Making the Most of Lateral Flow Immunochromatographic Tests: An Efficient Protocol to Recover DNA
Abstract
1. Introduction
2. Experimental Design
2.1. Materials
- Fresh human saliva samples directly collected by spitting in an Eppendorf tube. Based on our current experiments, other body fluids like semen and blood can be used.
- Sterile, wood-shaft, cotton-tipped applicators from McKesson (Richmond, Virginia).
- SERATEC® Amylase test (SERATEC®, Göttingen, Germany).
- SERATEC® extraction buffer (SERATEC®, Göttingen, Germany). Components: 8.0 g of NaCl; 0.2 g of KCl; 1.44 g of Na2HPO4·2H2O; 0.24 g of KH2PO4; 0.1 mL of 10 wt% NaN3; pH 7.4 in 1 L of distilled water.
- DNeasy Blood & Tissue kit (Qiagen®, Hilden, Germany).
- Molecular biology-grade ethanol, 99% (Sigma-Milipore, Saint Louis, MO, USA).
- Qubit dsDNA HS (high-sensitivity) Assay Kit (Life Technologies, Carlsbad, CA, USA).
- Thin-wall, clear, 0.5 mL PCR tubes (Genessee Scientific, Morrisville, NC, USA).
- 96-well plates (Thermo Fisher Scientific, Waltham, MA, USA).
- PowerQuant System (Promega Corporation, Madison, WI, USA).
- Promega PowerPlex Fusion 6C System (Promega Corporation, Madison, WI, USA).
- Hi-Di formamide (Thermo Fisher Scientific, Waltham, MA, USA).
2.2. Equipment
- Biological safety cabinet (Thermo Fisher Scientific, Waltham, MA, USA).
- Vortex (Thermo Fisher Scientific, Waltham, MA, USA).
- Thermal mixer with blocks (Thermo Fisher Scientific, Waltham, MA, USA).
- Centrifuge (Thermo Fisher Scientific, Waltham, MA, USA).
- Qubit Fluorometer 3.0 (Thermo Fisher Scientific, Waltham, MA, USA).
- QuantStudio 5 (Thermo Fisher Scientific, Waltham, MA, USA).
- ProFlex PCR system (Thermo Fisher Scientific, Waltham, MA, USA).
- SeqStudio (Thermo Fisher Scientific, Waltham, MA, USA).
2.3. Software
- Promega PowerQuant Analysis Software Version: 4.8.0.0 (Promega Corporation, Madison, WI, USA).
- Microsatellite Analysis Software on Thermo Fisher Cloud (Thermo Fisher Scientific, Waltham, MA, USA).
3. Procedure
3.1. DNA Extraction from the Swab and Extraction Buffer (Figure 1)
- Keep the swab on the Eppendorf tube. Complete to 400 µL with SERATEC® extraction buffer.
- From the DNeasy Blood & Tissue kit, add to the buffer 20 µL of Proteinase K and 400 µL of Reagent AL. Vortex for 15 s.
- Incubate and shake at 56 °C for 10 min on the thermal mixer with blocks.
- Add 400 µL of Ethanol. Vortex for 15 s.
- Transfer 700 µL of the mixture to the DNeasy Blood and Tissue kit column. Centrifuge at 6000× g (8000 rpm) for 1 min. Discard the flow-through liquid. Repeat this step with the rest of the mixture. The DNA is now attached to the column.
- Add to the column 500 µL of Buffer AW1. Centrifuge at 6000× g (8000 rpm) for 1 min. Discard the flow-through liquid.
- Add to the column 500 µL of Buffer AW2. Centrifuge at 20,000× g (14,000 rpm) for 3 min. Discard the flow-through liquid.
- In order to evaporate all ethanol, centrifuge the column at 20,000× g (14,000 rpm) for 1 min.
- After cleaning the DNA, perform the elution/release of DNA from the column by adding to the column 50 µL of buffer AE (or nuclease-free water). Incubate for 1 min at RT. Then, centrifuge at 6000× g (8000 rpm) for 1 min.
- The DNA is released.

