuspend cell pellet with 1550 µL of cold D-PBS in the 15 mL conical tube;
- 7.
- Add 450 µL of debris removal solution and mix well;
- 8.
- Gently overlay 2 mL of D-PBS into tube. Be careful not to mix phases;
- OPTIONAL STEP Slowly pipette D-PBS with the conical tube at a 45° angle to prevent phase mixing;
- 9.
- Centrifuge samples at 300× g for 10 min at 4 °C;
- 10.
- After three phases are formed, aspirate the top two phases gently and completely discard;
CRITICAL STEP Slowly remove at a 45° angle to increase accuracy;
- 11.
- With one phase remaining, fill tube with cold D-PBS to a final volume of 10 mL;
- 12.
- Gently invert three times;
- 13.
- Centrifuge at 1000× g for 10 min at 4 °C. Aspirate remaining supernatant completely;
- 14.
- Prepare 1X Red Blood Cell Removal Solution (see Table 3);
- 15.
- Resuspend pellet in 500 µL of cold 1X Red Blood Cell Removal Solution, and incubate for 10 min at 4 °C;
- 16.
- Add 5 mL of PB buffer and centrifuge at 300× g for 10 min at 4 °C. Remove remaining supernatant completely and proceed to microglial magnetic beads labelling.
3.5. Isolation of Mouse Microglia (Figure 4)
- Magnetic Beads Incubation (CD11b+ microglia labeling):
- 1.
- Prepare PB buffer solution (see Table 3);
- 2.
- Resuspend pellet in 90 µL cold PB buffer by slowly pipetting up and down;
- 3.
- Add 10 µL CD11b (Microglia) MicroBeads, human and mouse mix well;
- 4.
- Incubate in the dark for 15 min at 4 °C;
- 5.
- Add 1 mL of cold PB buffer and mix well;
- 6.
- Centrifuge at 300× g for 5 min at 4 °C;
- 7.
- Remove supernatant and resuspend cells in 500 µL PB buffer;
- Magnetic Separation with MS Columns for Microglia
- 8.
- Place MS columns in the magnetic field of MACS Separator and prepare column by rinsing through 500 µL PB buffer. Discard flow through;
CRITICAL STEP Wait until column is completely empty before proceeding to next step;
- OPTIONAL STEP To remove air bubbles from column after 500 µL rinse, use a needle syringe to pop bubbles;
- 9.
- Prepare 5 mL flow through collection tubes underneath MS columns for negative flow through collection;
- 10.
- Apply the 500 µL of CD11b+ labeled cells (from step 7 above) into MS column attached to magnetic separator;
CRITICAL STEP To increase purity of microglia, apply negative flow through from each collection tube back into corresponding MS column for a second time;
- 11.
- Wash MS column with 500 µL PB buffer three times;
- 12.
- After wash is complete, remove 5 mL collection tubes (containing negative fraction);
- 13.
- Place 5 mL collection tubes (containing negative fraction) into a 15 mL falcon to allow for centrifugation;
CRITICAL STEP
- 14.
- Centrifuge 15 mL falcon containing 5 mL collection tubes with negative flow though at 300× g for 10 min at 4 °C. Set aside for astrocyte labelling in Section 3.6;
- 15.
- Remove MS column containing CD11b+ bound cells from the separator, place it on corresponding collection tube, add 1 mL PB buffer, and immediately flush out to elute CD11b+ magnetically labeled cells with provided plunger;
- 16.
- Repeat step 14 with an additional 1 mL of PB buffer to increase yield;
CRITICAL STEP
- 17.
- Proceed to RNA extraction of eluted Cd11b+ microglia.

3.6. Isolation of Mouse Astrocytes (Figure 4)
- Resuspend pellet in 80 µL AstroMACS Separation Buffer (or PB buffer alternatively);
- Add 10 µL of FcR Blocking Reagent, mix well, and incubate in the dark for 10 min at 4 °C;
- Add 10 µL of Anti-ACSA-2 MicroBeads, mix well, and incubate in the dark for 10 min at 4 °C;
- Add 1 mL of AstroMACS Separation buffer and place 5 mL reagent tubes back into 15 mL conical tubes and centrifuge at 300× g for 5 min at 4 °C;
- Remove 5 mL reagent tubes, completely remove supernatant, and resuspend cells in 500 µL AstroMACS Separation Buffer;
- Magnetic Separation with MS Columns for Astrocytes
- 6.
- Place new MS columns in the magnetic field of MACS Separator and prepare column by rinsing through 500 µL PB buffer. Discard flow through;
CRITICAL STEP Wait until column is completely empty before proceeding to next step;
- OPTIONAL STEP To remove air bubbles from column after 500 µL rinse, use a needle syringe to pop bubbles;
- 7.
