Establishing the Signal above the Noise: Accounting for an Environmental Background in the Detection and Quantification of Salmonid Environmental DNA
Abstract
:1. Introduction
2. Materials and Methods
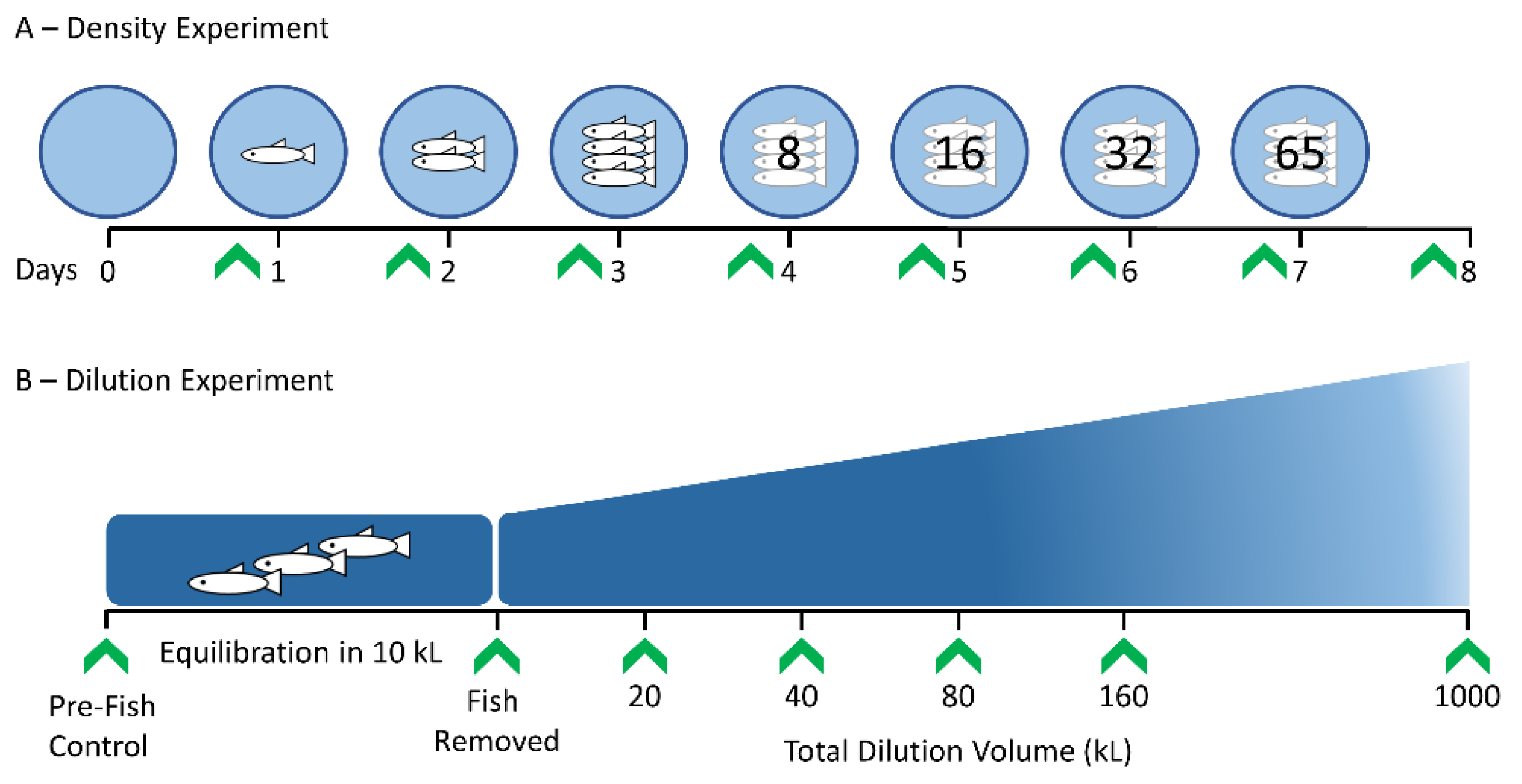
2.1. Experimental Study System and Design
2.2. eDNA Sample Collection and Filtration
2.3. Lab Sample Processing
2.4. Coho Salmon eDNA Assay (eONKI4) Design and Validation
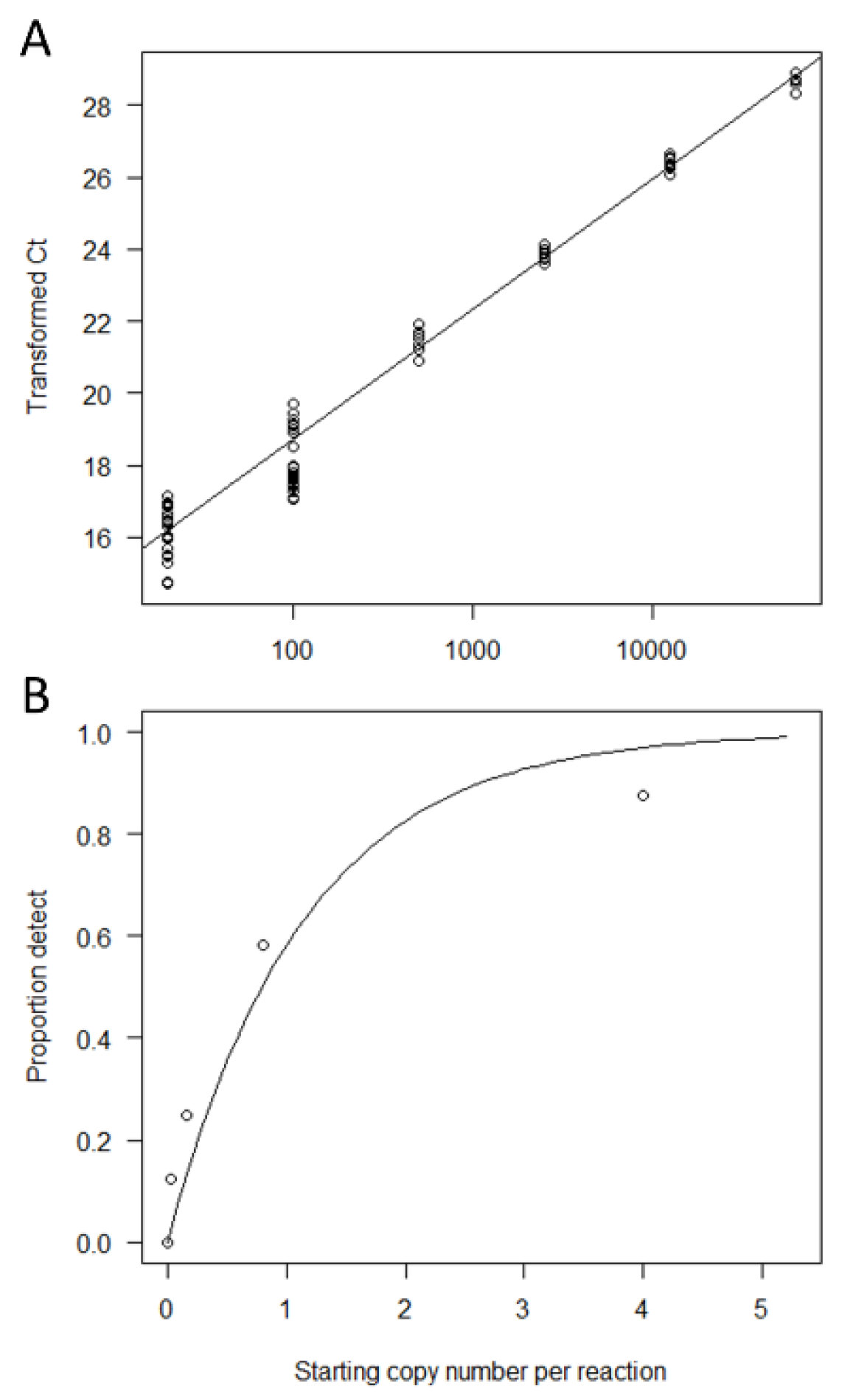
2.5. Generation of Standard Curves
2.6. Statistical Analyses of Fish Density and Dilution Experiments
3. Results
3.1. qPCR Salmon eDNA Assay Design and Calibration to Copy Number
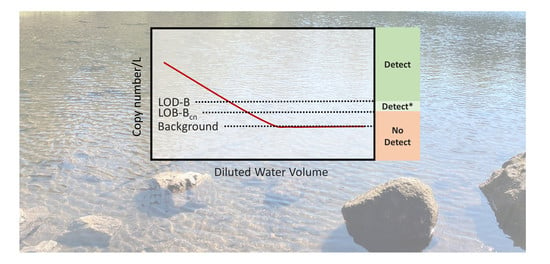
3.2. Determination of the LOB-B and LOD-B
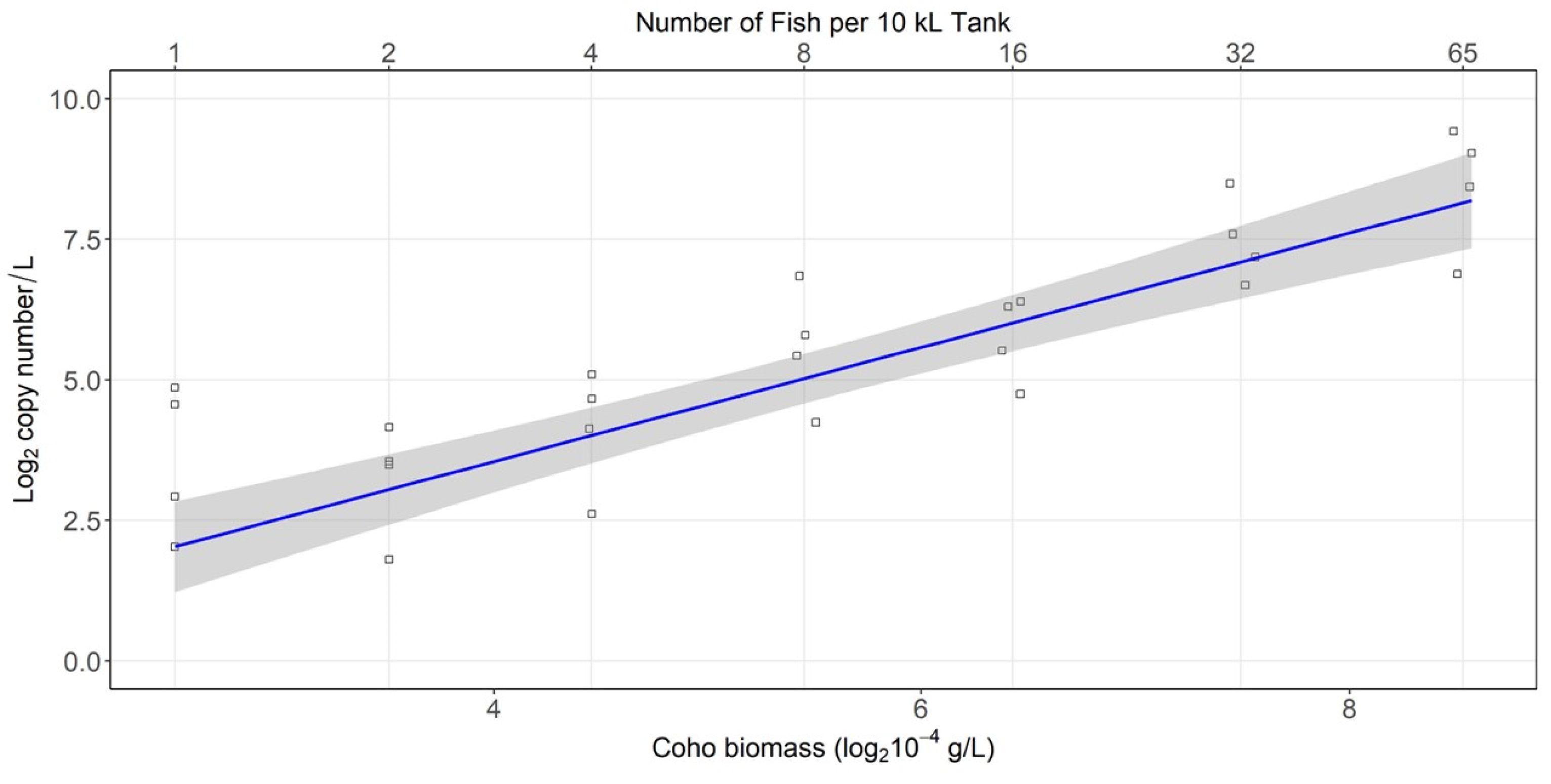
3.3. Density Experiment Results
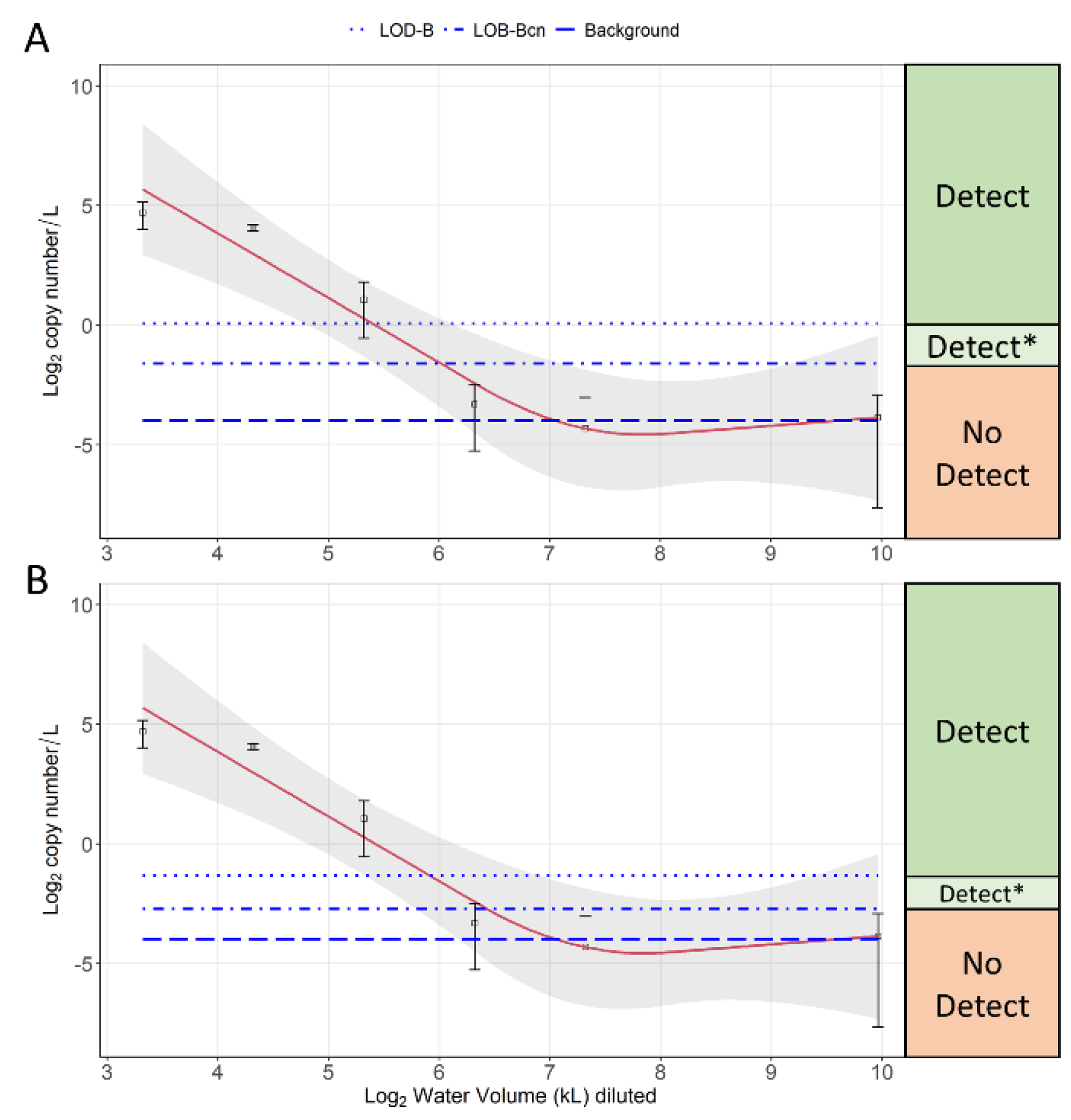
3.4. Dilution Experiment Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Beja-Pereira, A.; Oliveira, R.; Alves, P.C.; Schwartz, M.K.; Luikart, G. Advancing ecological understandings through technological transformations in noninvasive genetics. Mol. Ecol. Resour. 2009, 9, 1279–1301. [Google Scholar] [CrossRef] [PubMed]
- Imbery, J.J.; Buday, C.; Miliano, R.C.; Shang, D.; Round, J.M.; Kwok, H.; Van Aggelen, G.; Helbing, C.C. Evaluation of gene bioindicators in the liver and caudal fin of juvenile Pacific coho salmon in response to low sulfur marine diesel seawater-accommodated fraction exposure. Environ. Sci. Technol. 2019, 53, 1627–1638. [Google Scholar] [CrossRef]
- Ficetola, G.F.; Miaud, C.; Pompanon, F.; Taberlet, P. Species detection using environmental DNA from water samples. Biol. Lett. 2008, 4, 423–425. [Google Scholar] [CrossRef] [PubMed]
- Levi, T.; Allen, J.M.; Bell, D.; Joyce, J.; Russell, J.R.; Tallmon, D.A.; Vulstek, S.C.; Yang, C.; Yu, D.W. Environmental DNA for the enumeration and management of Pacific salmon. Mol. Ecol. Resour. 2019, 19, 597–608. [Google Scholar] [CrossRef] [PubMed]
- Piaggio, A.J.; Engeman, R.M.; Hopken, M.W.; Humphrey, J.S.; Keacher, K.L.; Bruce, W.E.; Avery, M.L. Detecting an elusive invasive species: A diagnostic PCR to detect Burmese python in Florida waters and an assessment of persistence of environmental DNA. Mol. Ecol. Resour. 2014, 14, 374–380. [Google Scholar] [CrossRef]