3.2. Quantification of Total DNA with Qubit dsDNA HS
- Bring all the kit components and samples to room temperature (RT).
- Prepare a Working Solution (WS) composed of Qubit® dsDNA HS Buffer and Qubit® dsDNA HS Reagent (concentrated 200×), considering that each sample will be diluted in approximately 200 µL of this solution.
- The kit includes two DNA standards, Standard #1, 0 ng/µL DNA in TE buffer, and Standard #2, 10 ng/µL DNA in TE buffer.
- Mix 190 µL of the WS with 10 µL of each standard in thin-wall, clear, 0.5-well tubes.
- For the samples, although the manufacturer’s protocol indicates that it is possible to use as little as 1 µL of the sample (between 1 and 10 µL), from our experience, 2 µL works better. Thus, mix 2 µL of each sample with 198 µL of the WS in thin-wall, clear, 0.5-well tubes.
- Mix all the tubes by vortexing.
- Incubate at room temperature for 2 min.
- Read the sample on the Qubit Fluorometer 3.0, selecting dsDNA HS. Clean the tubes before introducing them on the instrument.
- After the reading, indicate how much sample you used to obtain the final concentration of your sample.
3.3. Specific Human Quantification with the PowerQuant System
- The day before the quantification, defrost the standard (PowerQuant male gDNA standard) and the dilution buffer and keep them at 4 °C.
- Prepare serial dilutions of the PowerQuant male gDNA standard with the PowerQuant dilution buffer, starting with the original 50 ng/µL, 2 ng/µL, 0.08 ng/µL, and 0.0032 ng/µL (Table 1).
- 3.
- Prepare the Master Mix considering the number of samples and the standards. Both samples and standards should be tested per duplicate. The Master Mix includes the PowerQuant Master Mix (concentrated 2×), the PowerQuant Primer/Probe/IPC Mix (concentrated 20×), and water amplification grade, totaling a volume of 18 µL, as depicted in Table 2.
- 4.
- Vortex the Master Mix and distribute 18 µL on a 96-well plate.
- 5.
- Add 2 µL of the standards/samples to the plate.
- 6.
- Seal the plate and centrifuge briefly.
- 7.
- Load the plate onto the QuantStudio.
- 8.
- Introduce the appropriate parameters to the QuantStudio: block type, 96-well 0.2 mL block; experiment type, standard curve; chemistry, TaqMan Reagents; run mode, standard.
- 9.
- In Experiment Method, enter 20 uL for “Volume” and set up the parameters for the thermocycler: Hold Stage, 98 °C 2 min; PCR stage: Step 1: 98 °C 15 s. and Step 2: 62 °C 35 s for 39 cycles. The ramp rate for all three steps is 2.44 °C/s.
- 10.
- In “Create a Run Template”, assign the following targets to all samples: Autosomal, with a reporter PQ_FAM and quencher NFQ-MGB; Y, with a reporter PQ_CFG540 and quencher NFQ-MGB; Degradation, with a reporter PQ_Q670 and quencher NFQ-MGB; and IPC, with a reporter PQ_TMR and quencher NFQ-MGB. The Task for autosomal, Y, and degradation targets should be “S”, and for IPC, it should be “U”.
- 11.
- In the Samples section, add the standards’ and the samples’ names and select the appropriate wells.
- 12.
- In the Quick Setup tab, in Plate Attributes, select PQ_CXR as the passive reference.
- 13.
- In the CT Settings tab, select each target and uncheck the Default Setting box and the Automatic Threshold box. Enter the following values for the Threshold: Autosomal, 0.2; Degradation, 0.2; IPC, 0.03; Y, 0.2. Select Apply.
- 14.
- When all the parameters are set up, click on Next, and then Start Run.
- 15.
- Data analyses will be carried out with the PowerQuant Analysis Software.
- 16.
- For the standard curve, the minimum acceptable value for R-Squared for the three targets is 0.99; for the Slope, it is −3.6, and the maximum acceptable value is −3.1. Including the Y-intercept is optional.
- 17.