- Prepare 5 mL flow through collection tubes underneath MS columns for negative flow through collection;
- 8.
- Apply the 500 µL of ACSA2+ labeled cells (from step 5 above) into MS column attached to magnetic separator;
CRITICAL STEP To increase purity of astrocytes, apply negative selection back through MS column a second time;
- OPTIONAL STEP To remove air bubbles from column entry disruption, use a needle syringe to remove obstructions;
- 9.
- Wash MS column with 500 µL PB buffer three times;
- 10.
- Remove MS column containing ACSA2+ bound cells from the separator, place it on corresponding collection tube, add 1 mL AstroMACS Separation buffer, and immediately flush out to elute ACSA2+ magnetically labeled cells with provided plunger.
- 11.
- Repeat step 11 with an additional 1 mL of AstroMACS separation buffer to increase yield;
CRITICAL STEP
- 12.
- Proceed to RNA extraction of eluted ACSA2+ astrocytes.
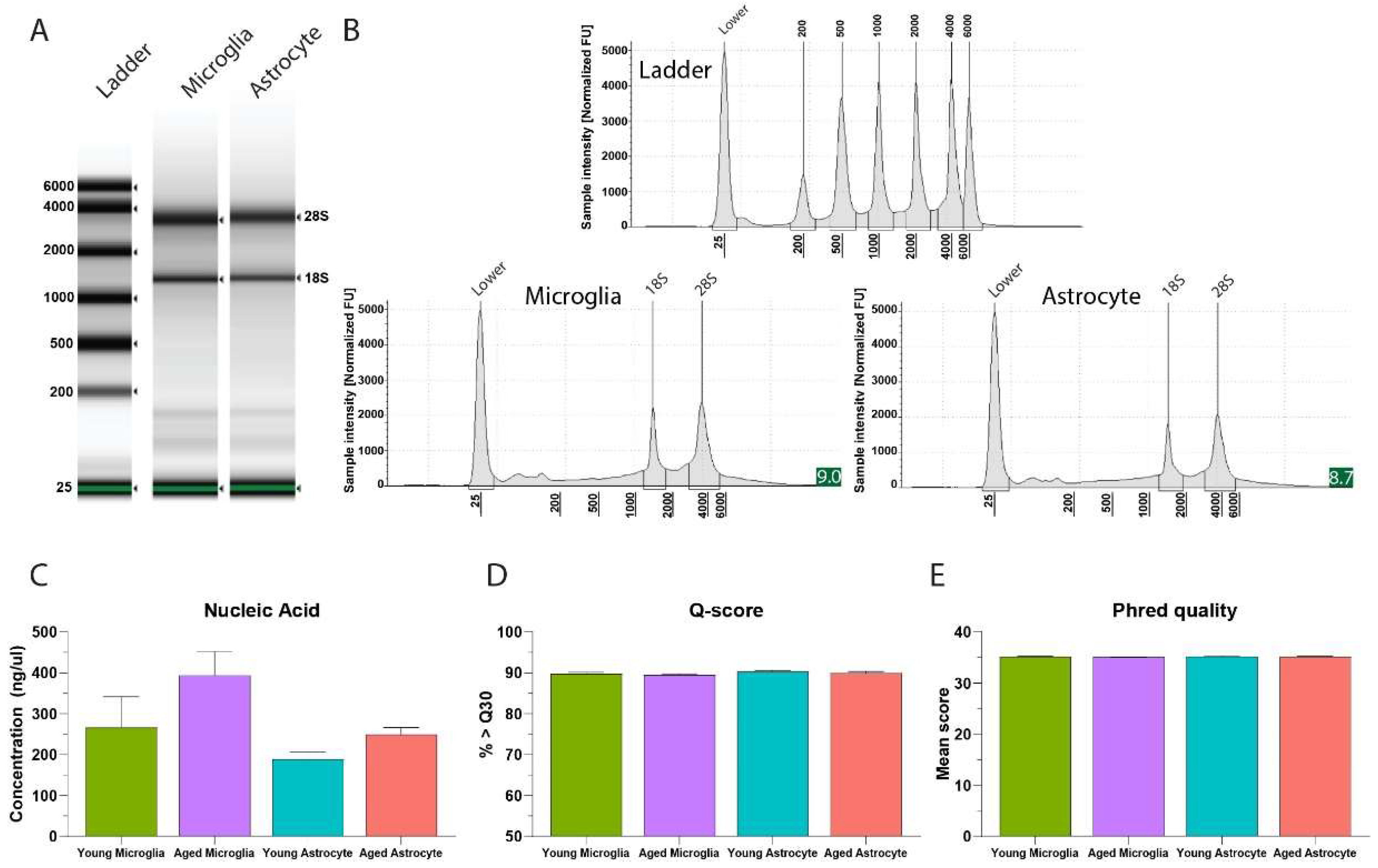
3.7. RNA Extraction from Microglial and Astrocyte Samples (Figure 5, Figure 6 and Figure 7)
- After magnetic separation of microglia or astrocytes, centrifuge samples at 1000× g for 5 min at 4 °C and remove supernatant (~2 mL);
- Add 500 µL TRIzol, sonicate pellet completely, and transfer solution to 1.5 mL Eppendorf tubes. Proceed with subsequent RNA extraction. Samples can be stored at −20 °C for later extraction;
- 3.
