Co-Culture of Gut Bacteria and Metabolite Extraction Using Fast Vacuum Filtration and Centrifugation
Abstract
:1. Introduction
2. Experimental Design
2.1. Materials for Isolation of Gut Bacteria from Human Faecal Samples
- Hungate Anaerobic tubes, 16 × 125 mm (Chemglass Life Sciences, Vineland, NJ, USA, Cat. No. CLS-4208-10);
- AnaeroGenTM 2.5 L sachets (Thermo Scientific, OXOIDTM, Basingstoke, UK, Cat. No. AN0025A);
- AnaeroJar, 2.5 L (Thermo Scientific™ AG0025A, OXOID, Basingstoke, UK);
- Fastidious Anaerobe broth (FAA broth) (E&O Labs, Bonnybridge, UK, Cat. No. KM0188-500G-500);
- Fastidious anaerobe agar with 7% horse blood (FAA + 7% HB) plates (E&O Labs, Bonnybridge, UK, Cat. No. PP1560-P090);
- Fastidious anaerobe agar with 7% horse blood and neomycin plates (FAA + 7% HB+ NEO) (E&O Labs, Bonnybridge, UK, Cat. No. PP0140-P090);
- Fastidious anaerobe agar with 7% horse blood, neomycin, and Aztreonam plates (FAA + 7% HB+ AZT) (E&O Labs, Bonnybridge, UK, Cat. No. PP0144-P090);
- de Man, Rogosa, and Sharpe (MRS) agar (OXOID, Basingstoke, UK, Cat. No. CM0361);
- Brain heart infusion (BHI) agar (OXOID, UK, Cat. No. CM1136);
- Phosphate-buffer saline tablets (Thermo Scientific™, OXOID, Basingstoke, UK Cat. No. 10209252);
- Sterile L-shaped plastic spreaders (Scientific labs, Nottingham, UK, Cat. No. 222088);
- Sterile plastic disposable inoculation loops (Fischer Scientific, Horsham, UK, Cat. No. 12870155).
Equipment
- Anaerobic chamber (Don Whitley Scientific, Bingley, UK);
- Digital microbiology incubator with shaker (Labnet, Edison, NJ, USA, Cat. No. I-5311-DS);
- Digital Microscope VHX-7000N (Keyence, Milton Keynes, UK).
2.2. Materials for Metabolite and Secretome Extractions
- 99.8% Methanol (HPLC grade, Sigma Aldrich, Dorset, UK, CAS. No. 67-56-1);
- Acetonitrile (HPLC grade, Sigma Aldrich, Dorset, UK, CAS. No. 75-05-8);
- Sintered glass funnel and vacuum filter holder kits. 47 mm (Merck, Feltham, UK, Cat. No. 15711719);
- Buchner filtering Flask (Sigma Aldrich, Dorset, UK, Cat. No. Z740686-6EA);
- 0.22 µm, 25 mm, hydrophilic membrane filters (Merck, Feltham, UK, Cat. No. GSWP02500);
- Dry ice;
- Petri dishes (sterile, Scientific laboratory supplies, Nottingham, UK, SLS2000);
- 5 mL, 1 mL sterile pipette tips and pipettes;
- Deionised water;
- Eppendorf tubes (1.5 mL, scientific laboratory supplies, Nottingham, UK);
- Sterile falcon tubes (15 mL and 50 mL, scientific laboratory supplies, Nottingham, UK).
Equipment
- Class 2 Biological safety cabinet (Monmouth Scientific, Dorset, UK, Cat. No. T1800);
- High pressure portable vacuum pump (KNF N035, 30 L/min flow rate, 13 mbar ultimate vacuum, Sigma Aldrich, Dorset, UK);
- Temperature controlled Centrifuge for falcon tubes (Eppendorf, Hamburg, Germany, Cat. No. 5804 R);
- Temperature controlled Centrifuge for Eppendorf’s (Eppendorf, Hamburg, Germany, Cat. No. 5430 R);
- Gas chromatography (5977 B) mass spectrometer (Agilent 8860, Manchester, UK).