- Rourke, M.L.; Fowler, A.M.; Hughes, J.M.; Broadhurst, M.K.; DiBattista, J.D.; Fielder, S.; Wilkes Walburn, J.; Furlan, E.M. Environmental DNA (eDNA) as a tool for assessing fish biomass: A review of approaches and future considerations for resource surveys. Environ. DNA 2022, 4, 3–33. [Google Scholar] [CrossRef]
- Spear, M.J.; Embke, H.S.; Krysan, P.J.; Vander Zanden, M.J. Application of eDNA as a tool for assessing fish population abundance. Environ. DNA 2020, 3, 83–91. [Google Scholar] [CrossRef]
- Thomsen, P.F.; Kielgast, J.; Iversen, L.L.; Wiuf, C.; Rasmussen, M.; Gilbert, M.T.; Orlando, L.; Willerslev, E. Monitoring endangered freshwater biodiversity using environmental DNA. Mol. Ecol. 2012, 21, 2565–2573. [Google Scholar] [CrossRef]
- Tillotson, M.D.; Kelly, R.P.; Duda, J.J.; Hoy, M.; Kralj, J.; Quinn, T.P. Concentrations of environmental DNA (eDNA) reflect spawning salmon abundance at fine spatial and temporal scales. Biol. Conserv. 2018, 220, 1–11. [Google Scholar] [CrossRef]
- Evans, N.T.; Shirey, P.D.; Wieringa, J.G.; Mahon, A.R.; Lamberti, G.A. Comparative cost and effort of fish distribution detection via environmental DNA analysis and electrofishing. Fisheries 2017, 42, 90–99. [Google Scholar] [CrossRef]
- Minamoto, T.; Yamanaka, H.; Takahara, T.; Honjo, M.N.; Kawabata, Z.i. Surveillance of fish species composition using environmental DNA. Limnology 2011, 13, 193–197. [Google Scholar] [CrossRef]
- Barnes, M.A.; Turner, C.R.; Jerde, C.L.; Renshaw, M.A.; Chadderton, W.L.; Lodge, D.M. Environmental conditions influence eDNA persistence in aquatic systems. Environ. Sci Technol. 2014, 48, 1819–1827. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, C.S.; Turner, C.R.; Deiner, K.; Klymus, K.E.; Thomsen, P.F.; Murphy, M.A.; Spear, S.F.; McKee, A.; Oyler-McCance, S.J.; Cornman, R.S.; et al. Critical considerations for the application of environmental DNA methods to detect aquatic species. Methods Ecol. Evol. 2016, 7, 1299–1307. [Google Scholar] [CrossRef]
- Strickler, K.M.; Fremier, A.K.; Goldberg, C.S. Quantifying effects of UV-B, temperature, and pH on eDNA degradation in aquatic microcosms. Biol. Conserv. 2015, 183, 85–92. [Google Scholar] [CrossRef]
- Yates, M.C.; Cristescu, M.E.; Derry, A.M. Integrating physiology and environmental dynamics to operationalize environmental DNA (eDNA) as a means to monitor freshwater macro-organism abundance. Mol. Ecol. 2021, 30, 6531–6550. [Google Scholar] [CrossRef]
