Accurate Wound and Lice Detection in Atlantic Salmon Fish Using a Convolutional Neural Network
Abstract
1. Introduction
Limitations
- Prepare an image dataset of underwater live salmon fish containing lice and wounds under various lighting conditions.
- Develop a convolutional neural network (CNN) based on fish lice and wound detection techniques using 2-D images.
- Design the convolutional neural network (CNN) to work efficiently in underwater scenarios and can able to classify fish as having wounds, lice, or an absence of wounds (nonwounded and non-lice) which performs better than standard models.
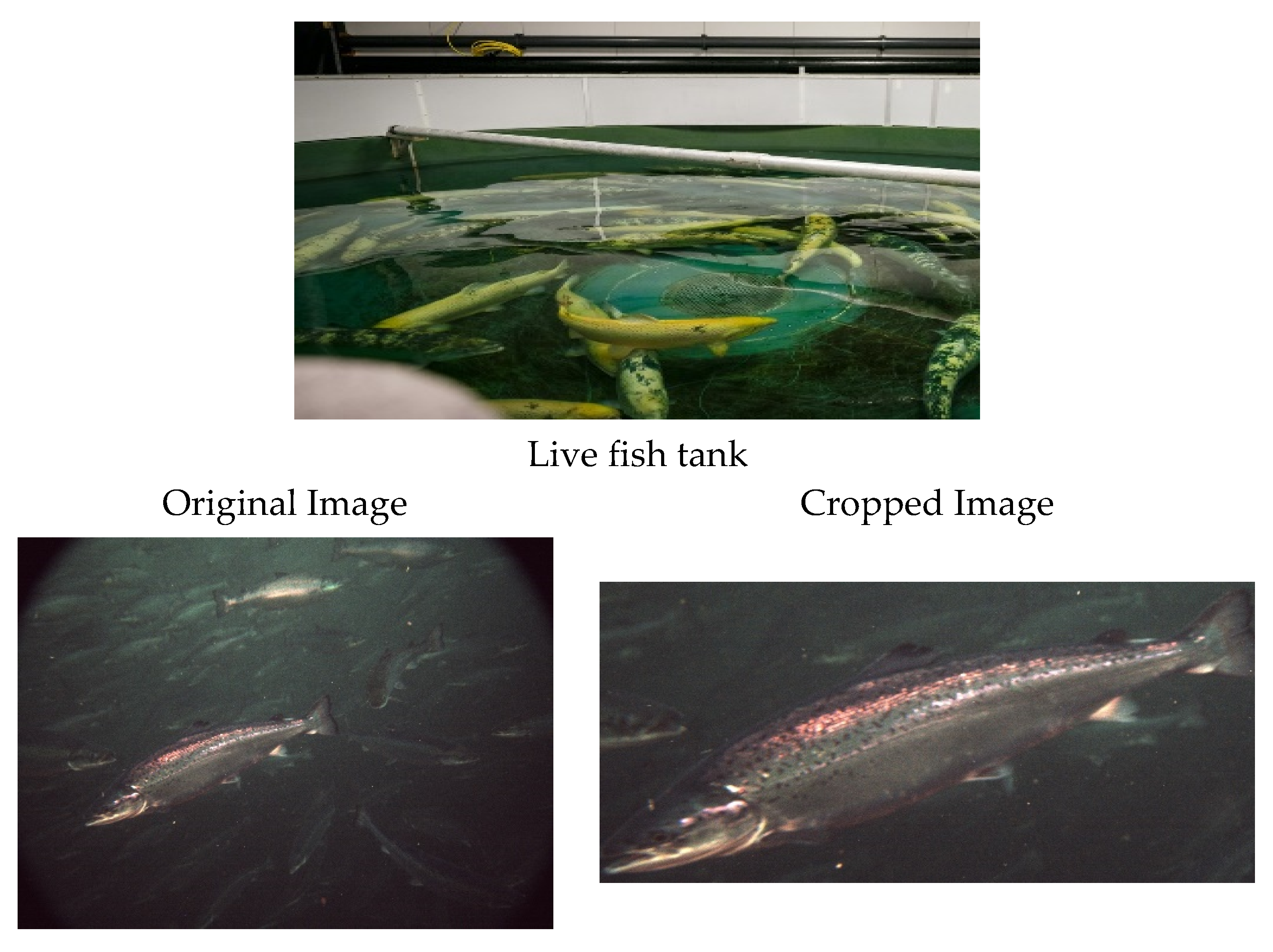
2. Dataset
Datasets Creation
3. Results
3.1. Preprocessing
3.2. Convolutional Neural Network Model
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Knutsen, H. Norwegian Agriculture Status and Trends 2019. NIBIO POP 2020, 6, 1–12. [Google Scholar]
- FAO. 2020: The State of World Fisheries and Aquaculture 2020; Sustainability in Action; FAO: Rome, Italy, 2022. [Google Scholar] [CrossRef]
- Abolofia, J.; Asche, F.; Wilen, J.E. The cost of lice: Quantifying the impacts of parasitic sea lice on farmed salmon. Mar. Resour. Econ. 2017, 32, 329–349. [Google Scholar] [CrossRef]
- Gallardo-Escárate, C.; Arriagada, G.; Carrera, C.; Gonçalves, A.T.; Nuñez-Acuña, G.; Valenzuela-Miranda, D.; Valenzuela-Muñoz, V. The race between host and sea lice in the Chilean salmon farming: A genomic approach. Rev. Aquac. 2019, 11, 325–339. [Google Scholar] [CrossRef]
- Coates, A.; Phillips, B.L.; Bui, S.; Oppedal, F.; Robinson, N.A.; Dempster, T. Evolution of salmon lice in response to management strategies: A review. Rev. Aquac. 2021, 13, 1397–1422. [Google Scholar] [CrossRef]
- Jensen, L.B.; Provan, F.; Larssen, E.; Bron, J.E.; Obach, A. Reducing sea lice (L epeophtheirus salmonis) infestation of farmed Atlantic salmon (S almo salar L.) through functional feeds. Aquac. Nutr. 2015, 21, 983–993. [Google Scholar] [CrossRef]
- Barrett, L.T.; Oppedal, F.; Robinson, N.; Dempster, T. Prevention not cure: A review of methods to avoid sea lice infestations in salmon aquaculture. Rev. Aquac. 2020, 12, 2527–2543. [Google Scholar] [CrossRef]
- ABB Enables First Remote-Controlled Submersible Fish Farm in the Arctic Ocean. 2019. Available online: https://new.abb.com/news/detail/24385/abb-enables-first-remote-controlled-submersible-fish-farm-in-the-arctic-ocean (accessed on 1 June 2022).
- Kabir, S.R.; Kousher, K.; Al, A. Fish Disease Detection System Using Machine Learning. Bachelor’s Thesis, Daffodil International University, Dhaka, Bangladesh, 2021. [Google Scholar]
