Guidelines to Analyze Preclinical Studies Using Perinatal Derivatives
Abstract
:1. Introduction
2. Materials and Methods
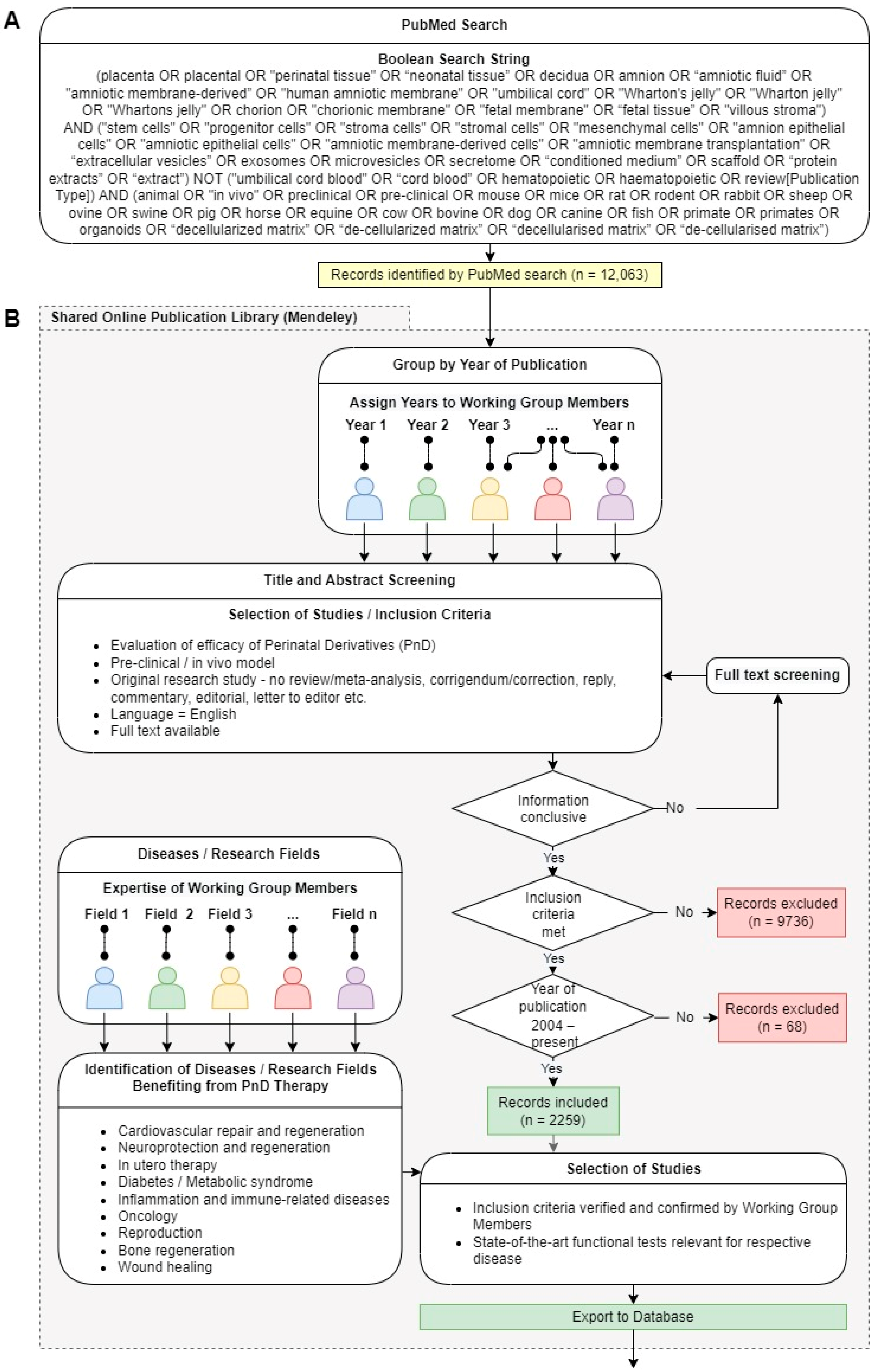
2.1. Search Strategy and Data Mining
2.2. Selection of Studies and Inclusion Criteria
2.3. Data Management
2.4. Data Extraction
2.5. Data Synthesis
3. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Silini, A.R.; Di Pietro, R.; Lang-Olip, I.; Alviano, F.; Banerjee, A.; Basile, M.; Borutinskaite, V.; Eissner, G.; Gellhaus, A.; Giebel, B.; et al. Perinatal Derivatives: Where Do We Stand? A Roadmap of the Human Placenta and Consensus for Tissue and Cell Nomenclature. Front. Bioeng. Biotechnol. 2020, 8, 1438. [Google Scholar] [CrossRef] [PubMed]
- Aziz, J.; Liao, G.; Adams, Z.; Rizk, M.; Shorr, R.; Allan, D.S. Systematic Review of Controlled Clinical Studies Using Umbilical Cord Blood for Regenerative Therapy: Identifying Barriers to Assessing Efficacy. Cytotherapy 2019, 21, 1112–1121. [Google Scholar] [CrossRef] [PubMed]
- Lange-Consiglio, A.; Capra, E.; Herrera, V.; Lang-Olip, I.; Ponsaerts, P.; Cremonesi, F. Application of Perinatal Derivatives in Ovarian Diseases. Front. Bioeng. Biotechnol. 2022, 10, 811875. [Google Scholar] [CrossRef] [PubMed]
- Norte-Muñoz, M.; Botelho, M.F.; Schoeberlein, A.; Chaves, J.; Neto Murta, J.; Ponsaerts, P.; Agudo-Barriuso, M.; Costa, E. Insights and Future Directions for the Application of Perinatal Derivatives in Eye Diseases: A Critical Review of Preclinical and Clinical Studies. Front. Bioeng. Biotechnol. 2022, 10, 2047. [Google Scholar] [CrossRef] [PubMed]
- Pichlsberger, M.; Jerman, U.D.; Obradović, H.; Tratnjek, L.; Macedo, A.S.; Mendes, F.; Fonte, P.; Hoegler, A.; Sundl, M.; Fuchs, J.; et al. Systematic Review of the Application of Perinatal Derivatives in Animal Models on Cutaneous Wound Healing. Front. Bioeng. Biotechnol. 2021, 9, 742858. [Google Scholar] [CrossRef] [PubMed]
- Teixo, R.; Pires, A.S.; Pereira, E.; Serambeque, B.; Marques, I.A.; Laranjo, M.; Mojsilović, S.; Gramignoli, R.; Ponsaerts, P.; Schoeberlein, A.; et al. Application of Perinatal Derivatives on Oncological Preclinical Models: A Review of Animal Studies. Int. J. Mol. Sci. 2022, 23, 8570. [Google Scholar] [CrossRef] [PubMed]
- Russell, W.M.S.; Burch, R.L. The Principles of Humane Experimental Technique; Methuen & Co. Limited: London, UK, 1959. [Google Scholar]