- Burian, A.; Mauvisseau, Q.; Bulling, M.; Domisch, S.; Qian, S.; Sweet, M. Improving the reliability of eDNA data interpretation. Mol. Ecol Resour 2021, 21, 1422–1433. [Google Scholar] [CrossRef]
- Kelly, R.P.; Port, J.A.; Yamahara, K.M.; Martone, R.G.; Lowell, N.; Thomsen, P.F.; Mach, M.E.; Bennett, M.; Prahler, E.; Caldwell, M.R.; et al. Environmental monitoring. Harnessing DNA to improve environmental management. Sci. Am. Assoc. Adv. Sci. 2014, 344, 1455–1456. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Forootan, A.; Sjoback, R.; Bjorkman, J.; Sjogreen, B.; Linz, L.; Kubista, M. Methods to determine limit of detection and limit of quantification in quantitative real-time PCR (qPCR). Biomol. Detect. Quantif. 2017, 12, 1–6. [Google Scholar] [CrossRef]
- Klymus, K.E.; Merkes, C.M.; Allison, M.J.; Goldberg, C.S.; Helbing, C.C.; Hunter, M.E.; Jackson, C.A.; Lance, R.F.; Mangan, A.M.; Monroe, E.M.; et al. Reporting the limits of detection and quantification for environmental DNA assays. Environ. DNA 2020, 2, 271–282. [Google Scholar] [CrossRef] [Green Version]
- Lesperance, M.L.; Allison, M.J.; Bergman, L.C.; Hocking, M.D.; Helbing, C.C. A statistical model for calibration and computation of detection and quantification limits for low copy number environmental DNA samples. Environ. DNA 2021, 3, 970–981. [Google Scholar] [CrossRef]
- Gagné, N.; Bernatchez, L.; Bright, D.; Côté, G.; Coulson, M.; Gurney, K.; Hanner, R.; Helbing, C.C.; Hobbs, J.; Hocking, M.; et al. Environmental DNA (eDNA) Reporting Requirements and Terminology; Canadian Standards Association: Toronto, ON, Canada, 2021; CSA W214:21; 31p. [Google Scholar]
- Doi, H.; Inui, R.; Akamatsu, Y.; Kanno, K.; Yamanaka, H.; Takahara, T.; Minamoto, T. Environmental DNA analysis for estimating the abundance and biomass of stream fish. Freshw. Biol. 2017, 62, 30–39. [Google Scholar] [CrossRef]
- Dejean, T.; Valentini, A.; Duparc, A.; Pellier-Cuit, S.; Pompanon, F.; Taberlet, P.; Miaud, C. Persistence of environmental DNA in freshwater ecosystems. PLoS ONE 2011, 6, e23398. [Google Scholar] [CrossRef] [PubMed]
- Kelly, R.P.; Port, J.A.; Yamahara, K.M.; Crowder, L.B. Using environmental DNA to census marine fishes in a large mesocosm. PLoS ONE 2014, 9, e86175. [Google Scholar] [CrossRef] [PubMed]
