Quantitative Image Analysis of Axonal Morphology in In Vivo Model
Abstract
:1. Introduction
2. Materials and Methods
2.1. Zebrafish Line and Breeding
2.2. Embryo Manipulation and Immnostaining
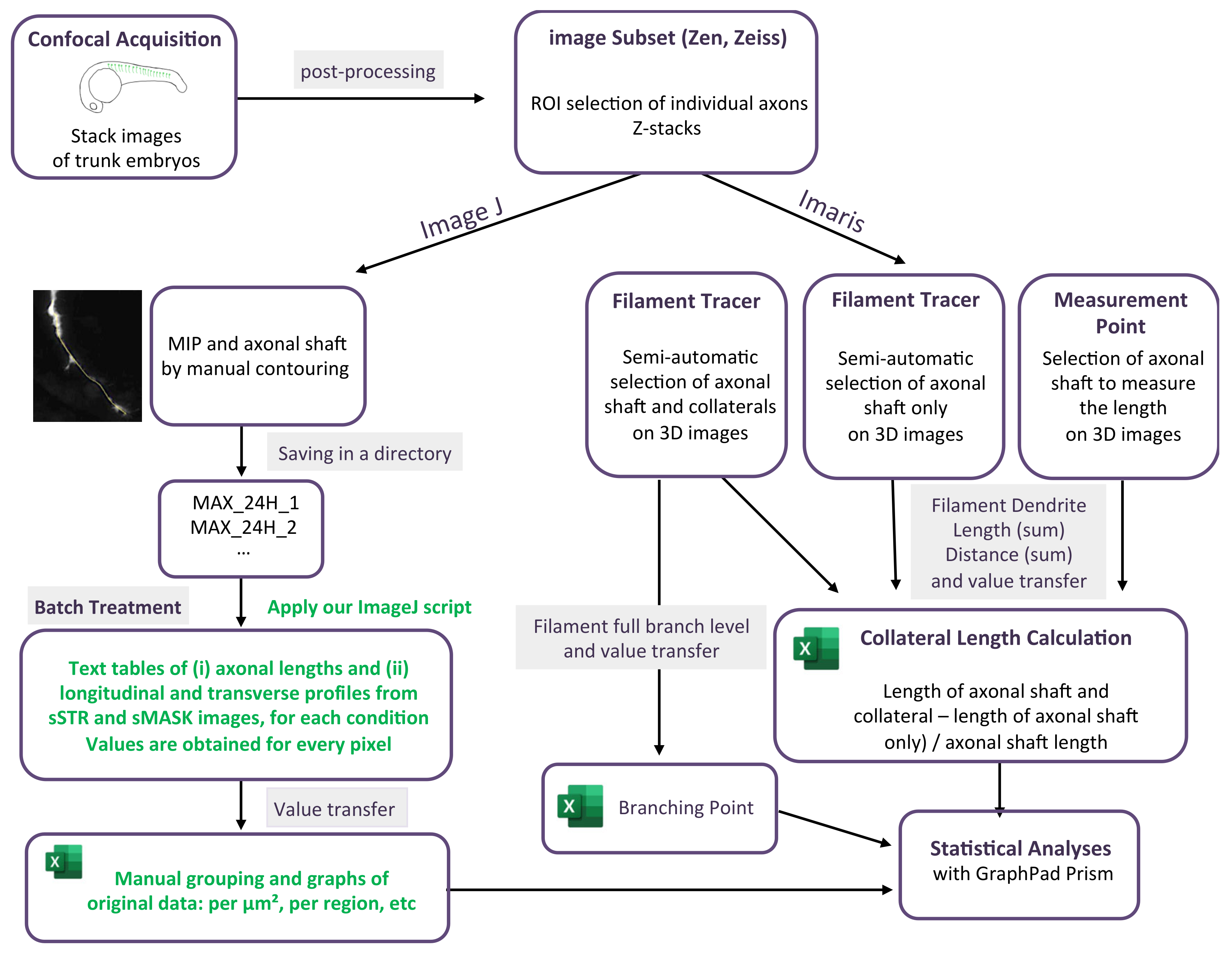
2.3. Confocal Acquisition and Image Post-Processing
2.4. ImageJ Analysis
2.5. Raw Data Processing
2.6. Imaris Analysis
2.7. Statistical Analysis
3. Results
3.1. Model of the Study
3.2. Branching Quantification Using a Home-Made Script with ImageJ
3.3. Branching Quantification Using 3D Analysis Imaris Software
3.4. Length Measurement of Growing Motor Axons
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Milner, L.D.; Rafuse, V.F.; Landmesser, L.T. Selective Fasciculation and Divergent Pathfinding Decisions of Embryonic Chick Motor Axons Projecting to Fast and Slow Muscle Regions. J. Neurosci. 1998, 18, 3297–3313. [Google Scholar] [CrossRef] [PubMed]
- Hao, L.T.; Duy, P.Q.; Jontes, J.D.; Wolman, M.; Granato, M.; Beattie, C.E. Temporal Requirement for SMN in Motoneuron Development. Hum. Mol. Genet. 2013, 22, 2612–2625. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Walder-Christensen, K.K.; Kim, N.; Wu, D.; Lorenzo, D.N.; Badea, A.; Jiang, Y.-H.; Yin, H.H.; Wetsel, W.C.; Bennett, V. ANK2 Autism Mutation Targeting Giant Ankyrin-B Promotes Axon Branching and Ectopic Connectivity. Proc. Natl. Acad. Sci. USA 2019, 116, 15262–15271. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.; Wang, X.; Zhu, C.; Dong, X.; Zhang, Q.; Wang, X.; Duan, X.; Qian, F.; Shi, Y.; Gao, Y.; et al. Insm1a Regulates Motor Neuron Development in Zebrafish. Front. Mol. Neurosci. 2017, 10, 274. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Suo, G.; Liu, D.; Li, H.; Wu, Y.; Ni, H. Nexmifa Regulates Axon Morphogenesis in Motor Neurons in Zebrafish. Front. Mol. Neurosci. 2022, 15, 848257. [Google Scholar] [CrossRef] [PubMed]
- Zschätzsch, M.; Oliva, C.; Langen, M.; De Geest, N.; Özel, M.N.; Williamson, W.R.; Lemon, W.C.; Soldano, A.; Munck, S.; Hiesinger, P.R.; et al. Regulation of Branching Dynamics by Axon-Intrinsic Asymmetries in Tyrosine Kinase Receptor Signaling. eLife 2014, 3, e01699. [Google Scholar] [CrossRef] [PubMed]
- Sainath, R.; Granato, M. Plexin A3 and Turnout Regulate Motor Axonal Branch Morphogenesis in Zebrafish. PLoS ONE 2013, 8, e54071. [Google Scholar] [CrossRef] [PubMed]
- Tymanskyj, S.R.; Yang, B.; Falnikar, A.; Lepore, A.C.; Ma, L. MAP7 Regulates Axon Collateral Branch Development in Dorsal Root Ganglion Neurons. J. Neurosci. 2017, 37, 1648–1661. [Google Scholar] [CrossRef] [PubMed]
- Beattie, C.E. Control of Motor Axon Guidance in the Zebrafish Embryo. Brain Res. Bull. 2000, 53, 489–500. [Google Scholar] [CrossRef] [PubMed]
- Flanagan-Steet, H.; Fox, M.A.; Meyer, D.; Sanes, J.R. Neuromuscular Synapses Can Form in Vivo by Incorporation of Initially Aneural Postsynaptic Specializations. Development 2005, 132, 4471–4481. [Google Scholar] [CrossRef] [PubMed]
