Mapping Gene Expression in Whole Larval Brains of Bicyclus anynana Butterflies
Abstract
:1. Introduction
2. Experimental Design
2.1. Materials
2.1.1. Consumables
- Blade holder (Swann-Morton, Sheffield, England; Cat. No.: 0934)
- Blades (Swann-Morton, Sheffield, England; Cat. No.: 0115)
- Dissection silicone plate: Dragon Skin 30 Mold Making Silicone Rubber (Smooth-On, Macungie, PA, USA; Cat. No.: 0751635278417), Petri dish (Sigma-Aldrich, Burlington, MA, USA; Cat. No.: P5981)
- Filter pipette tips, 10 µL (Biotix, San Diego, CA, USA; Cat. No.: 63300041)
- Filter pipette tips, 20 µL (Biotix, San Diego, CA, USA; Cat. No.: 63300042)
- Filter pipette tips, 300 µL (Biotix, San Diego, CA, USA; Cat. No.: 63300045)
- Filter pipette tips, 1250 µL (Biotix, San Diego, CA, USA; Cat. No.: 63300047)
- Glass spot plate (Corning, Corning, NY, USA; Cat. No.: 7220-85)
- Insect pins (BioQuip Products, Rancho Dominguez, CA, USA; Cat. No.: 1208B2)
- Lint-free tissue, Kimtech Science™ Kimwipes™ Delicate Task Wipes (Kimberly-Clark Professional, Roswell, GA, USA; Cat No.: 34120)
- Microcentrifuge tubes, 1.5 mL (Eppendorf, Hamburg, Germany; Cat. no.: T9661-500EA)
- RNaseZap™ RNase Decontamination Solution (ThermoFisher Scientific, Waltham, MA, USA, Cat. No.: AM9780)
- Straight tweezers, fine (Dumont Switzerland, Montignez, Jura, Switzerland; Cat. No.: 11254-20)
- Straight tweezers, regular (Dumont Switzerland, Montignez, Jura, Switzerland; Cat. No.: 0203-5-PO)
- Superfine Vannas scissors, 8 cm (World Precision Instruments, Sarasota, FL, USA; Cat. No.: 501778)
2.1.2. Reagents
- Acrylamide/bis-acrylamide, 19:1 (Sigma-Aldrich, Burlington, MA, USA; Cat. No.: A2917)
- Ammonium persulfate (APS) (Sigma-Aldrich, Burlington, MA, USA; Cat. No.: A7460)
- Calcium chloride (CaCl2) (Sigma-Aldrich, Burlington, MA, USA; Cat. No.: C4901)
- Citric acid (Sigma-Aldrich, Burlington, MA, USA; Cat. No.: 251275)
- DAPI (Sigma-Aldrich, Burlington, MA, USA; Cat. No.: D9542)
- Denhardt’s solution, 50× (ThermoFisher, Massachusetts, USA; Cat. No.: 750018)
- Dextran sulfate (Sigma-Aldrich, Burlington, MA, USA; Cat. No.: D6001)
- Diethyl pyrocarbonate (DEPC) (Sigma-Aldrich, Burlington, MA, USA; Cat. No.: D5758)
- Dimethyl sulfoxide (DMSO) (Sigma-Aldrich, Burlington, MA, USA; Cat. No.: D8418)
- DNase I, 1 U/µL (ThermoFisher Scientific, Waltham, MA, USA, Cat. No.: EN0521)
- Ethylenediaminetetraacetic acid (EDTA) (Sigma-Aldrich, Burlington, MA, USA; Cat. No.: E9884)
- Formaldehyde, 37% (Sigma-Aldrich, Burlington, MA, USA; Cat. No.: F8775)
- Formamide (Sigma-Aldrich, Burlington, MA, USA; Cat. No.: F7503)
- Glycerol (Sigma-Aldrich, Burlington, MA, USA; Cat. No.: G7893)
- H1, H2 hairpins (Molecular Instruments, Los Angeles, CA, USA)
- Heparin (Sigma-Aldrich, Burlington, MA, USA; Cat. No.: H3393)
- Magnesium chloride (MgCl2) (Sigma-Aldrich, Burlington, MA, USA; Cat. No.: M8266)
- N,N,N′,N′-Tetramethylethylenediamine (TEMED) (Sigma-Aldrich, Burlington, MA, USA; Cat. No.: T9281)
- Primary oligos, 100 µM in IDTE, pH 8.0 (Molecular Instruments, CA, USA; Integrated DNA Technologies, Coralville, IA, USA)
- Potassium phosphate dibasic (K2HPO4) (Sigma-Aldrich, Burlington, MA, USA; Cat. No.: P3786)
- Potassium phosphate monobasic (KH2PO4) (Sigma-Aldrich, Burlington, MA, USA; Cat. No.: P0662)
- Sodium chloride (NaCl) (Sigma-Aldrich, Burlington, MA, USA; Cat. No.: S9888)
- Sodium dodecyl sulfate (SDS) (Sigma-Aldrich, Burlington, MA, USA; Cat. No.: 436143)
- Sodium hydroxide (NaOH) pellets (Sigma-Aldrich, Burlington, MA, USA; Cat. No.: 221465)
- Tris-hydrochloride (Tris-HCl), pH 7.5 (Sigma-Aldrich, Burlington, MA, USA; Cat. No.: 10812846001)
- Trisodium citrate (Sigma-Aldrich, Burlington, MA, USA; Cat. No.: S1804)
- TWEEN 20 (Sigma-Aldrich, Burlington, MA, USA; Cat. No.: P1379)
2.2. Equipment
- Autoclave (HIRAYAMA, Saitama, Japan; Product ID: HV-110)
- Confocal microscope, Olympus FLUOVIEW FV3000 confocal LSM (Olympus Life Science, Waltham, MA, USA; Product ID: FV3000)
- Incubating rocking shaker (OHAUS, Parsippany, NJ, USA; Product ID: ISRK04HDG)
- Micropipette, 0.1–2.5 µL (Eppendorf, Hamburg, Germany; Cat. no.: 3123000012)
- Micropipette, 0.5–10 µL (Eppendorf, Hamburg, Germany; Cat. no.: 3123000020)
- Micropipette, 20–200 µL (Eppendorf, Hamburg, Germany; Cat. no.: 3123000055)
- Micropipette, 100–1000 µL (Eppendorf, Hamburg, Germany; Cat. no.: 3123000063)
- Milli-Q® Ultrapure Water Systems (Merck Millipore, Burlington, MA, USA)
- Stereo microscope, ZEISS Stemi 305 (ZEISS, Oberkochen, Germany; Product ID: Stemi 305)
3. Procedure
3.1. Probe Design
3.2. Dissection and Preparation of Larval Brains for HCR
3.2.1. Dissection and Fixation (~3 h for 1 Sample, +20 Min for Every Additional Sample)
- In a 1.5 mL microcentrifuge tube, prepare 500 µL of fresh 4% phosphate-buffered formaldehyde (PFA). Prepare one tube for every three larval brains processed.