- Barnes, M.A.; Turner, C.R. The ecology of environmental DNA and implications for conservation genetics. Conserv. Genet. 2015, 17, 1–17. [Google Scholar] [CrossRef]
- Thomsen, P.F.; Willerslev, E. Environmental DNA—An emerging tool in conservation for monitoring past and present biodiversity. Biol. Conserv. 2015, 183, 4–18. [Google Scholar] [CrossRef]
- McKelvey, K.S.; Young, M.K.; Knotek, W.L.; Carim, K.J.; Wilcox, T.M.; Padgett-Stewart, T.M.; Schwartz, M.K. Sampling large geographic areas for rare species using environmental DNA: A study of bull trout Salvelinus confluentus occupancy in western Montana. J. Fish Biol. 2016, 88, 1215–1222. [Google Scholar] [CrossRef]
- Rees, H.C.; Baker, C.A.; Gardner, D.S.; Maddison, B.C.; Gough, K.C. The detection of great crested newts year round via environmental DNA analysis. BMC Res. Notes 2017, 10, 327. [Google Scholar] [CrossRef]
- Darling, J.A.; Jerde, C.L.; Sepulveda, A.J. What do you mean by false positive? Environ. DNA 2021, 3, 879–883. [Google Scholar] [CrossRef]
- Ficetola, G.F.; Taberlet, P.; Coissac, E. How to limit false positives in environmental DNA and metabarcoding? Mol. Ecol. Resour. 2016, 16, 604–607. [Google Scholar] [CrossRef] [Green Version]
- Lahoz-Monfort, J.J.; Guillera-Arroita, G.; Tingley, R. Statistical approaches to account for false-positive errors in environmental DNA samples. Mol. Ecol. Resour. 2016, 16, 673–685. [Google Scholar] [CrossRef] [PubMed]
- Sepulveda, A.J.; Hutchins, P.R.; Forstchen, M.; McKeefry, M.N.; Swigris, A.M. The Elephant in the Lab (and Field): Contamination in Aquatic Environmental DNA Studies. Front. Ecol. Evol. 2020, 8, 609973. [Google Scholar] [CrossRef]
- Prince, A.M.; Andrus, L. PCR: How to kill unwanted DNA. Biotechniques 1992, 12, 358–360. [Google Scholar] [PubMed]
- Walsh, D.A.; Zaikova, E.; Hallam, S.J. Large volume (20L+) filtration of coastal seawater samples. J. Vis. Exp. 2009, 28, 1161. [Google Scholar] [CrossRef] [PubMed]
- Jerde, C.L.; Mahon, A.R.; Chadderton, W.L.; Lodge, D.M. “Sight-unseen” detection of rare aquatic species using environmental DNA. Conserv. Lett. 2011, 4, 150–157. [Google Scholar] [CrossRef]