- McKenzie, J.E.; Brennan, S.E.; Ryan, R.E.; Thomson, H.J.; Johnston, R.V.; Thomas, J. Defining the Criteria for Including Studies and How They Will Be Grouped for the Synthesis. In Cochrane Handbook for Systematic Reviews of Interventions Version 6.2 (Updated February 2021); Higgins, J.P.T., Thomas, J., Chandler, J., Cumpston, M., Li, T., Page, M.J., Welch, V.A., Eds.; Cochrane: London, UK, 2021. [Google Scholar]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef] [PubMed]
- Page, M.J.; Moher, D.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. PRISMA 2020 Explanation and Elaboration: Updated Guidance and Exemplars for Reporting Systematic Reviews. BMJ 2021, 372, n160. [Google Scholar] [CrossRef] [PubMed]
- PubMed. Search String (Placenta OR Placental OR “Perinatal Tissue” OR “Neonatal Tissue” OR Decidua OR Amnion OR “Amniotic Fluid” OR “Amniotic Membrane-Derived” OR “Human”; United States National Library of Medicine, National Institutes of Health: Bethesda, MA, USA, 2019. [Google Scholar]
- Clarivate. Cites Journal Citation Report; Clarivate: Boston, MA, USA, 2020. [Google Scholar]
- Hutchins, B.I.; Yuan, X.; Anderson, J.M.; Santangelo, G.M. Relative Citation Ratio (RCR): A New Metric That Uses Citation Rates to Measure Influence at the Article Level. PLoS Biol. 2016, 14, e1002541. [Google Scholar] [CrossRef] [PubMed]
- Hutchins, B.I.; Hoppe, T.A.; Meseroll, R.A.; Anderson, J.M.; Santangelo, G.M. Additional Support for RCR: A Validated Article-Level Measure of Scientific Influence. PLoS Biol. 2017, 15, e2003552. [Google Scholar] [CrossRef] [PubMed]
- Hutchins, B.I.; Davis, M.T.; Meseroll, R.A.; Santangelo, G.M. Predicting Translational Progress in Biomedical Research. PLoS Biol. 2019, 17, e3000416. [Google Scholar] [CrossRef] [PubMed]
- Hutchins, B.I.; Baker, K.L.; Davis, M.T.; Diwersy, M.A.; Haque, E.; Harriman, R.M.; Hoppe, T.A.; Leicht, S.A.; Meyer, P.; Santangelo, G.M. The NIH Open Citation Collection: A Public Access, Broad Coverage Resource. PLoS Biol. 2019, 17, e3000385. [Google Scholar] [CrossRef] [PubMed]
- Hook, D.W.; Porter, S.J.; Herzog, C. Dimensions: Building Context for Search and Evaluation. Front. Res. Metr. Anal. 2018, 3, 23. [Google Scholar] [CrossRef]
- Dimensions. Digital Science & Research Solutions Inc.: London, UK. 2020. Available online: https://app.dimensions.ai/discover/publication (accessed on 11 December 2020).