2.3. Methods for Sample Collection, Co-Culture, Prior to Extractions
2.3.1. Sample Collection and Ethics
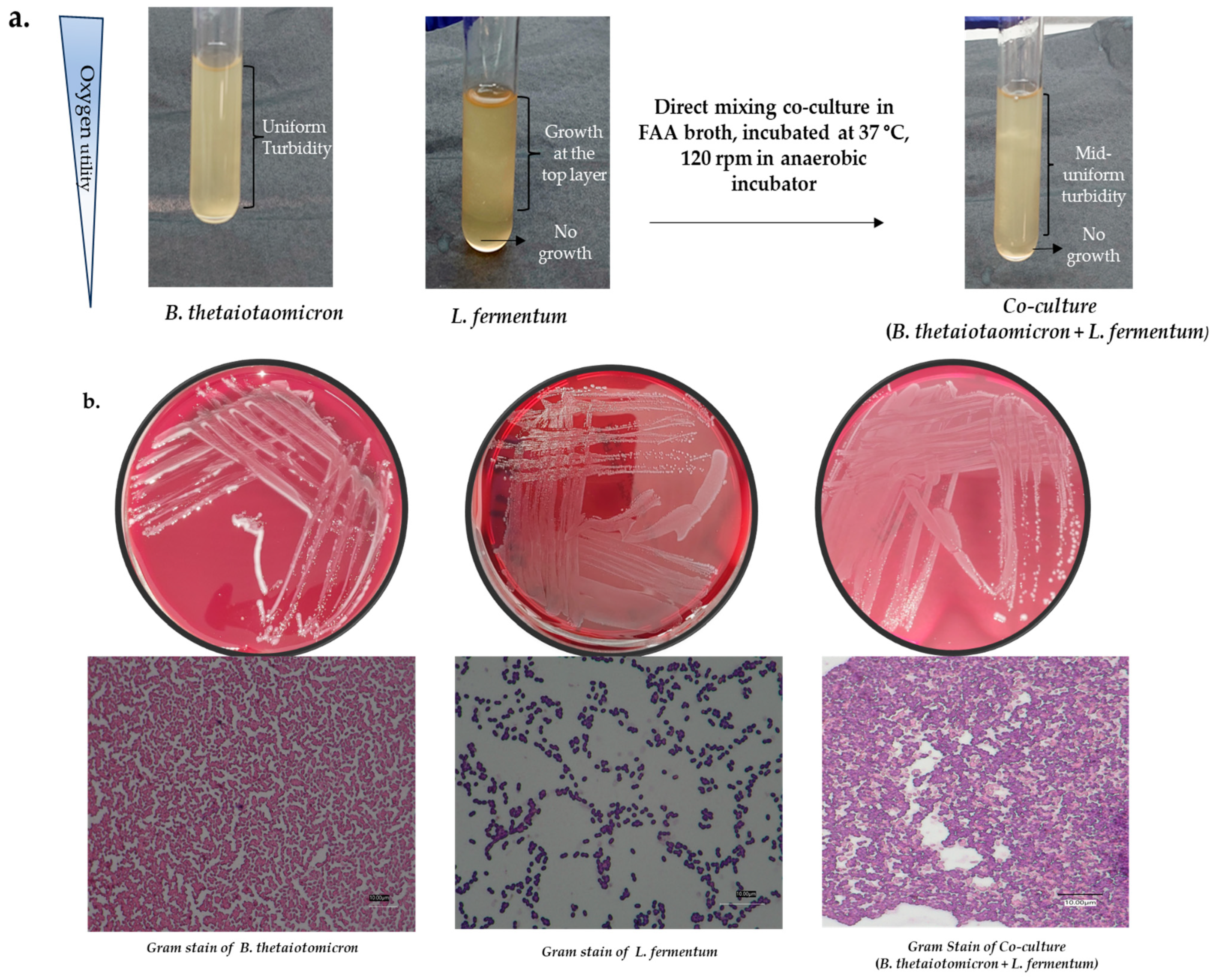
2.3.2. The Isolation and Co-Culture of Gut Bacterium: Direct Mixing Method
- Culture and isolation of gut bacteria
- Identification
- Co-culture of identified bacterium
3. Procedure
3.1. Day 1
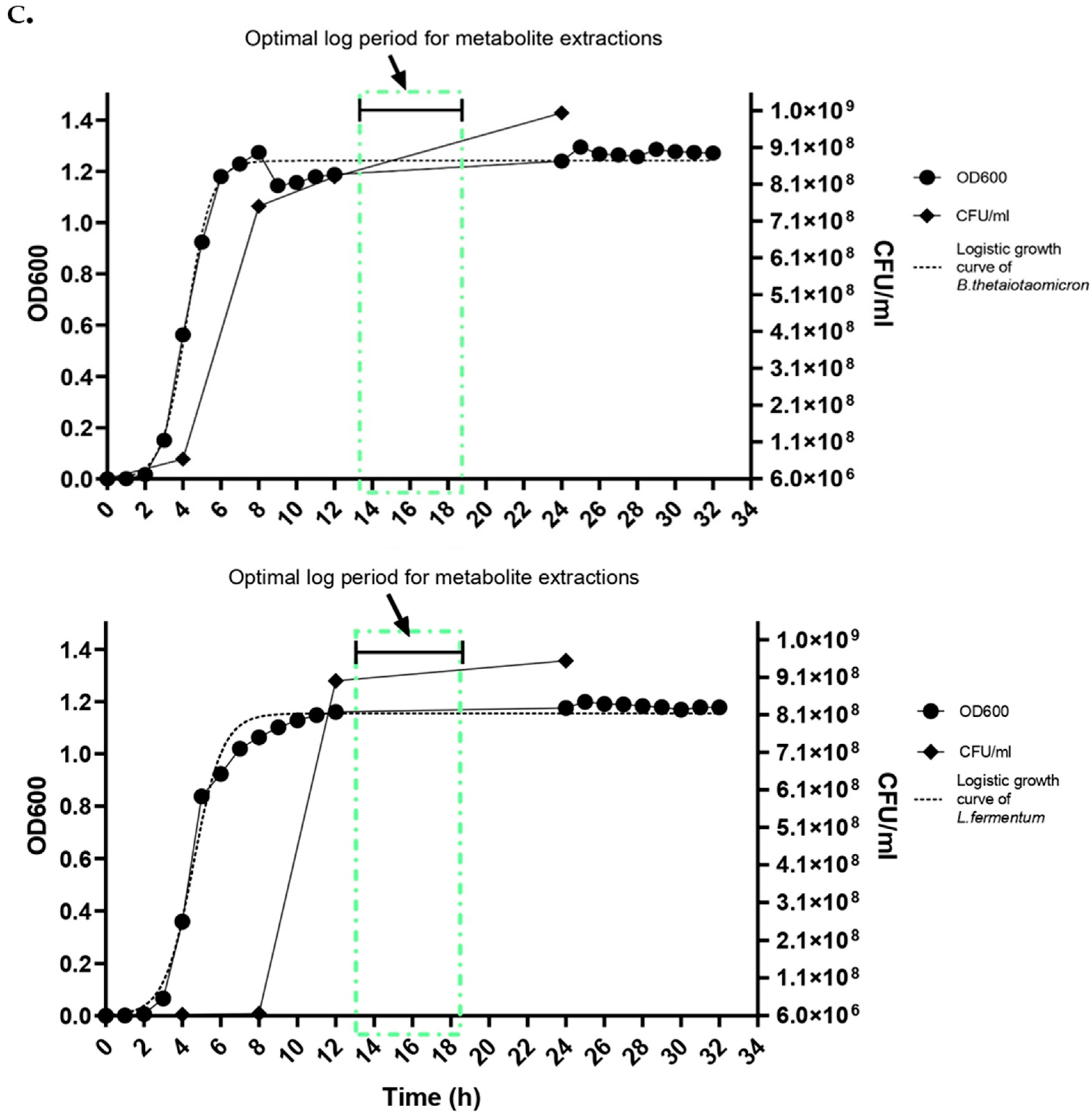
- Inoculate the isolated monocultures and co-culture (B. thetaiotaomicron and L. fermentum) in liquid broth FAA media and incubate at 37 °C in anaerobic conditions to reach the mid-log phase with a biomass yield of 108 CFU/mL. This is determined by performing a growth curve to understand the optimal log phase prior to the experiments.
- CRITICAL STEP: Cultures reaching 108 CFU/mL, OD600 = 0.9 is crucial as it tends to release a higher response in metabolomic profiling.
- Prepare extraction solvents: 80% Methanol (v/v) is used for denaturing and solubilizing bacterial molecules and volatile organic compounds. Acetonitrile–Methanol–Water (v/v) in a 2:2:1 ratio, which dissolve lipophilic substances, hydrophobic substances, and non-polar substances such as amino acids. These solvents should be stored at −20 °C overnight before extractions.
- CRITICAL STEP: Solvents should ice chilled in −20 °C before and during the experiments to ensure the stability, reproducibility, and reliability of the obtained metabolomes.
- Required consumables and filtration equipment should be autoclaved.
3.2. Day 2
- Before removing the culture media from −20 °C, set up the filtration equipment by inserting the glass funnel into the vacuum flask, which is attached to the vacuum pump using autoclavable tubing.
- CRITICAL STEP: Make sure everything is secure here and place this filtration apparatus in an ice box. This will minimise the loss of metabolites, proteins, and other biochemical compound yields during the procedure.
- Using forceps, place the 0.22 µm membrane filter paper carefully on the fritted glass base. Next, place the glass funnel on top to cover the porous surface of the membrane filter paper and seal it tightly before the vacuum pump is turned on.