- Establishing a Threshold value and Sample Assessment Message should be customized based on internal validation studies conducted by the lab. The Inhibitor threshold indicates the minimum IPC shift value at which you may expect to encounter inhibition. The Male/Female threshold specifies the minimum Autosomal/Y ratio indicative of potential male/female mixture. The Degradation threshold specifies the minimum autosomal/degradation ratio indicative of a potentially degraded DNA sample.
- 18.
- Once the csv file from QuantStudio is loaded onto the PowerQuant Software, the data will be analyzed, according to the set-up parameters. The Quantity Map will give the concentration of autosomal, Y, and IPC in ng/µL. The Ratio Map will give the Sample Assessment parameters’ values. For proceeding to the next step of STR amplification, the concentration of autosomal DNA in ng/µL is chosen.
3.4. Amplification of STRs Using the PowerPlex Fusion 6C System
- Centrifuge pre-amplification component tubes briefly, and vortex for 15 s before each use.
- From the samples, between 0.5 and 1 ng DNA can be used. A positive control is included, the 2800M Control DNA, from this 1 ng of DNA should be amplified.
- Prepare the Master Mix considering the number of samples, positive and negative controls (with Water, Amplification Grade). Add 1 or 2 reactions to this number. This Master Mix consists of 5 µL of PowerPlex Fusion 6C 5× Master Mix, 5 µL PowerPlex Fusion 6C 5× Primer Pair Mix, and Water, Amplification Grade to a final volume of 25 uL. Vortex this mix for 5–10 s and then add to each tube.
- Add the template DNA (up to 15 µL) considering that the final volume is 25 µL.
- Briefly centrifuge the tubes.
- Set up the tubes in the Proflex and program the following parameters: 1 cycle of 96 °C 1 min; 29 cycles of 96 °C 5 s. and 60 °C 1 min; 1 cycle 60 °C 10 min; Hold 4 °C.
- After the reaction, the samples can be stored at −20 °C or directly proceed with fragment analysis.
3.5. Fragment Analysis on the SeqStudio
- Centrifuge post-amplification component tubes briefly, and vortex for 15 s before each use.
- Apart from the previous samples, two allelic ladders are included in the fragment analysis to be able to interpret the bins.
- Prepare a loading cocktail considering the number of samples, positive and negative controls, and two allelic ladders. Add 1 or 2 reactions to this number. This cocktail consists of 0.5 µL of WEN ILS 500 and 9.5 µL of Hi-Di formamide. Vortex the cocktail and pipet 10 µL in each tube.
- Add 1 µL of amplified sample or allelic ladder in the tubes. Centrifuge briefly.
- Denature samples at 95 °C for 3 min in the thermoblock, and chill on a freezer block for 3 min.
- Transfer the samples to the 96-well plate and cover the wells with appropriate septa.
- Introduce the plate into the SeqStudio.
- Create a new plate in the instrument, selecting Fragment Analysis as the application.
- Select the appropriate wells and include as a Run Module “FragAnalysis”; Size Standard “Promega ILS 500”; and Dye Set “Promega 6C”. Include the sample name and indicate if they are a sample, positive control, negative control, or allelic ladder.
- The running parameters are the following: 7 s injection time; 1200 volts injection voltage; 1440 s run time; 9000 volts run volt.
- Analyze the data in the Microsatellite Analysis software on Thermo Fisher Cloud. Set up the parameters based on the available panels, bins, and size standards from Promega online.