- Using a P1000 micropipette, gently aspirate the dissected brains and transfer them into the microcentrifuge tubes containing 4% PFA. Each tube should contain no more than three brains.
CRITICAL STEP Samples should be aspirated gently to prevent damage. One technique is to use the volume adjustment ring on the pipette instead of the plunger to aspirate samples gently. This is accomplished by first adjusting the set volume on the pipette to the lowest setting (100 µL), immersing the pipette tip into the solution next to the samples, and then smoothly rotating the volume adjustment ring toward a higher volume setting. The tip of the pipette can also be cut using a pair of scissors for a larger inlet diameter.
- 4.
- Incubate the samples on a shaker for 1.5 h at RT with gentle agitation (~60 rpm in a horizontal shaker). For smaller tissues, fixation time can be reduced.
- 5.
- Wash the samples three times with 500 µL of 1× phosphate-buffered saline with TWEEN 20 (1× PBST) for 10 min each, at RT.
PAUSE STEP The samples can be stored in 1× PBST at 4 °C for up to 3 days.
3.2.2. Permeabilization and Post-Fixation (~2.5 h)
- 6.
- Replace 1× PBST with 500 µL of detergent solution.
- 7.
- Incubate the samples on a shaker for 30 min at RT with gentle agitation.
- 8.
- Rinse the samples two times with 500 μL of 1× PBST each, at RT.
- 9.
- Transfer the samples into microcentrifuge tubes containing 500 μL of fresh 4% PFA.
- 10.
- Incubate the samples on a shaker for 20 min at RT with gentle agitation.
- 11.
- Wash the samples three times with 500 µL of 1× PBST for 10 min each, at RT.
- 12.
- Wash the samples two times with 500 µL of 5× saline–sodium citrate buffer with TWEEN 20 (SSCT) for 3 min each, at RT.
- 13.
- Transfer the samples into microcentrifuge tubes containing 500 μL of 30% probe hybridization buffer.
- 14.
- Incubate the samples on an incubating shaker for 30 min at 37 °C with gentle agitation.
PAUSE STEP The samples can be stored in 30% probe hybridization buffer at 4 °C for up to 3 weeks.
3.3. Embedment of Fixed Brains in Polyacrylamide Gel
3.3.1. Preparation for Gel Embedment (~30 min)
- In a 1.5 mL microcentrifuge tube, add 200 µL of 40% acrylamide solution, 400 µL of 2.0 M NaCl, 60 µL of 1.0 M Tris-HCl, pH 7.5, 325 µL of DEPC H2O, and 4 µL of TEMED for 1 mL of gel solution.
CRITICAL STEP The gel solution should be prepared fresh. A total of 1 mL of solution is sufficient for five gel castings. Invert or pipette gently to mix. Do not vortex the gel solution or any of the components.
- 2.
- Allow the gel solution and 10% APS stock solution to normalize to RT.
- 3.
- If more than one gel piece is to be cast using the stock gel solution, for every gel piece, prepare a 1.5 mL microcentrifuge tube containing 2 µL of 10% APS. Otherwise, skip this step.
- 4.
- Clean a microscope slide, five coverslips, tweezers, and scalpel blade with RNaseZap, using KimWipes.
- 5.
- Using a P2 micropipette, dispense 0.5 μL of DEPC H2O onto the microscope slide, approximately 30 mm apart (Figure 1A).
- 6.
- Place a coverslip on top of each droplet of water, with a clearance of about 12–16 mm between the coverslips (Figure 1B). Press the coverslips firmly down onto the droplets of water to temporarily adhere the coverslips to the microscope slide.
- 7.
- Dispense 0.5 μL of DEPC H2O using a P2 micropipette onto the center of each of the coverslips (Figure 1C).
- 8.
- Stack another coverslip onto each of the coverslips on the slide, pressing firmly down to adhere the coverslips to each other (Figure 1D).
3.3.2. Gel Embedment of Larval Brains (~30 min)
- 9.
- Using a P1000 micropipette, transfer the desired number of fixed brain samples onto the microscope slide in between the coverslips (Figure 1E).
- 10.
- Using a P200 micropipette, remove any excess buffer solution on the microscope slide.
- 11.
- If only one gel piece is to be cast from the stock gel solution, add 10 µL of 10% APS to the gel solution prepared in Step 1. If more than one gel pieces are needed, add 200 µL of the gel solution in Step 1 to a microcentrifuge tube containing 2 µL of 10% APS (prepared in Step 3). Minor fluctuations in volume do not affect performance.
CRITICAL STEP After APS is added to the gel solution, the gel polymerizes in the microcentrifuge tube within 1 min. Make sure to only proceed with this step after all other preparation steps are complete. Work quickly from this step onward until Step 18 to prevent the gel solution from polymerizing prematurely.
- 12.
- Invert or pipette three times to mix.
- 13.
- Immediately transfer up to 200 μL of gel solution onto the brain samples. The area between the coverslips should be mostly filled with gel solution (Figure 1F).
- 14.