- Add 500 µL of TRIzol Reagent (total 1 mL) and incubate for 5 min at room temperature (RT);
- 4.
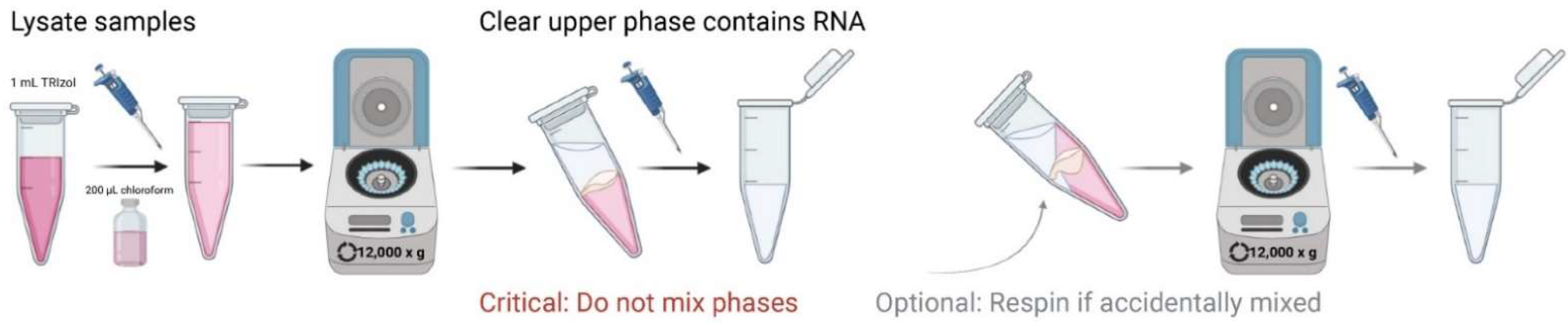
- Add 200 µL chloroform, thoroughly mix by shaking, and incubate at RT for 2 min.
- 5.
- Centrifuge samples at 4 °C for 15 min at 12,000× g;
- 6.
- The mixture separates into 3 layers which include a colorless upper phase, white interphase, and pink phenol-chloroform lower phase;
CRITICAL STEP In a 45° angle, being careful not to mix phases together, remove clear top layer containing RNA into a new tube. For optimal purity, closely view tube for phase movement and do not transfer contaminated mixed-phase solution (Figure 5);
- OPTIONAL STEP Should any of the three layers be accidentally mixed, centrifuge samples again at 4 °C for 15 min at 12,000× g and repeat the upper-phase extraction, which contains RNA (Figure 5);
- 7.
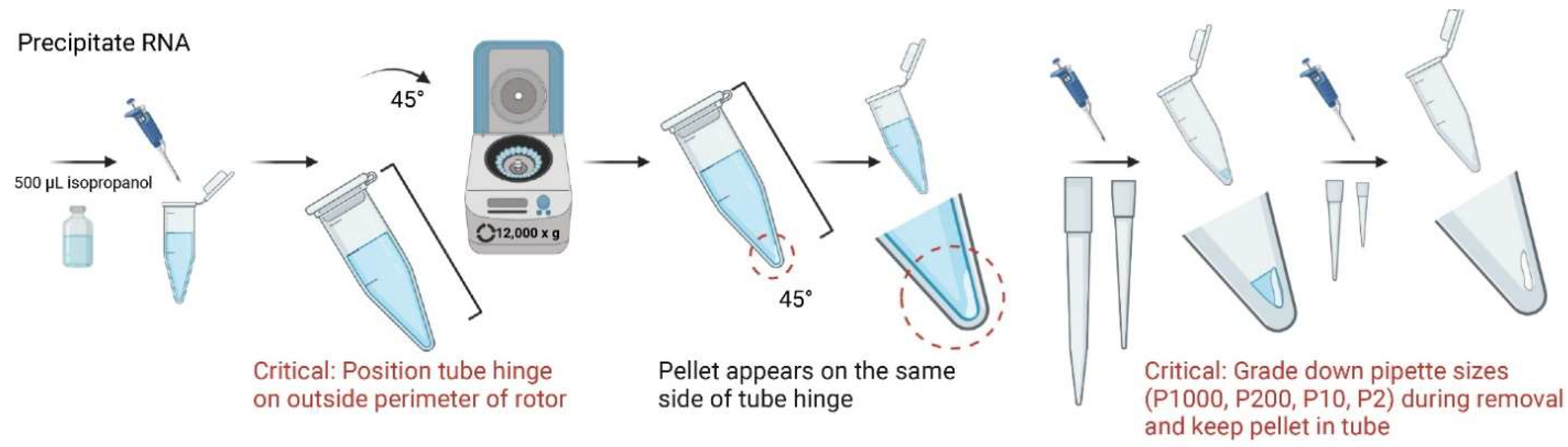
- Add 500 µL of 100% isopropanol to each sample, invert tubes three times, and incubate in 4 °C for 10 min;
- 8.