4. Expected Results
4.1. Reagents
4.2. Timeline
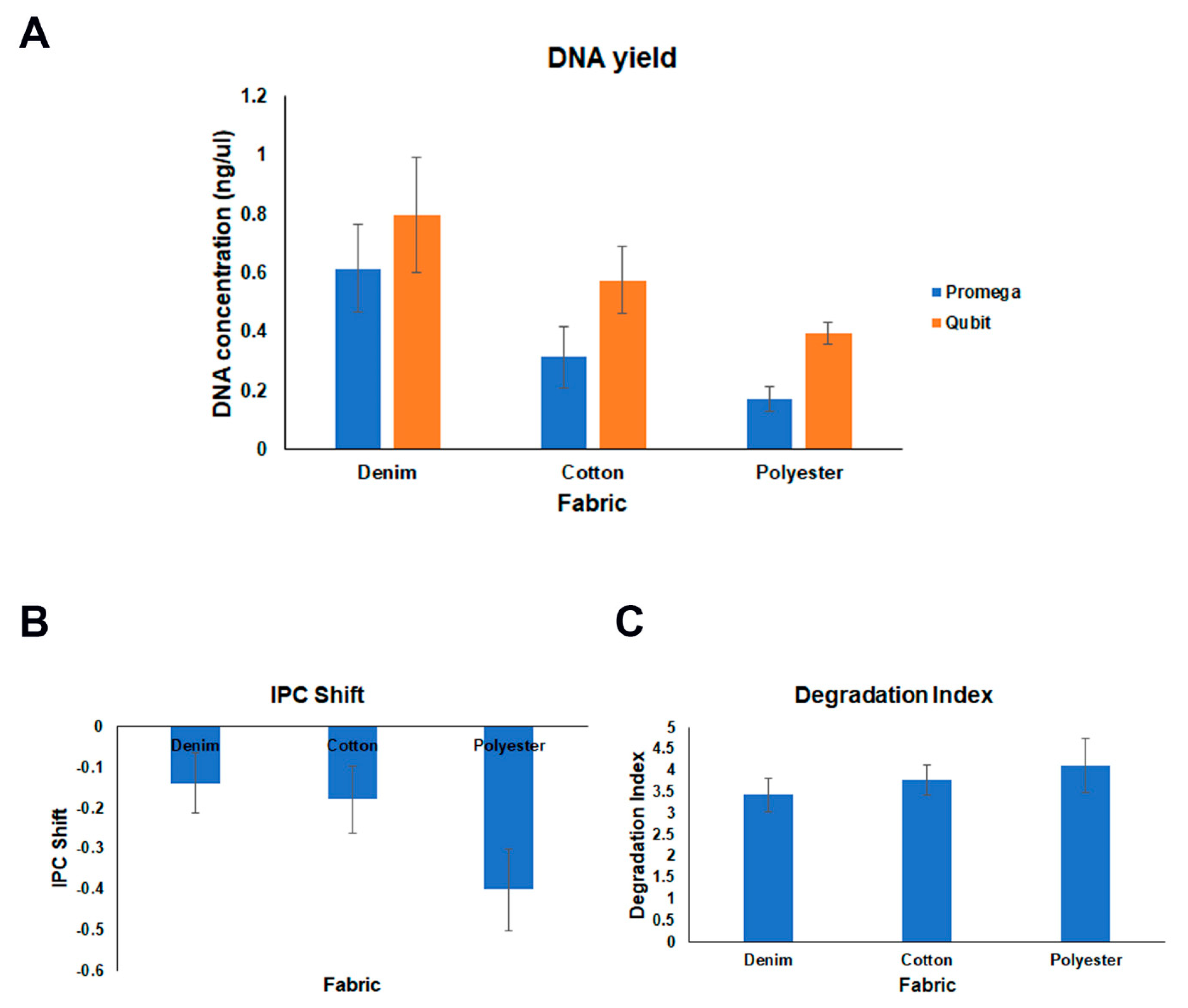
4.3. DNA Concentration and Quality
4.4. STR Profiles
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sakurada, K.; Watanabe, K.; Akutsu, T. Current Methods for Body Fluid Identification Related to Sexual Crime: Focusing on Saliva, Semen, and Vaginal Fluid. Diagnostics 2020, 10, 693. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, J.; Dunn, S.; Best, A.; Mirza, J.; Percival, B.; Mayhew, M.; Megram, O.; Ashford, F.; White, T.; Moles-Garcia, E.; et al. Validation testing to determine the sensitivity of lateral flow testing for asymptomatic SARS-CoV-2 detection in low prevalence settings: Testing frequency and public health messaging is key. PLoS Biol. 2021, 19, e3001216. [Google Scholar] [CrossRef] [PubMed]
- Peto, T.; UK COVID-19 Lateral Flow Oversight Team. COVID-19: Rapid antigen detection for SARS-CoV-2 by lateral flow assay: A national systematic evaluation of sensitivity and specificity for mass-testing. EClinicalMedicine 2021, 36, 100924. [Google Scholar] [CrossRef] [PubMed]
- Niemi, M.E.K.; Daly, M.J.; Ganna, A. The human genetic epidemiology of COVID-19. Nat. Rev. Genet. 2022, 23, 533–546. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, H.; Chagan-Yasutan, H.; Leano, P.S.; Koizumi, N.; Nakajima, C.; Taurustiati, D.; Hanan, F.; Lacuesta, T.L.; Ashino, Y.; Suzuki, Y.; et al. Combined antibody and DNA detection for early diagnosis of leptospirosis after a disaster. Diagn. Microbiol. Infect. Dis. 2016, 84, 287–291. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.H.; Lu, P.L.; Liu, E.Y.; Chen, Y.Y.; Lin, F.M.; Lin, Y.T.; Chang, F.Y.; Lin, J.C. Rapid identification of capsular serotype K1/K2 Klebsiella pneumoniae in pus samples from liver abscess patients and positive blood culture samples from bacteremia cases via an immunochromatographic strip assay. Gut Pathog. 2019, 11, 11. [Google Scholar] [CrossRef] [PubMed]
- Alharbi, A.; Toulah, F.H.; Wakid, M.H.; Azhar, E.; Farraj, S.; Mirza, A.A. Detection of Giardia lamblia by Microscopic Examination, Rapid Chromatographic Immunoassay Test, and Molecular Technique. Cureus 2020, 12, e10287. [Google Scholar] [CrossRef] [PubMed]
- Kishbaugh, J.M.; Cielski, S.; Fotusky, A.; Lighthart, S.; Maguire, K.; Quarino, L.; Conte, J. Detection of prostate specific antigen and salivary amylase in vaginal swabs using SERATEC® immunochromatographic assays. Forensic Sci. Int. 2019, 304, 109899. [Google Scholar]