- Under a stereomicroscope, separate and rearrange the brain samples to the desired configuration using precision tweezers.
- 15.
- Lower the last cleaned coverslip onto the samples so that the ends of the coverslip are supported by the stacked coverslips (Figure 1G).
- 16.
- Gently wipe away any excess gel solution using a lint-free tissue.
- 17.
- Gently tilt the microscope slide along the longitudinal axis to remove any trapped bubbles (Figure 1H).
- 18.
- Allow the gel to polymerize fully for 15 min.
- 19.
- Using a pair of tweezers, gently remove the coverslips surrounding the gel layer.
- 20.
- Cut the gel to the desired size using a scalpel blade. The gel should fit comfortably in the imaging area of the confocal dish.
- 21.
- Using a P200 micropipette, dislodge the gel from the microscope slide by flushing it with 5× SSCT. This can be accomplished by gently inserting the pipette tip between the edge of the gel and the microscope slide, then dispensing 5× SSCT into the gap between the gel layer and the microscope slide.
- 22.
- Transfer the gel layer to a confocal dish using a pair of tweezers (Figure 1I).
- 23.
- Rinse the gel two times with 500 μL of 5× SSCT each, at RT.
- 24.
- To cast more gel pieces, repeat Steps 4–23. Otherwise, proceed to HCR v3.0.
3.4. HCR v3.0 Signal Capture
- Incubate the samples in 500 μL of the desired primary oligo mixture for 20–24 h in RT, followed by 1 h in an incubating shaker at 37 °C with gentle agitation.
- Preheat 30% probe wash buffer to 37 °C.
- Wash the samples eight times with 500 μL of 30% probe wash buffer for 15 min each at 37 °C.
- Wash the samples two times with 500 μL of 5× SSCT for 3 min each at RT.
- Incubate the samples on a shaker in 500 µL of amplification buffer at RT with gentle agitation.
- Replace the amplification buffer with 200 µL of the corresponding secondary oligo mixture.
CRITICAL STEP From this step onward, until after confocal imaging, all steps should be performed in the dark as much as possible.
- 7.
- Incubate the samples for 20–24 h in RT, followed by 2 h in an incubating shaker at 37 °C with gentle agitation.
- 8.
- Wash the samples six times with 500 μL of 5× SSCT for 10 min each at 37 °C.
- 9.
- OPTIONAL STEP If DAPI staining is desired, add 500 μL of DAPI buffer and incubate for 20 min at RT with gentle agitation. If not, skip Steps 9 and 10.
- 10.
- OPTIONAL STEP Wash the samples two times with 500 μL of 5× SSCT for 3 min each, at RT.
- 11.
- Remove any excess buffer solution using a P200 pipette.
- 12.
- Add a small amount of 60% glycerol, at a sufficient volume to fully coat the gel surface but not enough to cause the gel to shift around in the confocal dish.
- 13.
- Wait for 15 min in RT for the samples to stabilize before imaging.
PAUSE STEP The samples can be stored in 1 mL of 60% glycerol at 4 °C for more than 2 weeks if not imaged immediately. Before imaging, allow the samples to come to RT for at least 30 min and remove any excess 60% glycerol to prevent the samples from shifting positions during imaging.
- 14.
- For imaging, the confocal dish with brains embedded in the polyacrylamide gel was mounted on an Olympus fv3000 microscope (Tokyo, Japan). Z-section (~5 µm) scans of the entire brain were carried out at 2k or 4k resolution using four channels: DAPI, AF488, AF546, and AF647. Images were later processed and stitched via Olympus floview or Imaris viewer.
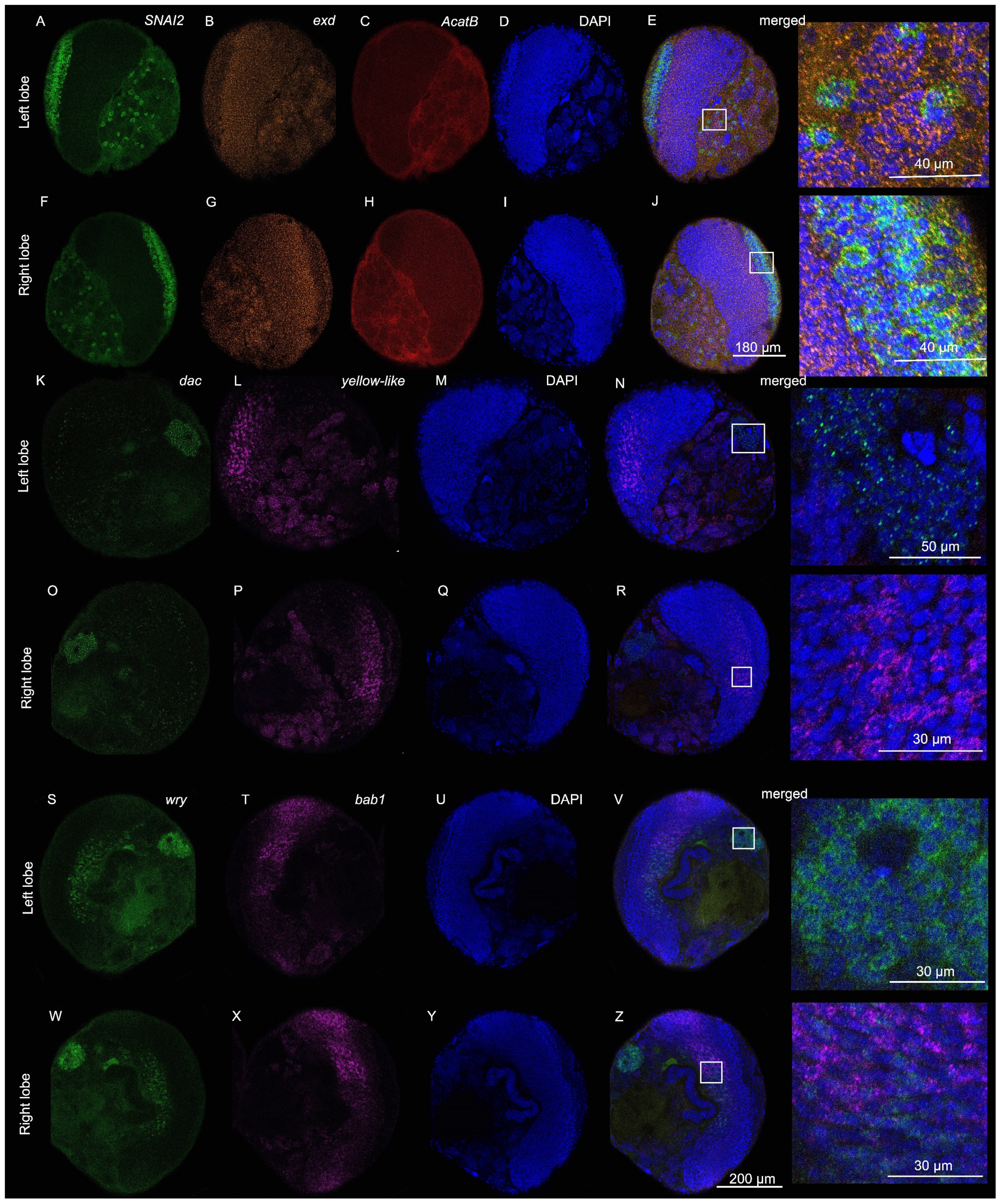
3.5. Signal Removal (from Banerjee et al., 2024 [34])
- Rinse the samples two times with 500 μL of 5× SSCT at RT.