- Centrifuge in 4 °C for 10 min. Tubes should be spun with the hinged cap side up, which will be used as a physical guideline for identifying pellets at later stages. It is important to maintain this tube angle in all centrifugation steps;
- 9.
- Due to use of a 45° rotor, a small, white translucent, gel-like pellet should appear after centrifugation along the bottom of the tube on the same side of the cap’s hinge. Carefully discard supernatant while maintaining pellet in the tube;
CRITICAL STEP It is recommended to grade down pipette sizes for increased precision of supernatant removal;
- OPTIONAL STEP For smaller samples, it is recommended to perform procedure alongside a larger tissue sample (50–100 mg) as a reference control for pellet location;
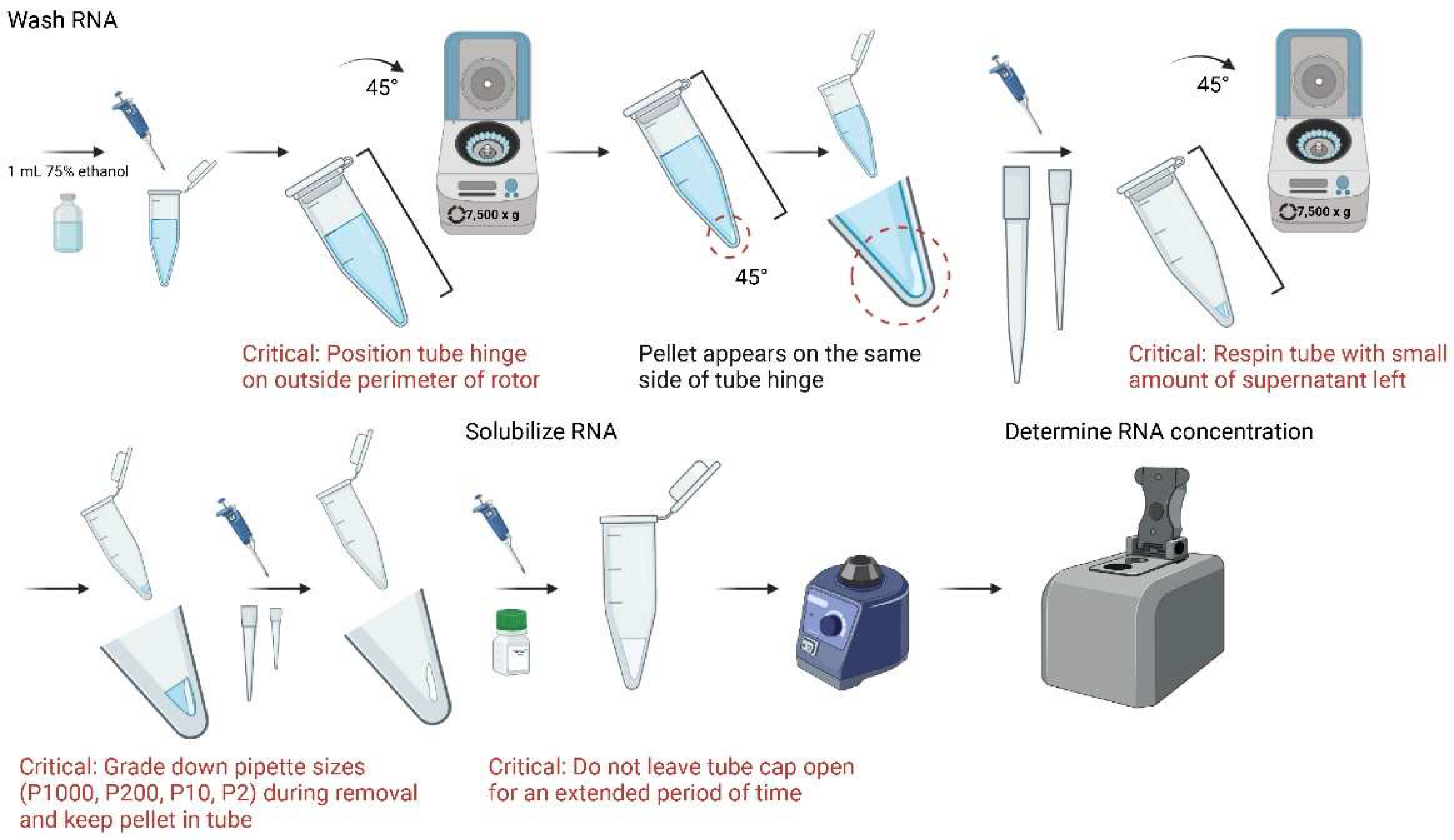
- 10.