- Aliquot 3 mL of any extraction solvent mentioned above based on the metabolite of interest in a Petri dish before placing it on dry ice.
- CRITICAL STEP: Using 3 mL of solvent during the extractions helps to solubilise highly volatile compounds and the desired metabolites while also helping to characterise them easily.
- Pipette 15 mL of bacterial culture onto the centre of the membrane filter paper during vacuum pressure.
- CRITICAL STEP: During this step, bacterial cells are filtered without adding any solvents to the culture, including deionised water on the membrane filter paper prior to filtration, to obtain pure extractions.
- Remove the membrane filter paper from the funnel and place it upside down onto a Petri dish containing extraction solvents.
- CRITICAL STEP: This step is carried out with the new filter papers to ensure extraction is performed with each solvent: (1) 80% Methanol (v/v) and (2) Acetonitrile–Methanol–Water (2:2:1 v/v) were used to ensure extraction is made individually.
- Lyse the bacterial cells by swirling the solvent across the membrane filter before evenly distributing the solvent using a 1 mL pipette 20 times.
- CRITICAL STEP: This should be performed as quickly as possible to minimise the amount of time that the culture spends outside of its growth condition, as the cellular metabolism changes rapidly. The speed of step is key to better metabolite recovery and yield.
- Immediately transfer the solvent with extracted metabolites into 1.5 mL Eppendorf tubes and place them in dry ice until proceeding to the next step.
- CRITICAL STEP: Dry ice helps to maintain the stability of the volatile nature of compounds extracted in the solvent.
- Repeat steps 5 to 9 for the rest of the bacterial cultures, if required, or discard the culture.
- CRITICAL STEP: It is advised to repeat the steps in order to maintain the reliability and reproducibility of the experiments.
- Once the filtration and extraction are completed, Eppendorf tubes are centrifuged at 4 °C maximum speed at 3000 rpm for 5 min.
- CRITICAL STEP: This step is carried out to remove the unwanted cellular debris from the culture and enhance the data analysis of metabolites using OMICS technologies and the quantification of proteins.
- After centrifugation, collect the supernatant without disrupting the pellet.
- PAUSE STEP: After collecting supernatants, store it in −80 °C for long-term storage and used immediately for metabolic profiling and quantification for proteins.
- Now, to extract the secretome, transfer the filtrate collected in the vacuum flask to individual 15 mL or 50 mL falcon tubes and centrifuge them for 10 min at 4 °C at 1000 rpm maximum speed.
- CRITICAL STEP: This filtrate is not exposed to any extraction solvents, which will aid in quantifying and gaining a more physiologically relevant understanding of the culture’s secretome analysis.
- Once centrifuged, collect the supernatant and pellet for the quantification of proteins using BCA assay; can perform Western blots immediately using the pellet as control.
- PAUSE STEP: Supernatants and pellets are stored separately in −80 °C for long-term usage.