- Hobbs, J.; Round, J.M.; Allison, M.J.; Helbing, C.C. Expansion of the known distribution of the coastal tailed frog, Ascaphus truei, in British Columbia, Canada, using robust eDNA detection methods. PLoS ONE 2019, 14, 16. [Google Scholar] [CrossRef]
- Veldhoen, N.; Hobbs, J.; Ikonomou, G.; Hii, M.; Lesperance, M.; Helbing, C.C. Implementation of novel design features for qPCR-based eDNA assessment. PLoS ONE 2016, 11, e0164907. [Google Scholar] [CrossRef]
- Lavagnini, I.; Magno, F. A statistical overview on univariate calibration, inverse regression, and detection limits: Application to gas chromatography/mass spectrometry technique. Mass Spectrom. Rev. 2007, 26, 1–18. [Google Scholar] [CrossRef]
- Toms, J.D.; Lesperance, M. Piecewise regression: A tool for identifying ecological thresholds. Ecology 2003, 84, 2034–2041. [Google Scholar] [CrossRef]
- Rosenfeld, J.; Porter, M.; Parkinson, E. Habitat factors affecting the abundance and distribution of juvenile cutthroat trout (Oncorhynchus clarki) and coho salmon (Oncorhynchus kisutch). Can. J. Fish Aquat. Sci. 2000, 57, 766–774. [Google Scholar] [CrossRef]
- Wilcox, T.M.; McKelvey, K.S.; Young, M.K.; Sepulveda, A.J.; Shepard, B.B.; Jane, S.F.; Whiteley, A.R.; Lowe, W.H.; Schwartz, M.K. Understanding environmental DNA detection probabilities: A case study using a stream-dwelling char Salvelinus fontinalis. Biol. Conserv. 2016, 194, 209–216. [Google Scholar] [CrossRef]
- Curtis, A.N.; Tiemann, J.S.; Douglass, S.A.; Davis, M.A.; Larson, E.R.; Yang, J. High stream flows dilute environmental DNA (eDNA) concentrations and reduce detectability. Divers. Distrib. 2020, 27, 1918–1931. [Google Scholar] [CrossRef]
- Troth, C.R.; Sweet, M.J.; Nightingale, J.; Burian, A. Seasonality, DNA degradation and spatial heterogeneity as drivers of eDNA detection dynamics. Sci. Total Environ. 2021, 768, 144466. [Google Scholar] [CrossRef] [PubMed]
- Collins, R.A.; Wangensteen, O.S.; O’Gorman, E.J.; Mariani, S.; Sims, D.W.; Genner, M.J. Persistence of environmental DNA in marine systems. Commun. Biol. 2018, 1, 185. [Google Scholar] [CrossRef] [PubMed]
- Saito, T.; Doi, H. A model and simulation of the influence of temperature and amplicon length on environmental DNA degradation rates: A meta-analysis approach. Front. Ecol. Evol. 2021, 9, 623831. [Google Scholar] [CrossRef]