- Resuspend pellet with 1 mL of 75% ethanol, vortex, and centrifuge in 4 °C for 5 min at 7500× g;
- 11.
- A thin gel-like pellet should appear after centrifugation in the same 45° location. Discard as much supernatant as possible while maintaining visible sight of pellet. If pellet is not clearly visible, use a larger tissue sample for reference of location;
CRITICAL STEP For optimal purity, leave a small amount of supernatant (~ 20 µL) in the tube and centrifuge at 7500× g in 4 °C for 5 min again. After spin, discard supernatant with P2 or P10 pipette;
CRITICAL STEP It is recommended to grade down pipette sizes for increased precision of supernatant removal and preservation of RNA;
- 12.
- Add 15 µL of nuclease-free H2O to each sample and vortex. Place samples in −80 °C for storage until use;
CRITICAL STEP To prevent RNA degradation and evaporation, do not leave tube cap open for an extended period of time;
- 13.
- Add 1 µL of extracted RNA to NanoDrop One/OneC Microvolume UV-Vis Spectrophotometer to determine concentration. Store at -80C until further use.
4. Expected Results and Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Escartin, C.; Galea, E.; Lakatos, A.; O’Callaghan, J.P.; Petzold, G.C.; Serrano-Pozo, A.; Steinhauser, C.; Volterra, A.; Carmignoto, G.; Agarwal, A.; et al. Reactive astrocyte nomenclature, definitions, and future directions. Nat. Neurosci. 2021, 24, 312–325. [Google Scholar] [CrossRef] [PubMed]
- Norris, G.T.; Kipnis, J. Immune cells and CNS physiology: Microglia and beyond. J. Exp. Med. 2019, 216, 60–70. [Google Scholar] [CrossRef]
- Kwon, H.S.; Koh, S.H. Neuroinflammation in neurodegenerative disorders: The roles of microglia and astrocytes. Transl. Neurodegener. 2020, 9, 42. [Google Scholar] [CrossRef]
- Greenhalgh, A.D.; David, S.; Bennett, F.C. Immune cell regulation of glia during CNS injury and disease. Nat. Rev. Neurosci. 2020, 21, 139–152. [Google Scholar] [CrossRef]
- Vainchtein, I.D.; Molofsky, A.V. Astrocytes and Microglia: In Sickness and in Health. Trends Neurosci. 2020, 43, 144–154. [Google Scholar] [CrossRef] [PubMed]
- Subhramanyam, C.S.; Wang, C.; Hu, Q.; Dheen, S.T. Microglia-mediated neuroinflammation in neurodegenerative diseases. Semin. Cell. Dev. Biol. 2019, 94, 112–120. [Google Scholar] [CrossRef]
- Tomlinson, M.J.; Tomlinson, S.; Yang, X.B.; Kirkham, J. Cell separation: Terminology and practical considerations. J. Tissue Eng. 2013, 4, 2041731412472690. [Google Scholar] [CrossRef] [PubMed]
- Guez-Barber, D.; Fanous, S.; Harvey, B.K.; Zhang, Y.; Lehrmann, E.; Becker, K.G.; Picciotto, M.R.; Hope, B.T. FACS purification of immunolabeled cell types from adult rat brain. J. Neurosci. Methods 2012, 203, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Gross, A.; Schoendube, J.; Zimmermann, S.; Steeb, M.; Zengerle, R.; Koltay, P. Technologies for Single-Cell Isolation. Int. J. Mol. Sci. 2015, 16, 16897–16919. [Google Scholar] [CrossRef] [PubMed]
- Hu, P.; Zhang, W.; Xin, H.; Deng, G. Single Cell Isolation and Analysis. Front. Cell Dev. Biol. 2016, 4, 116. [Google Scholar] [CrossRef]
- Marek, R.; Caruso, M.; Rostami, A.; Grinspan, J.B.; Das Sarma, J. Magnetic cell sorting: A fast and effective method of concurrent isolation of high purity viable astrocytes and microglia from neonatal mouse brain tissue. J. Neurosci. Methods 2008, 175, 108–118. [Google Scholar] [CrossRef]
- Zelenka, L.; Pagelow, D.; Kruger, C.; Seele, J.; Ebner, F.; Rausch, S.; Rohde, M.; Lehnardt, S.; van Vorst, K.; Fulde, M. Novel protocol for the isolation of highly purified neonatal murine microglia and astrocytes. J. Neurosci. Methods 2022, 366, 109420. [Google Scholar] [CrossRef]