- Wash the samples three times with 500 μL of signal wash buffer 1 for 3 min each at RT.
- Incubate the samples on an incubating shaker in 500 μL of signal removal solution for 50 min at 37 °C with gentle agitation.
- Wash the samples two times with 500 μL of signal wash buffer 2 for 15 min each at 37 °C.
- Wash the samples four times with 500 μL of signal wash buffer 1 for 3 min each at 37 °C.
- OPTIONAL STEP Image the samples to ensure all previous signals (except for DAPI staining, if any) have been removed. If signals are still present, repeat from Step 1, adjusting the incubation time in Step 3 as needed.
- Wash the samples two times with 500 µL of 5× SSCT for 3 min each, at RT.
- Incubate the samples on an incubating shaker in 500 μL of 30% probe hybridization buffer for 30 min at 37 °C with gentle agitation.
PAUSE STEP The samples can be stored in 30% probe hybridization buffer at 4 °C for up to 3 weeks.
- 9.
- For the next round of HCR, repeat from Section 3.2, Step 1.
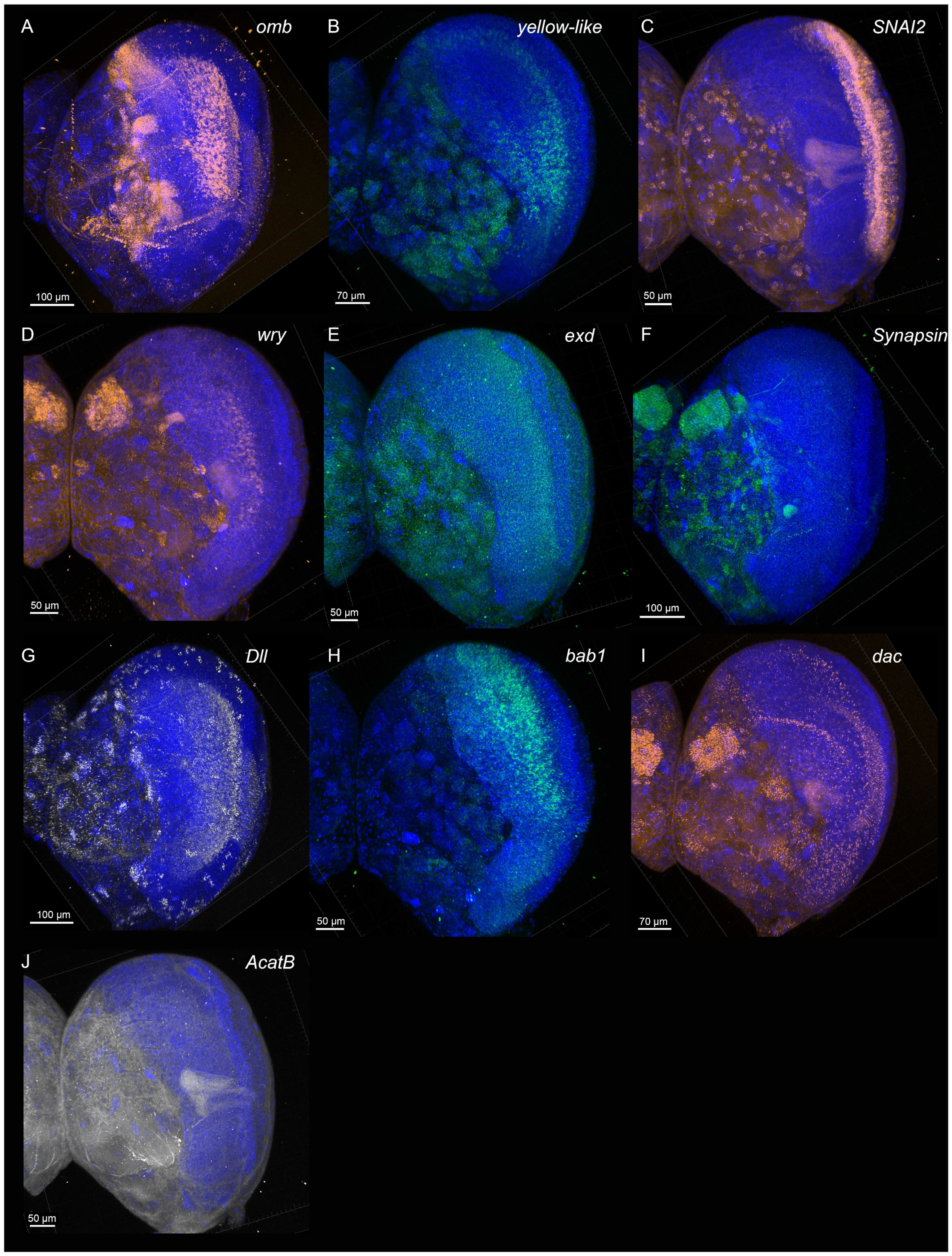
4. Expected Results
- optomotor-blind (omb)
- Distal-less (Dll)
- Synapsin
- Zinc finger protein SNAI2-like (SNAI2)
- Extradenticle (exd)
- Acetyl-CoA acetyltransferase B (AcatB)
- dachshund (dac)
- yellow-like
- bric à brac 1 (bab1)
- weary (wry)

5. Reagents Setup
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Menzel, R. Untersuchungen zum Erlernen von Spektralfarben durch die Honigbiene (Apis mellifica). Z. Vgl. Physiol. 1967, 56, 22–62. [Google Scholar] [CrossRef]
- Menzel, R. Das Gedächtnis der Honigbiene für Spektralfarben. Z. Vgl. Physiol. 1968, 60, 82–102. [Google Scholar] [CrossRef]
- Hammer, M.; Menzel, R. Learning and memory in the honeybee. J. Neurosci. 1995, 15, 1617–1630. [Google Scholar] [CrossRef]
- Nakano, R.; Takanashi, T.; Surlykke, A.; Skals, N.; Ishikawa, Y. Evolution of deceptive and true courtship songs in moths. Sci Rep. 2013, 3, 2003. [Google Scholar] [CrossRef]
- DeGennaro, M.; McBride, C.S.; Seeholzer, L.; Nakagawa, T.; Dennis, E.J.; Goldman, C.; Jasinskiene, N.; James, A.A.; Vosshall, L.B. orco mutant mosquitoes lose strong preference for humans and are not repelled by volatile DEET. Nature 2013, 498, 487–491. [Google Scholar] [CrossRef]
- Dekker, T.; Cardé, R.T. Moment-to-moment flight manoeuvres of the female yellow fever mosquito (Aedes aegypti L.) in response to plumes of carbon dioxide and human skin odour. J. Exp. Biol. 2011, 214, 3480–3494. [Google Scholar] [CrossRef]
- McMeniman, C.J.; Corfas, R.A.; Matthews, B.J.; Ritchie, S.A.; Vosshall, L.B. Multimodal Integration of Carbon Dioxide and Other Sensory Cues Drives Mosquito Attraction to Humans. Cell 2014, 156, 1060–1071. [Google Scholar] [CrossRef]
- Jové, V.; Gong, Z.; Hol, F.J.H.; Zhao, Z.; Sorrells, T.R.; Carroll, T.S.; Prakash, M.; McBride, C.S.; Vosshall, L.B. Sensory Discrimination of Blood and Floral Nectar by Aedes aegypti Mosquitoes. Neuron 2020, 108, 1163–1180.e1112. [Google Scholar] [CrossRef]
- Dorkenwald, S.; Matsliah, A.; Sterling, A.R.; Schlegel, P.; Yu, S.-C.; McKellar, C.E.; Lin, A.; Costa, M.; Eichler, K.; Yin, Y.; et al. Neuronal wiring diagram of an adult brain. Nature 2024, 634, 124–138. [Google Scholar] [CrossRef]
- Chen, S.; Li, M.; Liu, J.; Feng, Y.; Yao, J.; Shi, L.; Chen, X. Visual and olfactory sensory responses of the butterfly Papilio maackii during foraging and courtship. Entomol. Res. 2021, 51, 518–527. [Google Scholar] [CrossRef]
- Peftuloglu, D.; Bonestroo, S.; Lenders, R.; Smid, H.M.; Dicke, M.; van Loon, J.J.A.; Haverkamp, A. Olfactory learning in Pieris brassicae butterflies is dependent on the intensity of a plant-derived oviposition cue. Proc. R. Soc. B Biol. Sci. 2024, 291, 20240533. [Google Scholar] [CrossRef] [PubMed]
- Reppert, S.M.; Gegear, R.J.; Merlin, C. Navigational mechanisms of migrating monarch butterflies. Trends Neurosci. 2010, 33, 399–406. [Google Scholar] [CrossRef] [PubMed]