- CRITICAL STEP: Do not discard the pellet, which can also be used to analyse the influence of co-cultures interactions (positive or negative, i.e., 16S rRNA sequencing. This acts as a control for the secretome analysis. It is advised to use the pellet of 16S sequencing immediately to minimise DNA degradation.
4. Expected Results and Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cunningham, A.L.; Stephens, J.W.; Harris, D.A. Gut microbiota influence in type 2 diabetes mellitus (T2DM). Gut Pathog. 2021, 13, 50. [Google Scholar] [CrossRef]
- Vernocchi, P.; Del Chierico, F.; Putignani, L. Gut microbiota metabolism and interaction with food components. Int. J. Mol. Sci. 2020, 21, 3688. [Google Scholar] [CrossRef]
- Hills, R.D.; Pontefract, B.A.; Mishcon, H.R.; Black, C.A.; Sutton, S.C.; Theberge, C.R. Gut microbiome: Profound implications for diet and disease. Nutrients 2019, 11, 1613. [Google Scholar] [CrossRef] [PubMed]
- Jardon, K.M.; Canfora, E.E.; Goossens, G.H.; Blaak, E.E. Dietary macronutrients and the gut microbiome: A precision nutrition approach to improve cardiometabolic health. Gut 2022, 71, 1214–1226. [Google Scholar] [CrossRef] [PubMed]
- Cani, P.D. Human gut microbiome: Hopes, threats and promises. Gut 2018, 67, 1716–1725. [Google Scholar] [CrossRef] [PubMed]
- Ogunrinola, G.A.; Oyewale, J.O.; Oshamika, O.O.; Olasehinde, G.I. The human microbiome and its impacts on health. Int. J. Microbiol. 2020, 2020, 8045646. [Google Scholar] [CrossRef]
- Silva, Y.P.; Bernardi, A.; Frozza, R.L. The Role of Short-Chain Fatty Acids from Gut Microbiota in Gut-Brain Communication. Front. Endocrinol. 2020, 11, 25. [Google Scholar] [CrossRef] [PubMed]
- Portincasa, P.; Bonfrate, L.; Vacca, M.; De Angelis, M.; Farella, I.; Lanza, E.; Khalil, M.; Wang, D.Q.H.; Sperandio, M.; Di Ciaula, A. Gut Microbiota and Short Chain Fatty Acids: Implications in Glucose Homeostasis. Int. J. Mol. Sci. 2022, 23, 1105. [Google Scholar] [CrossRef]
- Selegato, D.M.; Castro-Gamboa, I. Enhancing chemical and biological diversity by co-cultivation. Front. Microbiol. 2023, 14, 1117559. [Google Scholar] [CrossRef] [PubMed]
- Vidal-Veuthey, B.; Damariz González and Juan Pablo Cárdenas. Role of microbial secreted proteins in gut microbiota-host interactions. Front. Cell Infect. Microbiol. 2022, 12, 964710. [Google Scholar] [CrossRef]
- Bayer, G.; Ganobis, C.M.; Allen-Vercoe, E.; Philpott, D.J. Defined gut microbial communities: Promising tools to understand and combat disease. Microbes Infect. 2021, 23, 104816. [Google Scholar] [CrossRef] [PubMed]
- Ravikrishnan, A.; Blank, L.M.; Srivastava, S.; Raman, K. Investigating metabolic interactions in a microbial co-culture through integrated modelling and experiments. Comput. Struct. Biotechnol. J. 2020, 18, 1249–1258. [Google Scholar] [CrossRef]
- Kapoore, R.V.; Padmaperuma, G.; Maneein, S.; Vaidyanathan, S. Co-culturing microbial consortia: Approaches for applications in biomanufacturing and bioprocessing. Crit. Rev. Biotechnol. 2021, 42, 46–72. [Google Scholar] [CrossRef] [PubMed]
- Brenner, K.; You, L.; Arnold, F.H. Engineering microbial consortia: A new frontier in synthetic biology. Trends Biotechnol. 2008, 26, 483–489. [Google Scholar] [CrossRef]