- Eichmiller, J.J.; Best, S.E.; Sorensen, P.W. Effects of temperature and trophic state on degradation of environmental DNA in lake water. Environ. Sci. Technol. 2016, 50, 1859–1867. [Google Scholar] [CrossRef]
- Seymour, M.; Durance, I.; Cosby, B.J.; Ransom-Jones, E.; Deiner, K.; Ormerod, S.J.; Colbourne, J.K.; Wilgar, G.; Carvalho, G.R.; de Bruyn, M.; et al. Acidity promotes degradation of multi-species environmental DNA in lotic mesocosms. Commun. Biol. 2018, 1, 4. [Google Scholar] [CrossRef]
- Tsuji, S.; Ushio, M.; Sakurai, S.; Minamoto, T.; Yamanaka, H. Water temperature-dependent degradation of environmental DNA and its relation to bacterial abundance. PLoS ONE 2017, 12, e0176608. [Google Scholar] [CrossRef]
- Mauvisseau, Q.; Davy-Bowker, J.; Bulling, M.; Brys, R.; Neyrinck, S.; Troth, C.; Sweet, M. Combining ddPCR and environmental DNA to improve detection capabilities of a critically endangered freshwater invertebrate. Sci. Rep. 2019, 9, 14064. [Google Scholar] [CrossRef]
- Buxton, A.; Matechou, E.; Griffin, J.; Diana, A.; Griffiths, R.A. Optimising sampling and analysis protocols in environmental DNA studies. Sci. Rep. 2021, 11, 11637. [Google Scholar] [CrossRef]
- Ficetola, G.F.; Pansu, J.; Bonin, A.; Coissac, E.; Giguet-Covex, C.; De Barba, M.; Gielly, L.; Lopes, C.M.; Boyer, F.; Pompanon, F.; et al. Replication levels, false presences and the estimation of the presence/absence from eDNA metabarcoding data. Mol. Ecol. Resour. 2015, 15, 543–556. [Google Scholar] [CrossRef] [PubMed]
- Chambert, T.; Miller, D.A.W.; Nichols, J.D. Modeling false positive detections in species occurrence data under different study designs. Ecology 2015, 96, 332–339. [Google Scholar] [CrossRef] [PubMed]
| Sequence Type | Sequence (5′→3′) |
|---|---|
| Forward Primer | CCCTTTTGACCTCACAGAG |
| Reverse Primer | GGTTCAGGGCAGTTAGTTCT |
| Probe | FAM- TGCCCTCTTTTTCCTAGCCGAGTACG-ZEN/Iowa Black FQ |
| Amplicon | CCCTTTTGACCTCACAGAGGGGGAGTCAGAATTAGTCTCCGGATCAATGTAGAA TATGCTGGAGGGCCCTTTGCCCTCTTTTTCCTAGCCGAGTACGCTAACATCCTTCTAA TAAATACACTCTCAGCCATCCTATTCCTAGGCGCATCCCACATCCCCGCTTTAC CAGAACTAACTGCCCTGAACC |
| Species Name | Common Name | Species Abbreviation | Percent Detection by eONKI4 |
|---|---|---|---|
| Oncorhynchus kisutch | Coho Salmon | ONKI | 100% |
| Oncorhynchus tschawytscha | Chinook Salmon | ONTS | 0% |
| Oncorhynchus clarkii | Cutthroat Trout | ONCL | 0% |
| Oncorhynchus gorbuscha | Pink Salmon | ONGO | 0% |