- Jungblut, M.; Tiveron, M.C.; Barral, S.; Abrahamsen, B.; Knobel, S.; Pennartz, S.; Schmitz, J.; Perraut, M.; Pfrieger, F.W.; Stoffel, W.; et al. Isolation and characterization of living primary astroglial cells using the new GLAST-specific monoclonal antibody ACSA-1. Glia 2012, 60, 894–907. [Google Scholar] [CrossRef]
- Batiuk, M.Y.; de Vin, F.; Duque, S.I.; Li, C.; Saito, T.; Saido, T.; Fiers, M.; Belgard, T.G.; Holt, M.G. An immunoaffinity-based method for isolating ultrapure adult astrocytes based on ATP1B2 targeting by the ACSA-2 antibody. J. Biol. Chem. 2017, 292, 8874–8891. [Google Scholar] [CrossRef]
- Xue, Y.J.; Cui, S.S.; Guo, D.C.; Liu, J.S.; Yang, M.F.; Kang, H.T.; Jiang, Q.; Qu, L.D. Development of a method for the isolation and culture of astrocytes from the canine cerebral cortex. J. Neurosci. Methods 2022, 370, 109476. [Google Scholar] [CrossRef]
- Guneykaya, D.; Ivanov, A.; Hernandez, D.P.; Haage, V.; Wojtas, B.; Meyer, N.; Maricos, M.; Jordan, P.; Buonfiglioli, A.; Gielniewski, B.; et al. Transcriptional and Translational Differences of Microglia from Male and Female Brains. Cell Rep. 2018, 24, 2773–2783.e6. [Google Scholar] [CrossRef]
- Pan, J.; Wan, J. Methodological comparison of FACS and MACS isolation of enriched microglia and astrocytes from mouse brain. J. Immunol. Methods 2020, 486, 112834. [Google Scholar] [CrossRef]
- Sutermaster, B.A.; Darling, E.M. Considerations for high-yield, high-throughput cell enrichment: Fluorescence versus magnetic sorting. Sci. Rep. 2019, 9, 227. [Google Scholar] [CrossRef]
- Dincman, T.A.; Beare, J.E.; Ohri, S.S.; Whittemore, S.R. Isolation of cortical mouse oligodendrocyte precursor cells. J. Neurosci. Methods 2012, 209, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Holt, L.M.; Olsen, M.L. Novel Applications of Magnetic Cell Sorting to Analyze Cell-Type Specific Gene and Protein Expression in the Central Nervous System. PLoS ONE 2016, 11, e0150290. [Google Scholar] [CrossRef]
- He, Y.; Taylor, N.; Bhattacharya, A. Isolation and Culture of Astrocytes from Postnatal and Adult Mouse Brains. Methods Mol. Biol. 2019, 1938, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Holt, L.M.; Stoyanof, S.T.; Olsen, M.L. Magnetic Cell Sorting for In Vivo and In Vitro Astrocyte, Neuron, and Microglia Analysis. Curr. Protoc. Neurosci. 2019, 88, e71. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.R.S.; Tomiuk, S.; Rüberg, S.; Fekete, R.; Jungblut, M.; Bosio, A. Efficient Isolation of Viable Primary Neural Cells from Adult Murine Brain Tissue Based on a Novel Automated Tissue Dissociation Protocol; Program No. 674.04; 2016 Neuroscience Meeting Planner; Society for Neuroscience: San Diego, CA, USA, 2016. [Google Scholar]
- Bordt, E.A.; Block, C.L.; Petrozziello, T.; Sadri-Vakili, G.; Smith, C.J.; Edlow, A.G.; Bilbo, S.D. Isolation of Microglia from Mouse or Human Tissue. STAR Protoc. 2020, 1, 100035. [Google Scholar] [CrossRef]
- Ocanas, S.R.; Pham, K.D.; Blankenship, H.E.; Machalinski, A.H.; Chucair-Elliott, A.J.; Freeman, W.M. Minimizing the Ex Vivo Confounds of Cell-Isolation Techniques on Transcriptomic and Translatomic Profiles of Purified Microglia. eNeuro 2022, 9, 2. [Google Scholar] [CrossRef] [PubMed]
- Emrich, S.J.; Barbazuk, W.B.; Li, L.; Schnable, P.S. Gene discovery and annotation using LCM-454 transcriptome sequencing. Genome Res. 2007, 17, 69–73. [Google Scholar] [CrossRef]
- Lister, R.; O’Malley, R.C.; Tonti-Filippini, J.; Gregory, B.D.; Berry, C.C.; Millar, A.H.; Ecker, J.R. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 2008, 133, 523–536. [Google Scholar] [CrossRef] [PubMed]
- Stark, R.; Grzelak, M.; Hadfield, J. RNA sequencing: The teenage years. Nat. Rev. Genet. 2019, 20, 631–656. [Google Scholar] [CrossRef]
- Illumina. For All You Seq. Available online: https://emea.illumina.com/techniques/sequencing/ngs-library-prep/library-prep-methods.html (accessed on 16 September 2022).