- Hays, S.G.; Patrick, W.G.; Ziesack, M.; Oxman, N.; Silver, P.A. Better together: Engineering and application of microbial symbioses. Curr. Opin. Biotechnol. 2015, 36, 40–49. [Google Scholar] [CrossRef]
- Fuchs, A.P.; Tripet, B.; Cloud, B.; Ammons, M.; Copie, V. Optimization of metabolite extraction protocols for the identification and profiling of small molecule metabolites from planktonic and biofilm Pseudomonas aeruginosa cultures. Curr. Metabolomics 2016, 4, 141–147. [Google Scholar] [CrossRef]
- Gutiérrez Méndez, N.; Kamal, M.; Mohd, M.; Rahim, A.; Abdullah, H.; Azlan, A.K. Bacterial metabolomics: Sample preparation methods. Biochem. Res. Int. 2022, 2022, 9186536. [Google Scholar]
- Pinu, F.; Villas-Boas, S.; Aggio, R. Analysis of intracellular metabolites from microorganisms: Quenching and extraction protocols. Metabolites 2017, 7, 53. [Google Scholar] [CrossRef]
- Bell, H.N.; Das, N.K.; Shah, Y.M. Protocol for isolation and analysis of small volatile microbiome metabolites from human or mouse samples. STAR Protoc. 2022, 3, 101311. [Google Scholar] [CrossRef]
- Bordag, N.; Janakiraman, V.; Nachtigall, J.; González Maldonado, S.; Bethan, B.; Laine, J.P.; Fux, E. Fast Filtration of Bacterial or Mammalian Suspension Cell Cultures for Optimal Metabolomics Results. PLoS ONE 2016, 11, 0159389. [Google Scholar] [CrossRef]
- Wan, X.; Yang, Q.; Wang, X.; Bai, Y.; Liu, Z. Isolation and Cultivation of Human Gut Microorganisms: A Review. Microorganisms 2023, 11, 1080. [Google Scholar] [CrossRef] [PubMed]
- Ndeh, D.; Munoz Munoz, J.; Cartmell, A.; Bulmer, D.; Wills, C.; Henrissat, B.; Gray, J. The human gut microbe Bacteroides thetaiotaomicron encodes the founding member of a novel glycosaminoglycan-degrading polysaccharide lyase family PL29. J. Biol. Chem. 2018, 293, 17906–17916. [Google Scholar] [CrossRef]
- Wexler, H.M. Bacteroides: The Good, the Bad, and the Nitty-Gritty. Clin. Microbiol. Rev. 2007, 20, 593–621. [Google Scholar] [CrossRef]
- Lacerda, D.C.; da Costa, P.C.T.; Pontes, P.B.; Dos Santos, L.A.C.; Neto, J.P.R.C.; Luis, C.C.S.; de Sousa Brito, V.P.; de Brito Alves, J.L. Potential role of Limosilactobacillus fermentum as a probiotic with anti-diabetic properties: A review. World J. Diabetes 2022, 13, 717–728. [Google Scholar] [CrossRef] [PubMed]
- Liang, T.; Wu, L.; Xi, Y.; Li, Y.; Xie, X.; Fan, C.; Yang, L.; Yang, S.; Chen, X.; Zhang, J.; et al. Probiotics supplementation improves hyperglycemia, hypercholesterolemia, and hypertension in type 2 diabetes mellitus: An update of meta-analysis. Crit. Rev. Food Sci. Nutr. 2020, 61, 1670–1688. [Google Scholar] [CrossRef]
- Guo, X.; Xia, X.; Tang, R.; Zhou, J.; Zhao, H.; Wang, K. Development of a real-time PCR method for Firmicutes andBacteroidetes in faeces and its application to quantify intestinal population of obese and lean pigs. Lett. Appl. Microbiol. 2008, 47, 367–373. [Google Scholar] [CrossRef] [PubMed]
- Janda, J.M.; Abbott, S.L. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: Pluses, perils, and pitfalls. J. Clin. Microbiol. 2007, 45, 2761–2764. [Google Scholar] [CrossRef]