| Oncorhynchus keta | Chum Salmon | ONKE | 0% |
| Oncorhynchus mykiss | Rainbow Trout | ONMY | 0% |
| Oncorhynchus nerka | Sockeye Salmon | ONNE | 0% |
| Thymallus arcticus | Arctic Grayling | THAR | 0% |
| Lithobates (Rana) catesbeiana | American Bullfrog | LICA | 0% |
| Homo sapiens | Human | HOSA | 0% |
| eONKI4 Limit of Detection | Number of Technical Replicates | |||||||
|---|---|---|---|---|---|---|---|---|
| 3 | 8 | 16 | 24 | 32 | 48 | 64 | 96 | |
| LOD | 1.15 | 0.43 | 0.21 | 0.14 | 0.11 | 0.07 | 0.05 | 0.04 |
| LOD_lower | 0.85 | 0.32 | 0.16 | 0.11 | 0.08 | 0.05 | 0.04 | 0.03 |
| LOD_upper | 1.77 | 0.66 | 0.33 | 0.22 | 0.17 | 0.11 | 0.08 | 0.06 |
| Limits | Number of Technical Replicates | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 3 | 8 | 16 | 24 | 32 | 40 | 48 | 64 | 96 | |
| LOB-B 1 | 1 | 2 | 3 | 3 | 4 | 5 | 5 | 7 | 9 |
| LOB-Bcn 2 | 0.46 | 0.33 | 0.24 | 0.15 | 0.15 | 0.15 | 0.13 | 0.13 | 0.11 |
| LOD-B 2 | 2.29 | 1.05 | 0.62 | 0.40 | 0.35 | 0.32 | 0.27 | 0.25 | 0.20 |
| LOD-B_lower 2 | 1.69 | 0.78 | 0.46 | 0.29 | 0.26 | 0.24 | 0.20 | 0.18 | 0.15 |
| LOD-B_upper 2 | 3.55 | 1.62 | 0.96 | 0.61 | 0.54 | 0.50 | 0.41 | 0.39 | 0.30 |
| Dilution Volume (kL) Treatment | Tank Number | Transformed Ct Estimate | Estimated Number of Copies per Reaction | Number of Samples per Tank | Number of Technical Replicates per Tank | Number of Detects for All Technical Replicates | Tank Detection | Treatment Detection |
|---|---|---|---|---|---|---|---|---|
| 10 | 1 | 16.32 | 21.5 | 4 | 32 | 32 | Detect | Detect |
| 2 | 16.83 | 29.7 | 4 | 32 | 32 | Detect | ||
| 3 | 15.74 | 14.8 | 3 | 24 | 24 | Detect | ||
| 4 | 17.20 | 37.6 | 3 | 24 | 24 | Detect | ||
| 20 | 1 | 15.73 | 14.7 | 2 | 16 | 16 | Detect | Detect |
| 2 | 16.00 | 17.4 | 3 | 24 | 24 | Detect | ||
| 3 | 16.00 | 17.5 | 3 | 24 | 24 | Detect | ||
| 4 | 15.98 | 17.3 | 3 | 24 | 24 | Detect | ||
| 40 | 1 | 5.93 | 0.70 | 3 | 24 | 11 | Detect | Detect |
| 2 | 13.47 | 3.64 | 3 | 24 | 23 | Detect | ||
| 3 | 13.66 | 2.85 | 3 | 24 | 22 | Detect | ||
| 4 | 8.07 | 1.12 | 3 | 24 | 15 | Detect | ||
| 80 | 1 | 0.41 | 0.049 | 3 | 24 | 1 | No Detect | No Detect |
| 2 | 0.65 | 0.100 | 3 | 24 | 2 | No Detect | ||
| 3 | 1.99 | 0.209 | 3 | 24 | 4 | Detect* | ||
| 4 | 0.50 | 0.049 | 3 | 24 | 1 | No Detect | ||