- Kukurba, K.R.; Montgomery, S.B. RNA Sequencing and Analysis. Cold Spring Harb. Protoc. 2015, 2015, 951–969. [Google Scholar] [CrossRef]
- Zamanian, J.L.; Xu, L.; Foo, L.C.; Nouri, N.; Zhou, L.; Giffard, R.G.; Barres, B.A. Genomic analysis of reactive astrogliosis. J. Neurosci. 2012, 32, 6391–6410. [Google Scholar] [CrossRef] [PubMed]
- Bennett, M.L.; Bennett, F.C.; Liddelow, S.A.; Ajami, B.; Zamanian, J.L.; Fernhoff, N.B.; Mulinyawe, S.B.; Bohlen, C.J.; Adil, A.; Tucker, A.; et al. New tools for studying microglia in the mouse and human CNS. Proc. Natl. Acad. Sci. USA 2016, 113, E1738–E1746. [Google Scholar] [CrossRef]
- Clarke, L.E.; Liddelow, S.A.; Chakraborty, C.; Munch, A.E.; Heiman, M.; Barres, B.A. Normal aging induces A1-like astrocyte reactivity. Proc. Natl. Acad. Sci. USA 2018, 115, E1896–E1905. [Google Scholar] [CrossRef]
- Korthas, H.T.; Main, B.S.; Harvey, A.C.; Buenaventura, R.G.; Wicker, E.; Forcelli, P.A.; Burns, M.P. The Effect of Traumatic Brain Injury on Sleep Architecture and Circadian Rhythms in Mice—A Comparison of High-Frequency Head Impact and Controlled Cortical Injury. Biology 2022, 11, 1031. [Google Scholar] [CrossRef]



| Name | Source | Identifier | Location |
|---|---|---|---|
| Stock Solutions and Kits Phosphate Buffered Saline 10X Solution Adult Brain Dissociation Kit, mouse and rat | |||
| Fisher Scientific | BP399-20 | Fair Lawn, NJ, USA | |
| Miltenyi Biotec B.V. & Co. KG | 130-107-677 | Bergisch Gladbach, DE | |
| |||
| CD11b (Microglia) MicroBeads, mouse/human (1 mL) Anti-ACSA-2 MicroBead Kit mouse (2 × 1 mL)
| Miltenyi Biotec B.V. & Co. KG | 130-093-634 | Bergisch Gladbach, DE |
Miltenyi Biotec B.V. & Co. KG | 130-097-678 | Bergisch Gladbach, DE | |
| AstroMACS Separation Buffer (100 mL) Dulbecco’s Phosphate Buffered Saline (with calcium) MACS BSA Stock Solution TRIzol Reagent Chloroform Isopropanol Ethanol, Anhydrous Nuclease-Free Water (DEPC Treated) | Miltenyi Biotec B.V. & Co. KG Thermo Fischer Scientific | 130-117-336 14040117 | Bergisch Gladbach, DE Waltham, MA, USA |
| Miltenyi Biotec B.V. & Co. KG Invitrogen Fisher Scientific Fisher Scientific Fisher Scientific Invitrogen | 130-117-336 15596026 C298-500 A416-4 A405P-4 AM9906 | Bergisch Gladbach, DE Waltham, MA, USA Fair Lawn, NJ, USA Fair Lawn, NJ, USA Fair Lawn, NJ, USA Waltham, MA, USA |
| Name | Source | Identifier | Location |
|---|---|---|---|
| Tools 21 G x 1” needle Graefe forceps Light operating scissors Halsted mosquito forceps Operating scissors 6.5” Micro dissecting scissors 4.5” Scalpel handle Scalpel blades Equipment Fisherbrand Variable-Flow Peristaltic Pump Dissection cold plate OctoMACS Separator attached to MultiStand MS Columns 5 mL Tubes for MS Columns gentleMACS C Tubes gentleMACS Octo Dissociator with Heaters MACS SmartStrainers 70 µm 50 mL Conical Screw Cap Tubes, Grenier Bio-One 15 mL Conical Screw Cap Tubes, Grenier Bio-One 5 mL Falcon Round Bottom Polystyrene Test Tube Centrifuge (refrigerated) Vortex Mixer Sonicator 1.5 mL Microcentrifuge Tubes (clear) NanoDrop One Microvolume