- Rossi, M.; Hausmann, A.E.; Thurman, T.J.; Montgomery, S.H.; Papa, R.; Jiggins, C.D.; McMillan, W.O.; Merrill, R.M. Visual mate preference evolution during butterfly speciation is linked to neural processing genes. Nat. Commun. 2020, 11, 4763. [Google Scholar] [CrossRef] [PubMed]
- Tiong, G.J.L.; Naing, L.; Ng, E.; Dion, E.; Monteiro, A. Tympanal ears mediate male–male competition, courtship and mating success in Bicyclus anynana butterflies. R. Soc. Open Sci. 2024, 11, 231386. [Google Scholar] [CrossRef]
- Xu, W. How do moth and butterfly taste?—Molecular basis of gustatory receptors in Lepidoptera. Insect Sci. 2020, 27, 1148–1157. [Google Scholar] [CrossRef]
- Yack, J.E.; Yadav, C. Vibratory Sensing and Communication in Caterpillars. In Biotremology: Physiology, Ecology, and Evolution; Hill, P.S.M., Mazzoni, V., Stritih-Peljhan, N., Virant-Doberlet, M., Wessel, A., Eds.; Springer International Publishing: Cham, Switzerland, 2022; pp. 471–491. [Google Scholar]
- Kandori, I.; Yamaki, T. Reward and non-reward learning of flower colours in the butterfly Byasa alcinous (Lepidoptera: Papilionidae). Naturwissenschaften 2012, 99, 705–713. [Google Scholar] [CrossRef]
- Kandori, I.; Yamaki, T.; Okuyama, S.-I.; Sakamoto, N.; Yokoi, T. Interspecific and intersexual learning rate differences in four butterfly species. J. Exp. Biol. 2009, 212, 3810–3816. [Google Scholar] [CrossRef]
- Kandori, I.; Ohsaki, N. The learning abilities of the white cabbage butterfly, Pieris rapae, foraging for flowers. Res. Popul. Ecol. 1996, 38, 111–117. [Google Scholar] [CrossRef]
- Dell’Aglio, D.D.; Losada, M.E.; Jiggins, C.D. Butterfly Learning and the Diversification of Plant Leaf Shape. Front. Ecol. Evol. 2016, 4, 81. [Google Scholar] [CrossRef]
- Weiss, M.R.; Papaj, D.R. Colour learning in two behavioural contexts: How much can a butterfly keep in mind? Anim. Behav. 2003, 65, 425–434. [Google Scholar] [CrossRef]
- Jones, P.L.; Agrawal, A.A. Beyond preference and performance: Host plant selection by monarch butterflies, Danaus plexippus. Oikos 2019, 128, 1092–1102. [Google Scholar] [CrossRef]
- Westerman, E.L.; Hodgins-Davis, A.; Dinwiddie, A.; Monteiro, A. Biased learning affects mate choice in a butterfly. Proc. Natl. Acad. Sci. USA 2012, 109, 10948–10953. [Google Scholar] [CrossRef] [PubMed]
- Dion, E.; Pui, L.X.; Weber, K.; Monteiro, A. Early-exposure to new sex pheromone blends alters mate preference in female butterflies and in their offspring. Nat. Commun. 2020, 11, 53. [Google Scholar] [CrossRef] [PubMed]
- Dion, E.; Toh, Y.; Zhu, D.; Monteiro, A. Butterfly brains change in morphology and in gene splicing patterns after brief pheromone exposure. bioRxiv 2024. [Google Scholar] [CrossRef]
- Cope, A.J.; Vasilaki, E.; Minors, D.; Sabo, C.; Marshall, J.A.R.; Barron, A.B. Abstract concept learning in a simple neural network inspired by the insect brain. PLoS Comput. Biol. 2018, 14, e1006435. [Google Scholar] [CrossRef]
- de Croon, G.C.H.E.; Dupeyroux, J.J.G.; Fuller, S.B.; Marshall, J.A.R. Insect-inspired AI for autonomous robots. Sci. Robot. 2022, 7, eabl6334. [Google Scholar] [CrossRef]
- Chai, H.; Cheng, W.; Jin, D.; Miao, P. Recent Progress in DNA Hybridization Chain Reaction Strategies for Amplified Biosensing. ACS Appl. Mater. Interfaces 2021, 13, 38931–38946. [Google Scholar] [CrossRef]
- Dirks, R.M.; Pierce, N.A. Triggered amplification by hybridization chain reaction. Proc. Natl. Acad. Sci. USA 2004, 101, 15275–15278. [Google Scholar] [CrossRef]
- Sgammeglia, N.; Widmer, Y.F.; Kaldun, J.C.; Fritsch, C.; Bruggmann, R.; Sprecher, S.G. Memory phase-specific genes in the Mushroom Bodies identified using CrebB-target DamID. PLoS Genet. 2023, 19, e1010802. [Google Scholar] [CrossRef]
- Ferreira, A.A.; Sieriebriennikov, B.; Whitbeck, H. HCR RNA-FISH Protocol for the Whole-Mount Brains of Drosophila and Other Insects. Available online: http://doi.org/10.17504/protocols.io.bzh5p386 (accessed on 10 March 2025).