- Old, J.B.; Schweers, B.A.; Boonlayangoor, P.W.; Reich, K.A. Developmental validation of RSID-saliva: A lateral flow immunochromatographic strip test for the forensic detection of saliva. J. Forensic Sci. 2009, 54, 866–873. [Google Scholar] [CrossRef]
- Pandeshwar, P.; Das, R. Role of oral fluids in DNA investigations. J. Forensic. Leg. Med. 2014, 22, 45–50. [Google Scholar] [CrossRef]
- Holtkotter, H.; Schwender, K.; Wiegand, P.; Peiffer, H.; Vennemann, M. Improving body fluid identification in forensic trace evidence-construction of an immunochromatographic test array to rapidly detect up to five body fluids simultaneously. Int. J. Legal Med. 2018, 132, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Basset, P.; Blandin, P.; Grini, A.; Delemont, S.; Samie, L.; Castella, V. A simplified protocol for the detection of blood, saliva, and semen from a single biological trace using immunochromatographic tests. Forensic Sci. Med. Pathol. 2022, 18, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Kenna, J.; Smyth, M.; McKenna, L.; Dockery, C.; McDermott, S.D. The recovery and persistence of salivary DNA on human skin. J. Forensic Sci. 2011, 56, 170–175. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.; Cox, J.O.; Rhodes, C.; Lewis, C.; Koroma, M.; Hudson, B.C.; Cruz, T.D.; Seashols-Williams, S.J. Comparison of DNA typing success in compromised blood and touch samples based on sampling swab composition. J. Forensic Sci. 2021, 66, 1427–1434. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.; Panacek, E.; Green, W.; Kanthaswamy, S.; Hopkins, C.; Calloway, C. Recovery of salivary DNA from the skin after showering. Forensic Sci. Med. Pathol. 2015, 11, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Conte, J.; Ruddy, A.; Domonoski, L.; Shanahan, A.; Daley, N.; McDevitt, C.; Roca, G. Recovery of DNA from SERATEC® immunochromatographic PSA and saliva test strips. J. Forensic Sci. 2022, 67, 1176–1183. [Google Scholar] [CrossRef]
- Zapico, S.C.; Dytso, A.; Rubio, L.; Roca, G. The Perfect Match: Assessment of Sample Collection Efficiency for Immunological and Molecular Findings in Different Types of Fabrics. Int. J. Mol. Sci. 2022, 23, 10686. [Google Scholar] [CrossRef] [PubMed]
- Zapico, S.C.; Roca, G. A spit in time: Identification of saliva stains and assessment of total DNA recovery up to 180 days after deposition. Forensic Sci. Med. Pathol. 2023. [Google Scholar] [CrossRef]