Spectrophotometer | Covetrus Roboz Surgical Store Roboz Surgical Store Roboz Surgical Store Roboz Surgical Store Roboz Surgical Store Roboz Surgical Store Roboz Surgical Store Fisher Scientific Cell Path Miltenyi Biotec B.V. & Co. KG Miltenyi Biotec B.V. & Co. KG Miltenyi Biotec B.V. & Co. KG Miltenyi Biotec B.V. & Co. KG Miltenyi Biotec B.V. & Co. KG Miltenyi Biotec B.V. & Co. KG USA Scientific, Inc USA Scientific, Inc Corning Eppendorf, model 5810R Labnet International, Inc. QSonica, LLC USA Scientific, Inc Thermo Fisher Scientific | 060760 RS-5138 RS-6752 RS-7111L RS-6824 RS-5912 RS-9843 RS-9801-11 13-876-2 JRI-0100-00A 130-042-109 130-042-201 130-091-598 130-093-237 130-095-427 130-110-916 5622-7270 5618-8261 352054 022627040 S0100-VX100 Q55-100 1615-5510 ND-ONE-W | Dublin, OH, USA Gaithersburg, MD, USA Gaithersburg, MD, USA Gaithersburg, MD, USA Gaithersburg, MD, USA Gaithersburg, MD, USA Gaithersburg, MD, USA Gaithersburg, MD, USA Fair Lawn, NJ, USA Newtown, UK Bergisch Gladbach, DE Bergisch Gladbach, DE Bergisch Gladbach, DE Bergisch Gladbach, DE Bergisch Gladbach, DE Bergisch Gladbach, DE Ocala, FL, USA Ocala, FL, USA Glendale, AZ, USA Enfield, CT, USA Woodridge, NJ, USA Newtown, CT, USA Ocala, FL, USA Waltham, MA, USA |
| Name | Recipe |
|---|---|
| 1 × PBS Enzyme Mix 1 Enzyme Mix 2 D-PBS PB Buffer 1x Red Blood Cell Removal Solution | 1:10 dilution of Phosphate Buffered Saline 10X Solution with dH2O 50 µL Enzyme P and 1900 µL Buffer Z (prepare fresh) 20 µL Buffer Y and 10 µL Enzyme A (prepare fresh) 1:10 dilution of Dulbecco’s Phosphate Buffered Saline (with calcium) with dH2O and cool to 4 °C 1:20 dilution of MACS BSA Stock Solution with D-PBS and cool to 4 °C (or prepare a solution with pH of 7.2 with phosphate-buffered saline (PBS), 0.5% bovine serum albumin (BSA), and 2 mM EDTA) 1:10 dilution of Red Blood Cell Removal Solution (10X) with cold dH2O (prepare fresh) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Buenaventura, R.G.; Harvey, A.C.; Burns, M.P.; Main, B.S. Sequential Isolation of Microglia and Astrocytes from Young and Aged Adult Mouse Brains for Downstream Transcriptomic Analysis. Methods Protoc. 2022, 5, 77. https://doi.org/10.3390/mps5050077
Buenaventura RG, Harvey AC, Burns MP, Main BS. Sequential Isolation of Microglia and Astrocytes from Young and Aged Adult Mouse Brains for Downstream Transcriptomic Analysis. Methods and Protocols. 2022; 5(5):77. https://doi.org/10.3390/mps5050077
Chicago/Turabian StyleBuenaventura, Ruchelle G., Alex C. Harvey, Mark P. Burns, and Bevan S. Main. 2022. "Sequential Isolation of Microglia and Astrocytes from Young and Aged Adult Mouse Brains for Downstream Transcriptomic Analysis" Methods and Protocols 5, no. 5: 77. https://doi.org/10.3390/mps5050077
APA StyleBuenaventura, R. G., Harvey, A. C., Burns, M. P., & Main, B. S. (2022). Sequential Isolation of Microglia and Astrocytes from Young and Aged Adult Mouse Brains for Downstream Transcriptomic Analysis. Methods and Protocols, 5(5), 77. https://doi.org/10.3390/mps5050077