- Jost, T.; Lacroix, C.; Braegger, C.P.; Chassard, C. New insights in gut microbiota establishment in healthy breast fed neonates. PLoS ONE 2012, 7, e44595. [Google Scholar] [CrossRef]
- Wang, Z.; Zhu, H.; Xiong, W. Advances in mass spectrometry-based multi-scale metabolomic methodologies and their applications in biological and clinical investigation, Sci. Bull. 2023, 68, 2268–2284. [Google Scholar]
- Akhtar, M.; Chen, Y.; Ma, Z.; Zhang, X.; Shi, D.; Khan, J.A.; Liu, H. Gut microbiota-derived short chain fatty acids are potential mediators in gut inflammation. Anim. Nutr. 2022, 8, 350–360. [Google Scholar] [CrossRef]
- Layden, B.T.; Angueira, A.R.; Brodsky, M.; Durai, V.; Lowe, W.L. Short chain fatty acids and their receptors: New metabolic targets. Transl. Res. 2013, 161, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Doi, Y. Glycerol metabolism and its regulation in lactic acid bacteria. Appl. Microbiol. Biotechnol. 2019, 103, 5079–5093. [Google Scholar] [CrossRef] [PubMed]
- Hosomi, K.; Kiyono, H.; Kunisawa, J. Fatty acid metabolism in the host and commensal bacteria for the control of intestinal immune responses and diseases. Gut Microbes 2019, 11, 276–284. [Google Scholar] [CrossRef] [PubMed]
- Bofinger, M.R.; de Sousa, L.S.; Fontes, J.E.N.; Marsaioli, A.J. Diketopiperazines as cross-communication quorum-sensing signals between Cronobacter sakazakii and Bacillus cereus. ACS Omega 2017, 2, 1003–1008. [Google Scholar] [CrossRef]
- Ogilvie, C.E.; Czekster, C.M. Cyclic dipeptides and the human microbiome: Opportunities and challenges. Bioorg Med. Chem. 2023, 90, 117372. [Google Scholar] [CrossRef]
- Kapadia, C.; Kachhdia, R.; Singh, S.; Gandhi, K.; Poczai, P.; Alfarraj, S.; Ansari, M.J.; Gafur, A.; Sayyed, R.Z. Pseudomonas aeruginosa inhibits quorum-sensing mechanisms of soft rot pathogen Lelliottia amnigena RCE to regulate its virulence factors and biofilm formation. Front. Microbiol. 2022, 13, 977669. [Google Scholar] [CrossRef]
- Bojarska, J.; Mieczkowski, A.; Ziora, Z.M.; Skwarczynski, M.; Toth, I.; Shalash, A.O.; Parang, K.; El-Mowafi, S.A.; Mohammed, E.H.M.; Elnagdy, S.; et al. Cyclic dipeptides: The biological and structural landscape with special focus on the anti-cancer proline-based scaffold. Biomolecules 2021, 11, 1515. [Google Scholar] [CrossRef]
- Kado, D.M. Night-to-Night sleep duration variability and gut microbial diversity: More evidence for a brain-gut microbiome-sleep connection. Sleep 2024, 47, zsae005. [Google Scholar] [CrossRef]
- Mueller, G.P.; Driscoll, W.J. Biosynthesis of oleamide. Vitam. Horm. 2009, 81, 55–78. [Google Scholar]
- Needham, B.D.; Kaddurah-Daouk, R.; Mazmanian, S.K. Gut microbial molecules in behavioural and neurodegenerative conditions. Nat. Rev. Neurosci. 2020, 21, 717–731. [Google Scholar] [CrossRef]
- Rojo, F. Degradation of alkanes by bacteria. Environ. Microbiol. 2009, 11, 2477–2490. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Qi, Y.; Zheng, H. Dietary Polyphenol, Gut Microbiota, and Health Benefits. Antioxidants 2022, 11, 1212. [Google Scholar] [CrossRef] [PubMed]

| Parameter | Details |
|---|---|
| Extraction Solvent | 80% Ice cold Methanol |
| Analytical Technique | Untargeted characterisation using Agilent 8860 gas chromatography (5977B) mass spectrometry with inbuilt NIST database, single quadrupole |