- Rohatgi, A. WebPlotDigitizer (Version 4.4) [Computer software]. 2020. Available online: http://arohatgi.info/WebPlotDigitizer (accessed on 11 December 2020).
- Hooijmans, C.R.; Rovers, M.M.; de Vries, R.B.; Leenaars, M.; Ritskes-Hoitinga, M.; Langendam, M.W. SYRCLE’s Risk of Bias Tool for Animal Studies. BMC Med. Res. Methodol. 2014, 14, 43. [Google Scholar] [CrossRef] [PubMed]
- Sterne, J.A.C.; Savović, J.; Page, M.J.; Elbers, R.G.; Blencowe, N.S.; Boutron, I.; Cates, C.J.; Cheng, H.Y.; Corbett, M.S.; Eldridge, S.M.; et al. RoB 2: A Revised Tool for Assessing Risk of Bias in Randomised Trials. BMJ 2019, 366, l4898. [Google Scholar] [CrossRef] [PubMed]
- Sterne, J.A.; Hernán, M.A.; Reeves, B.C.; Savović, J.; Berkman, N.D.; Viswanathan, M.; Henry, D.; Altman, D.G.; Ansari, M.T.; Boutron, I.; et al. ROBINS-I: A Tool for Assessing Risk of Bias in Non-Randomised Studies of Interventions. BMJ 2016, 355, i4919. [Google Scholar] [CrossRef] [PubMed]
- Kassem, D.H.; Kamal, M.M. Therapeutic Efficacy of Umbilical Cord-Derived Stem Cells for Diabetes Mellitus: A Meta-Analysis Study. Stem Cell Res. Ther. 2020, 11, 484. [Google Scholar] [CrossRef] [PubMed]
- Maltais-Bilodeau, C.; Henckel, E.; Cobey, K.D.; Ahmadzai, N.; Skidmore, B.; Ferretti, E.; Thébaud, B. Efficacy of Mesenchymal Stromal Cells in Preclinical Models of Necrotizing Enterocolitis: A Systematic Review Protocol. Res. Sq. 2021, preprint. [Google Scholar] [CrossRef]
- Percie du Sert, N.; Hurst, V.; Ahluwalia, A.; Alam, S.; Avey, M.T.; Baker, M.; Browne, W.J.; Clark, A.; Cuthill, I.C.; Dirnagl, U.; et al. The ARRIVE Guidelines 2.0: Updated Guidelines for Reporting Animal Research. PLoS Biol. 2020, 18, e3000410. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pires, A.S.; Bollini, S.; Botelho, M.F.; Lang-Olip, I.; Ponsaerts, P.; Balbi, C.; Lange-Consiglio, A.; Fénelon, M.; Mojsilović, S.; Berishvili, E.; et al. Guidelines to Analyze Preclinical Studies Using Perinatal Derivatives. Methods Protoc. 2023, 6, 45. https://doi.org/10.3390/mps6030045
Pires AS, Bollini S, Botelho MF, Lang-Olip I, Ponsaerts P, Balbi C, Lange-Consiglio A, Fénelon M, Mojsilović S, Berishvili E, et al. Guidelines to Analyze Preclinical Studies Using Perinatal Derivatives. Methods and Protocols. 2023; 6(3):45. https://doi.org/10.3390/mps6030045
Chicago/Turabian StylePires, Ana Salomé, Sveva Bollini, Maria Filomena Botelho, Ingrid Lang-Olip, Peter Ponsaerts, Carolina Balbi, Anna Lange-Consiglio, Mathilde Fénelon, Slavko Mojsilović, Ekaterine Berishvili, and et al. 2023. "Guidelines to Analyze Preclinical Studies Using Perinatal Derivatives" Methods and Protocols 6, no. 3: 45. https://doi.org/10.3390/mps6030045
APA StylePires, A. S., Bollini, S., Botelho, M. F., Lang-Olip, I., Ponsaerts, P., Balbi, C., Lange-Consiglio, A., Fénelon, M., Mojsilović, S., Berishvili, E., Cremonesi, F., Gazouli, M., Bugarski, D., Gellhaus, A., Kerdjoudj, H., & Schoeberlein, A. (2023). Guidelines to Analyze Preclinical Studies Using Perinatal Derivatives. Methods and Protocols, 6(3), 45. https://doi.org/10.3390/mps6030045