| 160 | 1 | 0.00 | 0.000 | 3 | 24 | 0 | No Detect | No Detect |
| 2 | 0.34 | 0.049 | 3 | 24 | 1 | No Detect | ||
| 3 | 0.00 | 0.000 | 3 | 24 | 0 | No Detect | ||
| 4 | 1.49 | 0.153 | 3 | 24 | 3 | No Detect | ||
| 1000 | 1 | 0.00 | 0.000 | 4 | 32 | 0 | No Detect | No Detect |
| 2 | 0.46 | 0.049 | 3 | 24 | 1 | No Detect | ||
| 3 | 0.85 | 0.153 | 4 | 32 | 4 | No Detect | ||
| 4 | 0.44 | 0.074 | 4 | 32 | 2 | No Detect |
| Tank Number | Sample Number | Transformed Ct Estimate | Estimated Copy Numbers per Reaction | Number of Technical Replicates per Sample | Number of Detects per Sample | Sample Detection | Tank Detection |
|---|---|---|---|---|---|---|---|
| 1 | 1 | 6.64 | 0.79 | 8 | 4 | Detect* | Detect |
| 2 | 4.48 | 0.54 | 8 | 3 | Detect* | ||
| 3 | 6.66 | 0.79 | 8 | 4 | Detect* | ||
| 2 | 1 | 11.89 | 2.38 | 8 | 7 | Detect | Detect |
| 2 | 14.51 | 6.75 | 8 | 8 | Detect | ||
| 3 | 14.00 | 4.90 | 8 | 8 | Detect | ||
| 3 | 1 | 14.25 | 5.75 | 8 | 8 | Detect | Detect |
| 2 | 16.04 | 17.99 | 8 | 8 | Detect | ||
| 3 | 10.67 | 1.59 | 8 | 6 | Detect | ||
| 4 | 1 | 11.54 | 2.38 | 8 | 7 | Detect | Detect |
| 2 | 3.16 | 0.33 | 8 | 2 | Detect* | ||
| 3 | 9.51 | 1.59 | 8 | 6 | Detect |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hocking, M.D.; MacAdams, J.C.; Allison, M.J.; Bergman, L.C.; Sneiderman, R.; Koop, B.F.; Starzomski, B.M.; Lesperance, M.L.; Helbing, C.C. Establishing the Signal above the Noise: Accounting for an Environmental Background in the Detection and Quantification of Salmonid Environmental DNA. Fishes 2022, 7, 266. https://doi.org/10.3390/fishes7050266
Hocking MD, MacAdams JC, Allison MJ, Bergman LC, Sneiderman R, Koop BF, Starzomski BM, Lesperance ML, Helbing CC. Establishing the Signal above the Noise: Accounting for an Environmental Background in the Detection and Quantification of Salmonid Environmental DNA. Fishes. 2022; 7(5):266. https://doi.org/10.3390/fishes7050266
Chicago/Turabian StyleHocking, Morgan D., Jeffrey C. MacAdams, Michael J. Allison, Lauren C. Bergman, Robert Sneiderman, Ben F. Koop, Brian M. Starzomski, Mary L. Lesperance, and Caren C. Helbing. 2022. "Establishing the Signal above the Noise: Accounting for an Environmental Background in the Detection and Quantification of Salmonid Environmental DNA" Fishes 7, no. 5: 266. https://doi.org/10.3390/fishes7050266