- Li, D.; Du, L. Recent advances of deep learning algorithms for aquacultural machine vision systems with emphasis on fish. Artif. Intell. Rev. 2022, 55, 4077–4116. [Google Scholar] [CrossRef]
- Goodwin, M.; Halvorsen, K.T.; Jiao, L.; Knausgård, K.M.; Martin, A.H.; Moyano, M.; Thorbjørnsen, S.H. Unlocking the potential of deep learning for marine ecology: Overview, applications, and outlook. ICES J. Mar. Sci. 2022, 79, 319–336. [Google Scholar] [CrossRef]
- Gupta, A.; Bringsdal, E.; Salbuvik, N.; Knausgård, K.M.; Goodwin, M. An Accurate Convolutional Neural Networks Approach to Wound Detection for Farmed Salmon. In Proceedings of the International Conference on Engineering Applications of Neural Networks, Crete, Greece, 17–20 June 2022; Springer: Cham, Switzerland, 2022; pp. 139–149. [Google Scholar]
- Knausgård, K.M.; Wiklund, A.; Sørdalen, T.K.; Halvorsen, K.T.; Kleiven, A.R.; Jiao, L.; Goodwin, M. Temperate fish detection and classification: A deep learning based approach. Appl. Intell. 2022, 52, 6988–7001. [Google Scholar] [CrossRef]
- Noga, E.J. Fish Disease: Diagnosis and Treatment; John Wiley & Sons: Hoboken, NJ, USA, 2010. [Google Scholar]
- Ahmed, M.S.; Aurpa, T.T.; Azad, M.A.K. Fish disease detection using image based machine learning technique in aquaculture. J. King Saud Univ. Comput. Inform. Sci. 2022, 34, 5170–5182. [Google Scholar] [CrossRef]
- Malik, S.; Kumar, T.; Sahoo, A.K. Image processing techniques for identification of fish disease. In Proceedings of the 2017 IEEE 2nd International Conference on Signal and Image Processing (ICSIP), Singapore, 4–6 August 2017; pp. 55–59. [Google Scholar]
- Carrión, H.; Jafari, M.; Bagood, M.D.; Yang, H.Y.; Isseroff, R.R.; Gomez, M. Automatic wound detection and size estimation using deep learning algorithms. PLoS Comput. Biol. 2022, 18, e1009852. [Google Scholar] [CrossRef] [PubMed]
- Olsvik, E.; Trinh, C.; Knausgård, K.M.; Wiklund, A.; Sørdalen, T.K.; Kleiven, A.R.; Goodwin, M. Biometric fish classification of temperate species using convolutional neural network with squeeze-and-excitation. In Proceedings of the International Conference on Industrial, Engineering and Other Applications of Applied Intelligent Systems, Graz, Austria, 9–11 July 2019; Springer: Cham, Switzerland, 2019; pp. 89–101. [Google Scholar]
- Sture, Ø.; Øye, E.R.; Skavhaug, A.; Mathiassen, J.R. A 3D machine vision system for quality grading of Atlantic salmon. Comput. Electron. Agric. 2016, 123, 142–148. [Google Scholar] [CrossRef]