| GC Column | HP-5MS UI column (30 m × 250 µm × 0.25 μm) |
| GC Programme | Initial temperature ramp: 50 °C to 280 °C at 50 °C/min; subsequent ramps: 25 °C/min to 200 °C, then 15 °C/min to 280 °C; final hold: 280 °C for 5 min |
| Total GC Run Time | 20.33 min |
| Carrier Gas | Helium at 1.2 mL/min |
| Injection Mode | Splitless mode, 1 μL injection volume |
| Ion Source Temperature | 250 °C |
| Transfer Line Temperature | 280 °C |
| Ionisation Energy | 70 eV |
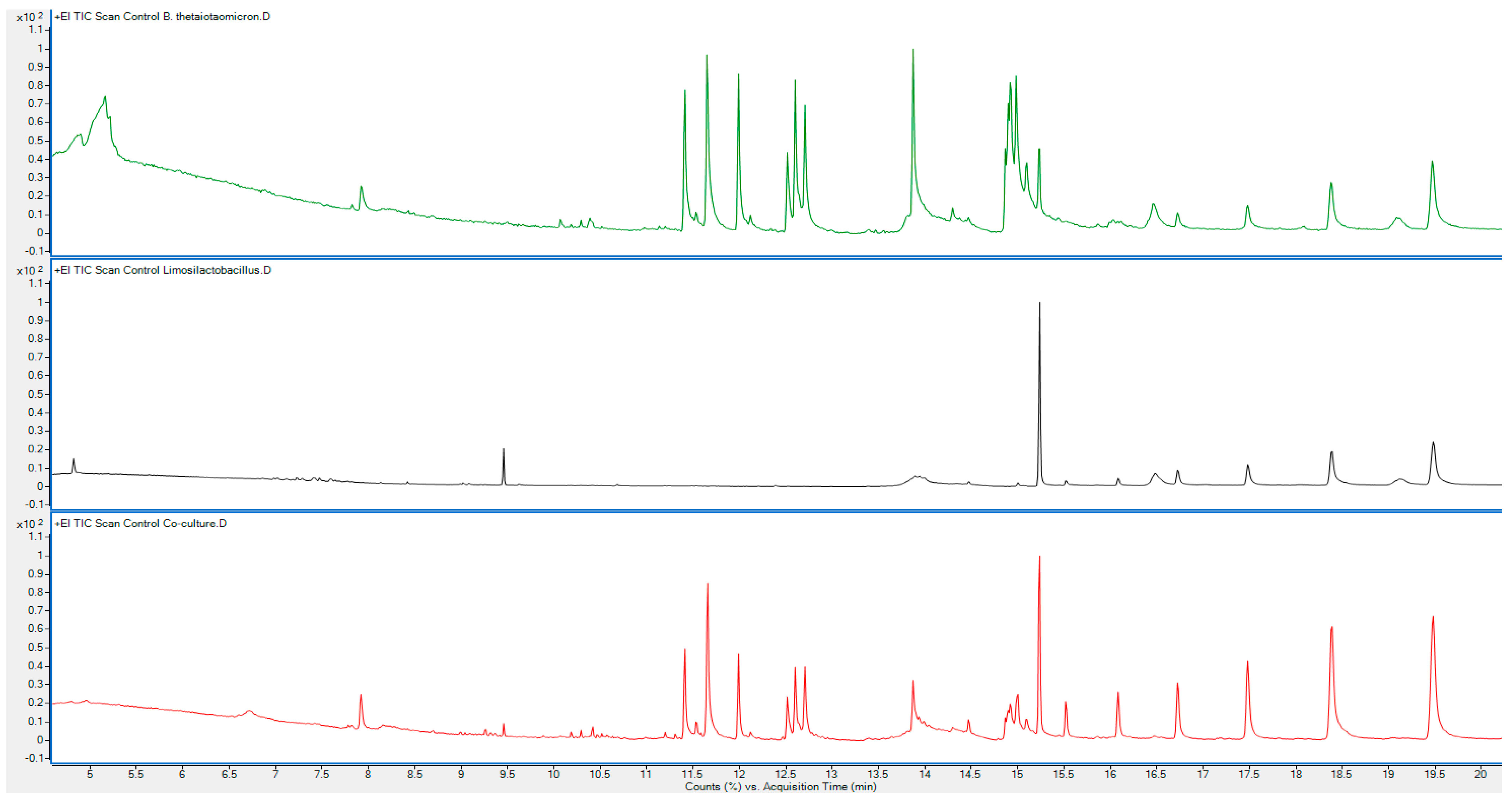
| Identified Compounds | Monoculture 1 B. thetaiotaomicron | Monoculture 2 L. fermentum | Coculture | Possible Pathways Linked to Gut Bacterial Metabolism | |||
|---|---|---|---|---|---|---|---|
| RT | MF | RT | MF | RT | MF | ||
| Isovaleric acid; butanoic acid, 3-methyl- | 4.897 | 867 | - | - | 4.779 | 750 | Short-chain fatty acid (SCFA) metabolism derived from amino acid catabolism [2,10,30,31]. |
| Hexanoic acid, 2-methyl- | 5.164 | 810 | - | - | 4.961 | 695 | Short-chain fatty acid metabolism (SCFA) [2,30,31]. |
| Glycerin | - | - | - | - | 6.724 | 619 | Central metabolism, glycerol is a play crucial intermediate [32]. |
| N-methylene-2-phenylethanamine | 7.92 | 884 | - | - | 7.92 | 912 | Amino acid metabolism and secondary metabolite biosynthesis [10,30]. |
| 5-keto-2,2-dimethyl heptanoic acid, ethyl(ester) | - | - | - | - | 9.459 | 579 | Fatty acid metabolism and degradation [33]. |
| Cyclo (L,prolyl-l-valine) | 11.991 12.119 | 760 641 | - | - | 11.991 | 916 | Diketopiperazine metabolism and peptide biosynthesis [34,35,36,37]. |
| 9-Octadecenamide, (Z)- or oleamide | 14.918 14.982 | 865 792 | - | - | 14.918 | 861 | Fatty acid amide metabolism [38,39,40]. |
| n-heptacosane | 16.456 16.723 17.482 18.379 19.469 | 707 621 675 775 801 | 16.082 16.723 | 790 844 | 15.003 15.516 16.082 16.723 | 734 828 843 859 | Alkane metabolism, derived from lipid degradation [41]. |
| Phenol, 2,2′-methylenebis(6-(1,1-dimethylethyl)-4-methyl- | 15.238 | 826 | 15.238 | 909 | 15.238 | 922 | Phenolic compound metabolism and secondary metabolite biosynthesis [42]. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guraka, A.; Duff, R.; Waldron, J.; Tripathi, G.; Kermanizadeh, A. Co-Culture of Gut Bacteria and Metabolite Extraction Using Fast Vacuum Filtration and Centrifugation. Methods Protoc. 2024, 7, 74. https://doi.org/10.3390/mps7050074
Guraka A, Duff R, Waldron J, Tripathi G, Kermanizadeh A. Co-Culture of Gut Bacteria and Metabolite Extraction Using Fast Vacuum Filtration and Centrifugation. Methods and Protocols. 2024; 7(5):74. https://doi.org/10.3390/mps7050074
Chicago/Turabian StyleGuraka, Asha, Richard Duff, Joe Waldron, Gyanendra Tripathi, and Ali Kermanizadeh. 2024. "Co-Culture of Gut Bacteria and Metabolite Extraction Using Fast Vacuum Filtration and Centrifugation" Methods and Protocols 7, no. 5: 74. https://doi.org/10.3390/mps7050074
APA StyleGuraka, A., Duff, R., Waldron, J., Tripathi, G., & Kermanizadeh, A. (2024). Co-Culture of Gut Bacteria and Metabolite Extraction Using Fast Vacuum Filtration and Centrifugation. Methods and Protocols, 7(5), 74. https://doi.org/10.3390/mps7050074