- Choi, H.M.T.; Schwarzkopf, M.; Fornace, M.E.; Acharya, A.; Artavanis, G.; Stegmaier, J.; Cunha, A.; Pierce, N.A. Third-generation in situ hybridization chain reaction: Multiplexed, quantitative, sensitive, versatile, robust. Development 2018, 145, dev165753. [Google Scholar] [CrossRef]
- Toh, Y.P.; Dion, E.; Monteiro, A. Dissections of Larval, Pupal and Adult Butterfly Brains for Immunostaining and Molecular Analysis. Methods Protoc. 2021, 4, 53. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, T.D.; Raine, J.; Mathuru, A.S.; Chen, K.H.; Monteiro, A. Spatial mRNA profiling using Rapid Amplified Multiplexed-FISH (RAM-FISH). bioRxiv 2024. [Google Scholar] [CrossRef]
- De Pasqual, C.; Groot, A.T.; Mappes, J.; Burdfield-Steel, E. Evolutionary importance of intraspecific variation in sex pheromones. Trends Ecol. Evol. 2021, 36, 848–859. [Google Scholar] [CrossRef] [PubMed]
- Ernst, D.A.; Westerman, E.L. Stage- and sex-specific transcriptome analyses reveal distinctive sensory gene expression patterns in a butterfly. BMC Genom. 2021, 22, 584. [Google Scholar] [CrossRef]
- Gowri, V.; Dion, E.; Viswanath, A.; Piel, F.M.; Monteiro, A. Transgenerational inheritance of learned preferences for novel host plant odors in Bicyclus anynana butterflies. Evolution 2019, 73, 2401–2414. [Google Scholar] [CrossRef]
- Poeck, B.; Hofbauer, A.; Pflugfelder, G.O. Expression of the Drosophila optomotor-blind gene transcript in neuronal and glial cells of the developing nervous system. Development 1993, 117, 1017–1029. [Google Scholar] [CrossRef]
- Plavicki, J.S.; Squirrell, J.M.; Eliceiri, K.W.; Boekhoff-Falk, G. Expression of the Drosophila homeobox gene, Distal-less, supports an ancestral role in neural development. Dev. Dyn. 2016, 245, 87–95. [Google Scholar] [CrossRef]
- Fornasiero, E.F.; Bonanomi, D.; Benfenati, F.; Valtorta, F. The role of synapsins in neuronal development. Cell. Mol. Life Sci. 2010, 67, 1383–1396. [Google Scholar] [CrossRef]
- Greengard, P.; Valtorta, F.; Czernik, A.J.; Benfenati, F. Synaptic Vesicle Phosphoproteins and Regulation of Synaptic Function. Science 1993, 259, 780–785. [Google Scholar] [CrossRef]
- Akbergenova, Y.; Bykhovskaia, M. Synapsin regulates vesicle organization and activity-dependent recycling at Drosophila motor boutons. Neuroscience 2010, 170, 441–452. [Google Scholar] [CrossRef]
- Pérez-Mancera, P.A.; González-Herrero, I.; Maclean, K.; Turner, A.M.; Yip, M.Y.; Sánchez-Martín, M.; García, J.L.; Robledo, C.; Flores, T.; Gutiérrez-Adán, A.; et al. SLUG (SNAI2) overexpression in embryonic development. Cytogenet. Genome Res. 2006, 114, 24–29. [Google Scholar] [CrossRef]
- Rembold, M.; Ciglar, L.; Yáñez-Cuna, J.O.; Zinzen, R.P.; Girardot, C.; Jain, A.; Welte, M.A.; Stark, A.; Leptin, M.; Furlong, E.E.M. A conserved role for Snail as a potentiator of active transcription. Genes Dev. 2014, 28, 167–181. [Google Scholar] [CrossRef]
- Rauskolb, C.; Peifer, M.; Wieschaus, E. extradenticle, a regulator of homeotic gene activity, is a homolog of the homeobox-containing human proto-oncogene pbx1. Cell 1993, 74, 1101–1112. [Google Scholar] [CrossRef]
- Rauskolb, C.; Smith, K.M.; Peifer, M.; Wieschaus, E. extradenticle determines segmental identities throughout Drosophila development. Development 1995, 121, 3663–3673. [Google Scholar] [CrossRef]
- Pinsonneault, J.; Florence, B.; Vaessin, H.; McGinnis, W. A model for extradenticle function as a switch that changes HOX proteins from repressors to activators. EMBO J. 1997, 16, 2032–2042. [Google Scholar] [CrossRef]
- Fujii, T.; Ito, K.; Katsuma, S.; Nakano, R.; Shimada, T.; Ishikawa, Y. Molecular and functional characterization of an acetyl-CoA acetyltransferase from the adzuki bean borer moth Ostrinia scapulalis (Lepidoptera: Crambidae). Insect Biochem. Mol. Biol. 2010, 40, 74–78. [Google Scholar] [CrossRef]