- Balaban, M.O.; Ünal Şengör, G.F.; Soriano, M.G.; Ruiz, E.G. Quantification of gaping, bruising, and blood spots in salmon fillets using image analysis. J. Food Sci. 2011, 76, 291–297. [Google Scholar] [CrossRef] [PubMed]
- Pate, M.; Jencic, V.; Zolnir-Dovc, M.; Ocepek, M. Detection of mycobacteria in aquarium fish in Slovenia by culture and molecular methods. Dis. Aquat. Org. 2005, 64, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Pizer, S.M.; Johnston, R.E.; Ericksen, J.P.; Yankaskas, B.C.; Muller, K.E. Contrast-limited adaptive histogram equalization: Speed and effectiveness. In Proceedings of the First Conference on Visualization in Biomedical Computing, Atlanta, GA, USA, 22–25 May 1990; IEEE Computer Society: Washington, DC, USA, 1990; pp. 337–338. [Google Scholar]
- Yussof, W.N.; Hitam, M.S.; Awalludin, E.A.; Bachok, Z. Performing contrast limited adaptive histogram equalization technique on combined color models for underwater image enhancement. Int. J. Interact. Digit. Media 2013, 1, 1–6. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Wang, K.; Thrampoulidis, C. Binary classification of Gaussian mixtures: Abundance of support vectors, benign overfitting and regularization. arXiv 2020, arXiv:2011.09148. [Google Scholar] [CrossRef]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Amin, M.A.; Yan, H. High speed detection of retinal blood vessels in fundus image using phase congruency. Soft Comput. 2011, 15, 1217–1230. [Google Scholar] [CrossRef]








| Parameter | FPR | TPR |
|---|---|---|
| Wound Detection | 0.0307 | 0.965 |
| Lice Detection | 0.0109 | 0.956 |
| Overall | 0.0212 | 0.97 |
| Model | Validation Accuracy |
|---|---|
| Proposed Model | 96.7% |
| VGG-16 | 91.2% |
| VGG-19 | 92.81% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gupta, A.; Bringsdal, E.; Knausgård, K.M.; Goodwin, M. Accurate Wound and Lice Detection in Atlantic Salmon Fish Using a Convolutional Neural Network. Fishes 2022, 7, 345. https://doi.org/10.3390/fishes7060345
Gupta A, Bringsdal E, Knausgård KM, Goodwin M. Accurate Wound and Lice Detection in Atlantic Salmon Fish Using a Convolutional Neural Network. Fishes. 2022; 7(6):345. https://doi.org/10.3390/fishes7060345
Chicago/Turabian StyleGupta, Aditya, Even Bringsdal, Kristian Muri Knausgård, and Morten Goodwin. 2022. "Accurate Wound and Lice Detection in Atlantic Salmon Fish Using a Convolutional Neural Network" Fishes 7, no. 6: 345. https://doi.org/10.3390/fishes7060345
APA StyleGupta, A., Bringsdal, E., Knausgård, K. M., & Goodwin, M. (2022). Accurate Wound and Lice Detection in Atlantic Salmon Fish Using a Convolutional Neural Network. Fishes, 7(6), 345. https://doi.org/10.3390/fishes7060345