- Zapico, S.C.; Lascano, V.; Sadik, T.; Paromita, P.; Amaya, J.; Stadler, C.; Roca, G. The killer outfit and timing: Impact of the fabric and time in body fluid identification and DNA profiling. Forensic Sci. Int. Genet. Suppl. Ser. 2022, 8, 248–250. [Google Scholar] [CrossRef]
- Rodpai, R.; Sadaow, L.; Sanpool, O.; Boonroumkaew, P.; Thanchomnang, T.; Laymanivong, S.; Janwan, P.; Limpanont, Y.; Chusongsang, P.; Ohmae, H.; et al. Development and Accuracy Evaluation of Lateral Flow Immunoassay for Rapid Diagnosis of Schistosomiasis Mekongi in Humans. Vector-Borne Zoonotic Dis. 2022, 22, 48–54. [Google Scholar]
- Zapico, S.C.; Ubelaker, D.H. Sex determination from dentin and pulp in a medicolegal context. JADA 2013, 144, 1379–1385. [Google Scholar] [CrossRef]
- Zapico, S.C.; Menéndez, S.T. Human mitochondrial DNA and nuclear DNA isolation from food bite marks. Arch. Oral Biol. 2016, 70, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Zapico, S.C.; Crucet, K.; Antevska, A.; Fernandez-Paradas, R.; Burns, C.; DeGaglia, C.; Ubelaker, D.H. From your eyes only: Efficiency of nuclear and mitochondrial DNA isolation from contact lenses at crime scenes. Electrophoresis 2021, 42, 122–125. [Google Scholar] [CrossRef]

| Standard Concentration | Volume of Male gDNA Standard | Volume of PowerQuant Dilution Buffer |
|---|---|---|
| 50 ng/µL | Undiluted standard | 0 µL |
| 2 ng/µL | 4 µL of undiluted standard | 96 µL |
| 0.08 ng/µL | 4 µL of 2 ng/µL dilution | 96 µL |
| 0.0032 ng/µL | 4 µL of 0.08 ng/µL dilution | 96 µL |
| Reaction Component | Volume per Reaction |
|---|---|
| Water Amplification Grade | 7 µL |
| PowerQuant 2× Master Mix | 10 µL |
| PowerQuant 20× Primer/Probe/IPC Mix | 1 µL |
| Final Volume | 18 µL |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zapico, S.C.; Roca, G. Making the Most of Lateral Flow Immunochromatographic Tests: An Efficient Protocol to Recover DNA. Methods Protoc. 2024, 7, 8. https://doi.org/10.3390/mps7010008
Zapico SC, Roca G. Making the Most of Lateral Flow Immunochromatographic Tests: An Efficient Protocol to Recover DNA. Methods and Protocols. 2024; 7(1):8. https://doi.org/10.3390/mps7010008
Chicago/Turabian StyleZapico, Sara C., and Gabriela Roca. 2024. "Making the Most of Lateral Flow Immunochromatographic Tests: An Efficient Protocol to Recover DNA" Methods and Protocols 7, no. 1: 8. https://doi.org/10.3390/mps7010008
APA StyleZapico, S. C., & Roca, G. (2024). Making the Most of Lateral Flow Immunochromatographic Tests: An Efficient Protocol to Recover DNA. Methods and Protocols, 7(1), 8. https://doi.org/10.3390/mps7010008