- Wu, C.-Y.; Hu, I.C.; Yang, Y.-C.; Ding, W.-C.; Lai, C.-H.; Lee, Y.-Z.; Liu, Y.-C.; Cheng, H.-C.; Lyu, P.-C. An essential role of acetyl coenzyme A in the catalytic cycle of insect arylalkylamine N-acetyltransferase. Commun. Biol. 2020, 3, 441. [Google Scholar] [CrossRef]
- Han, Q.; Robinson, H.; Ding, H.; Christensen, B.M.; Li, J. Evolution of insect arylalkylamine N-acetyltransferases: Structural evidence from the yellow fever mosquito, Aedes aegypti. Proc. Natl. Acad. Sci. USA 2012, 109, 11669–11674. [Google Scholar] [CrossRef]
- Tavsanli, B.C.; Ostrin, E.J.; Burgess, H.K.; Middlebrooks, B.W.; Pham, T.A.; Mardon, G. Structure–function analysis of the Drosophila retinal determination protein Dachshund. Dev. Biol. 2004, 272, 231–247. [Google Scholar] [CrossRef]
- Kurusu, M.; Nagao, T.; Walldorf, U.; Flister, S.; Gehring, W.J.; Furukubo-Tokunaga, K. Genetic control of development of the mushroom bodies, the associative learning centers in the Drosophila brain, by the eyeless, twin of eyeless, and dachshund genes. Proc. Natl. Acad. Sci. USA 2000, 97, 2140–2144. [Google Scholar] [CrossRef]
- Macias-Muñoz, A.; Smith, G.; Monteiro, A.; Briscoe, A.D. Transcriptome-Wide Differential Gene Expression in Bicyclus anynana Butterflies: Female Vision-Related Genes Are More Plastic. Mol. Biol. Evol. 2016, 33, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.-L.; Han, W.-K.; Ze, L.-J.; Peng, Y.-C.; Yang, Y.-L.; Zhang, J.; Yan, Q.; Dong, S.-L. Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR-Associated Protein 9 Mediated Knockout Reveals Functions of the yellow-y Gene in Spodoptera litura. Front. Physiol. 2020, 11, 615391. [Google Scholar] [CrossRef]
- Matsuoka, Y.; Monteiro, A. Melanin Pathway Genes Regulate Color and Morphology of Butterfly Wing Scales. Cell Rep. 2018, 24, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Connahs, H.; Tan, E.J.; Ter, Y.T.; Dion, E.; Matsuoka, Y.; Bear, A.; Monteiro, A. The yellow gene regulates behavioural plasticity by repressing male courtship in Bicyclus anynana butterflies. Proc. R. Soc. B Biol. Sci. 2022, 289, 20212665. [Google Scholar] [CrossRef]
- Zwarts, L.; Vanden Broeck, L.; Cappuyns, E.; Ayroles, J.F.; Magwire, M.M.; Vulsteke, V.; Clements, J.; Mackay, T.F.C.; Callaerts, P. The genetic basis of natural variation in mushroom body size in Drosophila melanogaster. Nat. Commun. 2015, 6, 10115. [Google Scholar] [CrossRef]
- Bahrampour, S.; Thor, S. The Five Faces of Notch Signalling During Drosophila melanogaster Embryonic CNS Development. In Notch Signaling in Embryology and Cancer: Notch Signaling in Embryology; Reichrath, J., Reichrath, S., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 39–58. [Google Scholar]
- Kim, I.-M.; Wolf, M.J.; Rockman, H.A. Gene Deletion Screen for Cardiomyopathy in Adult Drosophila Identifies a New Notch Ligand. Circ. Res. 2010, 106, 1233–1243. [Google Scholar] [CrossRef]
- Egger, B.; Gold, K.S.; Brand, A.H. Regulating the balance between symmetric and asymmetric stem cell division in the developing brain. Fly 2011, 5, 237–241. [Google Scholar] [CrossRef] [PubMed]
- Ito, K.; Shinomiya, K.; Ito, M.; Armstrong, J.D.; Boyan, G.; Hartenstein, V.; Harzsch, S.; Heisenberg, M.; Homberg, U.; Jenett, A.; et al. A Systematic Nomenclature for the Insect Brain. Neuron 2014, 81, 755–765. [Google Scholar] [CrossRef]
- Sehadová, H.; Podlahová, Š.; Reppert, S.M.; Sauman, I. 3D reconstruction of larval and adult brain neuropils of two giant silk moth species: Hyalophora cecropia and Antheraea pernyi. J. Insect Physiol. 2023, 149, 104546. [Google Scholar] [CrossRef]
- Wang, X.; Zhai, Y.; Zheng, H. Deciphering the cellular heterogeneity of the insect brain with single-cell RNA sequencing. Insect Sci. 2024, 31, 314–327. [Google Scholar] [CrossRef]


| Problem | Possible Reason | Solution |
|---|---|---|