- Kimmel, C.B.; Ballard, W.W.; Kimmel, S.R.; Ullmann, B.; Schilling, T.F. Stages of Embryonic Development of the Zebrafish. Dev. Dyn. 1995, 203, 253–310. [Google Scholar] [CrossRef] [PubMed]
- Guillon, E.; Bretaud, S.; Ruggiero, F. Slow Muscle Precursors Lay Down a Collagen XV Matrix Fingerprint to Guide Motor Axon Navigation. J. Neurosci. 2016, 36, 2663–2676. [Google Scholar] [CrossRef] [PubMed]
- Myers, P.; Eisen, J.; Westerfield, M. Development and Axonal Outgrowth of Identified Motoneurons in the Zebrafish. J. Neurosci. 1986, 6, 2278–2289. [Google Scholar] [CrossRef] [PubMed]
- Eisen, J.S.; Myers, P.Z.; Westerfield, M. Pathway Selection by Growth Cones of Identified Motoneurones in Live Zebra Fish Embryos. Nature 1986, 320, 269–271. [Google Scholar] [CrossRef] [PubMed]
- Ingham, P.W.; Kim, H.R. Hedgehog Signalling and the Specification of Muscle Cell Identity in the Zebrafish Embryo. Exp. Cell Res. 2005, 306, 336–342. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.R.; Ingham, P.W. The Extracellular Matrix Protein TGFBI Promotes Myofibril Bundling and Muscle Fibre Growth in the Zebrafish Embryo. Dev. Dyn. 2009, 238, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Popko, J.; Fernandes, A.; Brites, D.; Lanier, L.M. Automated Analysis of NeuronJ Tracing Data. Cytom. Pt A 2009, 75A, 371–376. [Google Scholar] [CrossRef] [PubMed]
- Longair, M.H.; Baker, D.A.; Armstrong, J.D. Simple Neurite Tracer: Open Source Software for Reconstruction, Visualization and Analysis of Neuronal Processes. Bioinformatics 2011, 27, 2453–2454. [Google Scholar] [CrossRef] [PubMed]
- Boulan, B.; Beghin, A.; Ravanello, C.; Deloulme, J.-C.; Gory-Fauré, S.; Andrieux, A.; Brocard, J.; Denarier, E. AutoNeuriteJ: An ImageJ Plugin for Measurement and Classification of Neuritic Extensions. PLoS ONE 2020, 15, e0234529. [Google Scholar] [CrossRef] [PubMed]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nemoz-Billet, L.; Brocard, J.; Ruggiero, F.; Bretaud, S. Quantitative Image Analysis of Axonal Morphology in In Vivo Model. Methods Protoc. 2023, 6, 116. https://doi.org/10.3390/mps6060116
Nemoz-Billet L, Brocard J, Ruggiero F, Bretaud S. Quantitative Image Analysis of Axonal Morphology in In Vivo Model. Methods and Protocols. 2023; 6(6):116. https://doi.org/10.3390/mps6060116
Chicago/Turabian StyleNemoz-Billet, Laurie, Jacques Brocard, Florence Ruggiero, and Sandrine Bretaud. 2023. "Quantitative Image Analysis of Axonal Morphology in In Vivo Model" Methods and Protocols 6, no. 6: 116. https://doi.org/10.3390/mps6060116
APA StyleNemoz-Billet, L., Brocard, J., Ruggiero, F., & Bretaud, S. (2023). Quantitative Image Analysis of Axonal Morphology in In Vivo Model. Methods and Protocols, 6(6), 116. https://doi.org/10.3390/mps6060116