| The gel is either too soft or does not polymerize. | Inhibition of polymerization reaction by oxygen. | Mix gel solution and gel components by gentle inversion or pipetting only. Work quickly while the gel solution on the microscope slide is exposed (i.e., without the top coverslip). |
| Regents have degraded in quality. | Refrain from refreezing thawed aliquots of 10% APS. Once thawed, store aliquots at 4 °C for up to 1 week. Alternatively, make 10% APS fresh. Check all reagents, especially TEMED and APS, to ensure that they have not degraded in quality. | |
| The number of initiators added is too low. | Titrate up the amount of TEMED and 10% APS added to the gel solution in increments of 1 μL per 1 mL of gel solution. | |
| Gel polymerizes too fast. | The number of initiators or the gel percentage is too high. | Titrate down the amount of 40% acrylamide solution added to the final gel solution in increments of 25 μL per 1 mL of gel solution (1% acrylamide decrease). Titrate down the amount of TEMED and 10% APS added to the gel solution in increments of 1 μL per 1 mL of gel solution. |
| Fluorescence is only detected on the surface of the brain. | The gel is too thick. | Reduce the stack height of supporting coverslips from 2 to 1. If this is not possible, attempt to reduce the gel concentration by titrating down the amount of 40% acrylamide solution added to the final gel solution in increments of 25 μL per 1 mL of gel solution (1% acrylamide decrease). |
| Reagents | Components | Details |
|---|---|---|
| Acrylamide solution, 40% |
| For 1 mL:
|
| Ammonium persulfate, 10%(10% APS) |
| For 1 mL:
|
| Amplification buffer |
| For 40 mL:
|
| DAPI buffer |
| For 1 mL:
|
| DAPI in DMSO, 1 mg/mL |
| For 1 mL:
|
| DEPC H2O |
| For 1 L:
|
| Detergent solution |
| For 50 mL:
|
| Dextran sulfate, 50% |
| For 40 mL:
|
| EDTA, 0.5 M, pH 8.0 |
| For 500 mL:
|
| Gel solution (8% polyacrylamide) |
| For 1 mL:
|
| Glycerol, 60% |
| For 10 mL:∙ 6 mL glycerol∙ Top up with DEPC H2O |
| Phosphate-buffered formaldehyde, 4% (4% PFA) |
| For 500 mL:
|
| Phosphate-buffered saline, 1×(1× PBS) |
| For 50 mL:
|
| Phosphate-buffered saline, 10×(10× PBS) |
| For 1 L:
|
| Phosphate-buffered saline with TWEEN 20, 1×(1× PBST) |
| For 50 mL:
|
| Primary oligo mixture |
| For each primary oligo set, up to 10 pairs of oligonucleotides, each at 100 μM, were pooled into a single master mix using 100 μL of each oligo. For 1 mL:
|
| Probe hybridization buffer, 30% |
| For 40 mL:
|
| Probe wash buffer, 30% |
| For 40 mL:
|
| Saline-sodium citrate buffer, 5× (5× SSC) |
| For 40 mL:
|
| Saline-sodium citrate buffer, 20×(20× SSC) |
| For 1 L:
|
| Saline-sodium citrate buffer with TWEEN 20, 5×(5× SSCT) |
| For 40 mL:
|
| SDS, 10% |
| For 50 mL:
|
| Secondary oligo mixture |
| Snap-cool each stock H1 and H2 hairpins separately by heating to 95 °C for 90 s, then allow to cool to RT in a dark environment for 30 min. For 600 μL:
|
| Signal removal solution |
| For 1 mL:
|
| Signal wash buffer 1 |
| For 10 mL:
|
| Signal wash buffer 2 |
| For 1 mL:
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Banerjee, T.D.; Zhang, L.; Monteiro, A. Mapping Gene Expression in Whole Larval Brains of Bicyclus anynana Butterflies. Methods Protoc. 2025, 8, 31. https://doi.org/10.3390/mps8020031
Banerjee TD, Zhang L, Monteiro A. Mapping Gene Expression in Whole Larval Brains of Bicyclus anynana Butterflies. Methods and Protocols. 2025; 8(2):31. https://doi.org/10.3390/mps8020031
Chicago/Turabian StyleBanerjee, Tirtha Das, Linwan Zhang, and Antónia Monteiro. 2025. "Mapping Gene Expression in Whole Larval Brains of Bicyclus anynana Butterflies" Methods and Protocols 8, no. 2: 31. https://doi.org/10.3390/mps8020031
APA StyleBanerjee, T. D., Zhang, L., & Monteiro, A. (2025). Mapping Gene Expression in Whole Larval Brains of Bicyclus anynana Butterflies. Methods and Protocols, 8(2), 31. https://doi.org/10.3390/mps8020031








